Abstract
Supramolecular self-assembly is an important process that enables the conception of complex structures mimicking biological motifs. Herein, we constructed helical fibrils through chiral self-assembly of nucleobase--peptide conjugates (NPCs), where achiral nucleobases are helically displayed on the surface of fibrils, comparable to polymerized nucleic acids. Selective binding between DNA and the NPC fibrils was observed with fluorescence polarization. Taking advantage of metal--nucleobase recognition, we highlight the possibility of deposition/assembly of plasmonic nanoparticles onto the fibrillar constructs. In this approach, the supramolecular chirality of NPCs can be adaptively imparted to metallic nanoparticles, covering them to generate structures with plasmonic chirality that exhibit significantly improved colloidal stability. The self-assembly of rationally designed NPCs into nanohelices is a promising way to engineer complex, optically diverse nucleobase-derived nanomaterials.
Keywords: Helical Structures, biomimetic chemistry, helical structures, peptides, plasmonic chirality, self-assembly
Chirality is an essential property of biological building blocks, including l-amino acids, DNA, and collagen helices, for example, with effects that scale from microscopic viruses to macroscopic sea shells and beyond. One example is the DNA double helix formed by Watson--Crick base pairing, which serves as the structural basis for genetic information storage. Advancements in oligonucleotide design have taken this biological motif and enabled the generation of programmable materials with controlled valency using directional DNA bonding.[1] Self-assembled DNA scaffolds have been utilized to precisely organize metallic particles, generating discrete periodic chiral nanostructure arrays and plasmonic chiroptical response.[2] Inspired by the intrinsic structural properties of the DNA helix, chemists have aimed to design DNA-mimicking molecules or structures with helically arranged nucleobases/nucleotides with sub-nanometer spatial precision.[3] For example, peptide nucleic acids (PNAs)[4] are DNA analogues that have a covalent pseudopeptide backbone with high chemical and enzymatic stability. Alternatively, non-covalent self-assembly of nucleotide-containing molecules into complex structures can partially lead to the functions of naturally occurring DNA.[5]
Herein, we seek to construct supramolecular helices by the chiral self-assembly[6] of nucleobase--peptide conjugates (NPCs) in which achiral nucleobases are helically presented on the surface (Figure 1). The amphiphilic NPCs, consisting of a hydrophobic diphenylalanine (Phe-Phe) conjugated alkyl tail and a hydrophilic peptide head group containing a terminal nucleobase, were synthesized by solid-phase peptide synthesis with Fmoc-protected amino acids and nucleobases (Figure 1a, see also the Supporting Information, Figures S1 and S2). The Phe-Phe moiety, inspired by the KLVFF sequence in amyloid-β, provides directional π-π stacking and hydrogen bonding, which promote the self-assembly of fibrillar structures.[7] The hydrocarbon tail strengthens the self-association of NPCs by hydrophobic collapse and chain crystallization.[8] Owing to their hydrophilic nature, the nucleobases are exposed on the surface of the fibrils, equivalent to a polymerized nucleic acid. Based on the metal--nucleobase recognition/affinity, such fibrils have been exploited as ideal templates for the deposition of plasmonically chiral nanoparticles.
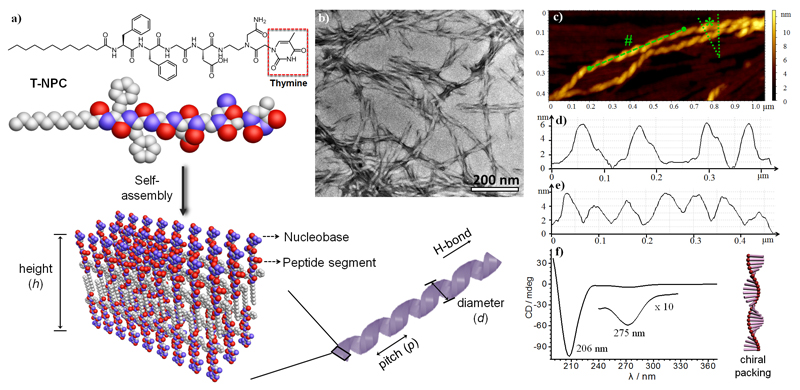
Figure 1.
a) Self-assembly of hybrid T-NPC into helical fibrils in the form of nanoribbons with nucleobases displayed at the surface. b) TEM and c) tapping-mode AFM images of T-NPC fibrils. d) The height profile across the marked section indicated by * in (c), showing the uniform height (h) of the fibrils. e) Cross-sectional analysis of the marked section indicated by # in (c), showing the uniform pitch (p) of the nanohelices. f) CD spectrum of the T-NPC fibrils, where the chirality signals from the peptide segment and thymine were observed at 206 nm and 275 nm, respectively.
NPC self-assembly occurred above a critical aggregation concentration (0.01-0.04 mm; Figures S3 and S4), and hydrogelation was noted above a concentration of 5 mm (Figure S5). Transmission electron microscopy (TEM) confirmed the formation of ribbons with a diameter (d) of approximately 18 nm and a regular pitch (p) of about 70 nm in a solution of thymine-containing NPCs (T-NPCs; Figures 1b and S6). Atomic force microscopy (AFM) showed the uniform structure of the helical ribbons with a thickness (h) of approximately 6^^nm, indicating a bilayer structure (Figure 1c,d). The pitches obtained from the cross-section profiles were measured to be about 75 nm (Figure 1e), which is in agreement with the TEM results. The supramolecular helicity was investigated by circular dichroism (CD); a negative peak was observed at 208-211 nm, which is due to π-π stacking of the aromatic side chains and to the distortion of the β-sheets (Figure 1f).[9] A weak peak was also observed at approximately 275^^nm, revealing the helical stacking of thymine.
Self-assembled helical fibrils from adenine-, cytosine-, and guanine-containing NPCs were also studied by TEM (Figures S6 and S7) and AFM (Figures S8-S11). The AFM height profiles showed that the fibrils had a thickness (h) of about 6 nm, suggesting a bilayer structure. All of the NPC fibrils presented CD peaks at 260—280 nm, confirming the chiral packing of the nucleobases (Figure S12). The presence of β-sheets and highly ordered amyloid structures was confirmed by a thioflavin T assay (Figure S13), NMR (Figure S14), and FTIR spectroscopy (Figure S15), where a strong amide I absorption at 1635 cm-1 was noted.[10]
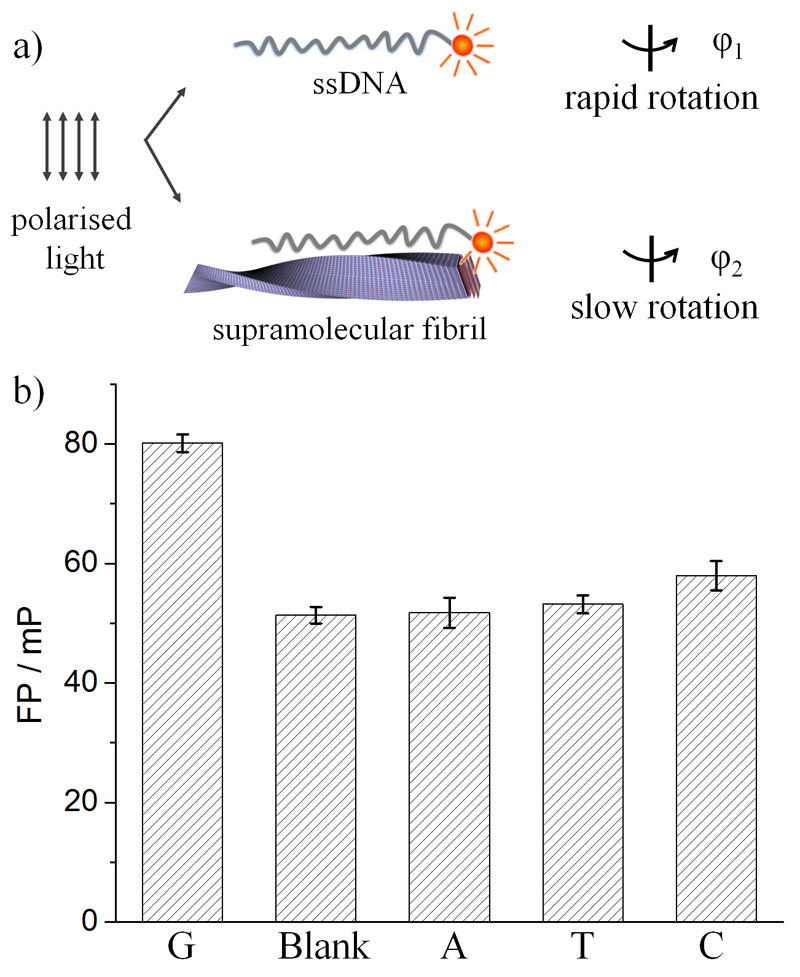
Considering the chiral stacking of the nucleobases, the NPC fibrils can be regarded as a versatile yet simple self-assembled mimic of DNA. The recognition of NPC fibrils by complementary DNA via base pairing was evaluated by fluorescence polarization (FP), which provides information on the molecular rotation dynamics of a dye. As shown in Figure 2a, when excited by polarized light, fluorescence depolarization of fluorophore-tagged ssDNA occurs by rapid molecular rotation while the adsorption of ssDNA to NPC fibrils restricted its rotation and therefore increased the FP. In Figure 2b, the FP value of fluorescein-labeled oligonucleotides containing twelve cytosines (FAM-C12) was found to be the highest in the presence of G-NPC fibrils, indicating the strongest binding of FAM-C12 onto guanine-containing fibrils via C-G interactions. The binding of FAM-T12 to A-NPC fibrils was not observed, possibly owing to the weakness of A-T interactions.
Figure 2.
a) Detection of ssDNA binding to NPC fibrils by fluorescence polarization (FP). When a dye is excited with polarized light, it will fluoresce with the same polarization. Depolarization occurs when the dye rotates during its emission lifetime. The adsorption of fluorophore-labeled ssDNA to NPC fibrils restricts the molecular rotation of the fluorophore and therefore decreases the FP. b) FP of FAM-C12 with or without NPC fibrils.
Nucleic acids exhibit sequence-specific binding affinities towards metal ions and particles, which have been exploited to mediate DNA-guided nucleation in the synthesis of fluorescent Ag clusters and plasmonic materials.[11] As nucleobases are repeatedly displayed at the fibril surface, we studied the ability of the NPC fibrils to nucleate and bind metallic nanoparticles (NPs). Indeed, AgNPs of 2—10 nm were observed to be deposited on T-NPC fibrils after reduction of the Ag ions with NaBH4 (Figures 3a,b and S16). Interestingly, the AgNPs were distributed at the edge of the fibrils following the helical arrangement of nucleobases where the particles exhibited the typical plasmonic response of non-aggregated AgNPs at 430 nm (Figure S17). Small angle X-ray scattering (SAXS) analysis showed that the scattered X-ray intensity of T-NPC fibrils as a function of the scattering vector Q could be effectively fitted using simulations of the scattering form factor of isotropically oriented infinite cylinders with a radius of 5.7 nm (Figure 3c). Following Ag deposition, the scattering profile exhibited a diffuse structure peak in the region of Q=0.12 Å-1, which corresponds to the inter-AgNP correlation length of 5 nm when bound to the planar surface of the fibrils, consistent with the TEM measurements.
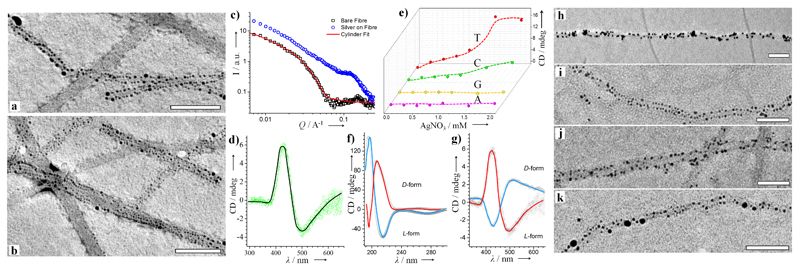
Figure 3.
a,b) TEM images of Ag-decorated T-NPC fibrils (Scale bar: 100^^nm). c) SAXS profiles for T-NPC fibrils and AgNP-loaded fibrils. d) CD spectrum showing the plasmonic chiral signal from the AgNPs. e) CD peak intensity (425 nm) for AgNPs on NPC fibrils. f) CD spectra highlighting the chirality inversion of T-NPC fibrils when using the l- and d- enantiomers of amino acids. g) Chiral signal of AgNPs synthesized on T-NPC fibrils with l/d-NPC. h-k) TEM images of metallic nanoparticles synthesized in situ on the NPC fibrils: h) Au on G-NPC; i) Au on C-NPC; j) Pd on C-NPC; k) Pd on G-NPC. Scale bars: 100 nm.
Interestingly, the helical patterning of AgNPs on T-NPC fibrils generates a plasmonic chirality in the CD spectrum, with a negative Cotton effect at 500^^nm and a positive Cotton effect at 425 nm (Figure 3d). Increasing the concentration of Ag ions progressively increased the CD signal before reaching a plateau (Figure 3e). Meanwhile a red shift of the peak was observed, indicating the growth of AgNPs (Figure S18). We also studied the chirality transfer to plasmonic AgNPs from supramolecular fibrils of A-NPC, C-NPC, and G-NPC (Figure 3e). We found plasmonic chirality for in situ synthesized AgNPs on T-NPC and C-NPC fibrils but not on G-NPC or A-NPC fibrils, which might be due to the stronger binding of cytosine and thymine with Ag. Moreover, the chirality of supramolecular fibrils can be readily switched by changing the molecular chirality, that is, by adjusting the stereochemistry of the amino acids. As shown in Figure 3f, the use of d-amino acids (i.e., aspartic acid and phenylalanine) in the T-NPC resulted in inversed CD signals for the fibrils. With d-form NPC fibrils as the templates, AgNPs with inversed plasmonic chirality were deposited (Figure 3g).
To highlight the versatility of the NPC templates, Au and Pd were nucleated at the surface of NPC fibrils (Figure 3h-k), and 2-5nm Au and Pd nanoparticles were helically deposited on the G- and C-NPC fibrils. The as-synthesized AuNPs were well dispersed as evident from the reddish color of the solution with an LSPR peak at 520 nm (Figure S19). The size of the metallic NPs was well controlled, and they were located in close proximity to the fibrils owing to fibril-mediated nucleation via metal-nucleobase affinity. We propose that the repetitive nucleobases at the surface tend to concentrate the cations (e.g., Ag, Au, and Pd), creating local supersaturation followed by oriented nucleation of the crystal. Although mineralization occurring on peptide systems has been investigated in other contexts,[12] the nucleation of metallic NPs by nucleobase patterns is less studied.
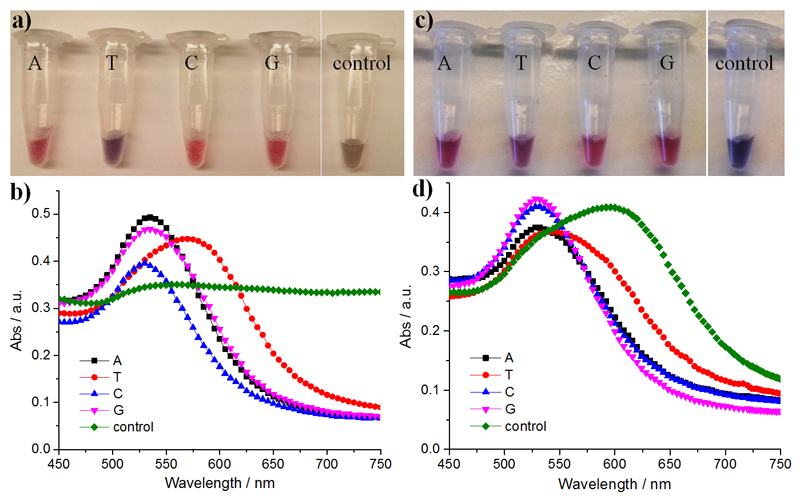
Physical adsorption of DNA onto Au by metal--nucleobase affinity stabilizes the nanoparticles, and this property has previously been utilized to design plasmonic biosensors for the detection of nucleic acids.[13] Herein, we demonstrate the binding of AuNPs (12 nm) to NPC fibrils to generate "pearl chain" structures (Figure S20), which enhanced the colloidal stability of the AuNPs. As shown in Figure 4a,b, the addition of 0.05m NaCl solution resulted in the aggregation of citrate-capped AuNPs as shown by the change in the ratio of the absorbance at 650 nm and 525 nm (A650/A525), which increased from 0.32 to 1.0, and the distinct color change to brown. In the presence of NPC fibrils, the AuNP solutions remain reddish (A650/A525=0.2). Surprisingly, the fibrils stabilized AuNPs even at a NaCl concentration of 0.75m (Figure S21). This is likely because multivalent gold--nucleobase binding can "lock" AuNPs to the fibril surface and prevent salt-induced aggregation. An exception was noted for T-NPC fibrils where an obvious peak red shift in the LSPR spectra was observed. Moreover, the immobilization of AuNPs on NPC fibrils helps overcome another challenge of AuNPs, which are known to irreversibly aggregate when frozen, as indicated by a red-to-blue color change (Figure 4c,d). Consistent with the salt stability investigations, fibrils from A-NPC, G-NPC, and C-NPC were found to stabilize AuNPs much better than T-NPC. We propose that the varied capacities of NPCs to stabilize AuNPs originate from the different gold nucleobase affinities; it was previously reported that homo-oligonucleotides adsorb on Au with the affinity decreasing in the order A>C≥G>T.[14] Current approaches to maintain colloidal stability typically involve surface modification with charged and often bulky ligands to ensure sufficient particle separation. Alternatively, NPC fibrils can serve as a soft dynamic scaffold to physically immobilize AuNPs by gold--nucleobase interactions and impede irreversible particle contact while maintaining the bulk mobility of the fibril-AuNP constructs.
Figure 4.
a,c) Macroscopic appearance and b,d) corresponding LSPR spectra of citrate-capped AuNPs with or without 0.2 mm NPCs. a,b) AuNPs with 50 mm NaCl. c,d) AuNPs after freeze-thaw cycles.
In conclusion, we have designed a new class of supramolecular fibrils that present nucleobases in helical patterns on the fibril surface. We capitalized on the chirality of NPC fibrils to organize plasmonic nanomaterials by both in situ nucleation and assembly of metallic nanoparticles (i.e., Ag, Au, Pd) through metal--nucleobase recognition. Multivalent gold nucleobase interactions at the interface of the NPC fibrils and AuNPs also offer a new strategy to increase the colloidal stability of AuNPs in a nucleobase-dependent manner. We expect that this system can be further engineered to create DNA-like structures that exert control over the crystal nucleation and growth of inorganic components. The recognition between NPC fibrils and nucleic acids could also be explored for DNA/RNA-templated self-assembly and the design of novel biomaterials.
Supplementary Material
Supporting information and the ORCID identification number(s) for the author(s) of this article can be found under: 10.1002/anie.201610976.
Amyloid fibrils were constructed through the self-assembly of nucleobase-peptide conjugates that were designed to repeatedly organize achiral nucleobases into helical patterns on the surface. The fibrils were utilized for the in situ nucleation and assembly of metallic nanoparticles through metal-nucleobase recognition.
Acknowledgements
M.M.S. gratefully acknowledges the ERC Seventh Framework Programme Consolidator grant "Naturale CG" (616417) and the grant "Bio-functionalised Nanomaterials for Ultrasensitive Biosensing" (EP/K020641/1) funded by the Engineering and Physical Sciences Research Council (EPSRC). M.R.T. thanks the i-sense EPSRC IRC in Early Warning Sensing Systems for Infectious Diseases (EP/K031953/1) for funding. We thank Prof. R.M. Richardson for assistance with SAXS measurements. The raw research data supporting this paper can be found at 10.5281/zenodo.159453.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Jones MR, Seeman NC, Mirkin CA. Science. 2015;347:840. doi: 10.1126/science.1260901. [DOI] [PubMed] [Google Scholar]
- 2a.Kuzyk A, Schreiber R, Fan Z, Pardatscher G, Roller E-M, Hogele A, Simmel FC, Govorov AO, Liedl T. Nature. 2012;483:311. doi: 10.1038/nature10889. [DOI] [PubMed] [Google Scholar]; 2b Zhao Y, Xu L, Ma W, Wang L, Kuang H, Xu C, Kotov NA. Nano Lett. 2014;14:3908. doi: 10.1021/nl501166m. [DOI] [PubMed] [Google Scholar]
- 3.Ura Y, Beierle JM, Leman LJ, Orgel LE, Ghadiri MR. Science. 2009;325:73. doi: 10.1126/science.1174577. [DOI] [PubMed] [Google Scholar]
- 4.Nielsen E, Egholm M, Berg RH, Buchardt O. Science. 1991;254:1497. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 5a.Li X, Kuang Y, Lin H-C, Gao Y, Shi J, Xu B. Angew Chem Int Ed. 2011;50:9365. doi: 10.1002/anie.201103641. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew Chem. 2011;123:9537. [Google Scholar]; 5b Yuan D, Du X, Shi J, Zhou N, Zhou J, Xu B. Angew Chem Int Ed. 2015;54:5705. doi: 10.1002/anie.201412448. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew Chem. 2015;127:5797. [Google Scholar]; 5c Moreau L, Barthelemy P, El Maataoui M, Grinstaff MW. J Am Chem Soc. 2004;126:7533. doi: 10.1021/ja039597j. [DOI] [PubMed] [Google Scholar]; 5d Banchelli M, Berti D, Baglioni P. Angew Chem Int Ed. 2007;46:3070. doi: 10.1002/anie.200604826. [DOI] [PubMed] [Google Scholar]; Angew Chem. 2007;119:3130. [Google Scholar]; 5e Gissot A, Camplo M, Grinstaff MW, Barthelemy P. Org Biomol Chem. 2008;6:1324. doi: 10.1039/b719280k. [DOI] [PMC free article] [PubMed] [Google Scholar]; 5f Iwaura R, Shimizu T. Angew Chem Int Ed. 2006;45:4601. doi: 10.1002/anie.200601173. [DOI] [PubMed] [Google Scholar]; Angew Chem. 2006;118:4717. [Google Scholar]; 5g Serpell CJ, Barlog M, Basu K, Fakhoury JF, Bazzi HS, Sleiman HF. Mater Horiz. 2014;1:348. [Google Scholar]; 5h Sivakova S, Rowan SJ. Chem Soc Rev. 2005;34:9. doi: 10.1039/b304608g. [DOI] [PubMed] [Google Scholar]
- 6a.Wang Y, Xu J, Wang Y, Chen H. Chem Soc Rev. 2013;42:2930. doi: 10.1039/c2cs35332f. [DOI] [PubMed] [Google Scholar]; 6b Liu M, Zhang L, Wang T. Chem Rev. 2015;115:7304. doi: 10.1021/cr500671p. [DOI] [PubMed] [Google Scholar]; 6c Liu W, Zhu Z, Deng K, Li Z, Zhou Y, Qiu H, Gao Y, Che S, Tang Z. J Am Chem Soc. 2013;135:9659. doi: 10.1021/ja312327m. [DOI] [PubMed] [Google Scholar]; 6d Querejeta-Fernández A, Chauve G, Methot M, Bouchard J, Kumacheva E. J Am Chem Soc. 2014;136:4788. doi: 10.1021/ja501642p. [DOI] [PubMed] [Google Scholar]; 6e Qiu H, Che S. Chem Soc Rev. 2011;40:1259. doi: 10.1039/c0cs00002g. [DOI] [PubMed] [Google Scholar]; 6f Wei Z, Laitinen T, Smarsly B, Ikkala O, Faul CFJ. Angew Chem. 2005;117:761. doi: 10.1002/anie.200460928. [DOI] [PubMed] [Google Scholar]; 6g Oda R, Huc I, Schmutz M, Candau SJ, MacKintosh FC. Nature. 1999;399:566. doi: 10.1038/21154. [DOI] [PubMed] [Google Scholar]; 6h Hirschberg JHKK, Brunsveld L, Ramzi A, Vekemans JAJM, Sijbesma RP, Meijer EW. Nature. 2000;407:167. doi: 10.1038/35025027. [DOI] [PubMed] [Google Scholar]; 6i Bae J, Choi J-H, Yoo Y-S, Oh N-K, Kim B-S, Lee M. J Am Chem Soc. 2005;127:966. doi: 10.1021/ja051961m. [DOI] [PubMed] [Google Scholar]; 6j Ajayaghosh A, Vijayakumar C, Varghese R, George SJ. Angew Chem Int Ed. 2006;45:456. doi: 10.1002/anie.200503258. [DOI] [PubMed] [Google Scholar]; Angew Chem. 2006;118:470. [Google Scholar]; 6k Lohr A, Lysetska M, Würthner F. Angew Chem Int Ed. 2005;44:5071. doi: 10.1002/anie.200500640. [DOI] [PubMed] [Google Scholar]; Angew Chem. 2005;117:5199. [Google Scholar]; 6l Toyofuku K, Alam MA, Tsuda A, Fujita N, Sakamoto S, Yamaguchi K, Aida T. Angew Chem Int Ed. 2007;46:6476. doi: 10.1002/anie.200701668. [DOI] [PubMed] [Google Scholar]; Angew Chem. 2007;119:6596. [Google Scholar]; 6m Danila I, Riobé F, Piron F, Puigmartí-Luis J, Wallis JD, Linares M, Ågren H, Beljonne D, Amabilino DB, Avarvari N. J Am Chem Soc. 2011;133:8344. doi: 10.1021/ja202211k. [DOI] [PubMed] [Google Scholar]
- 7a.Reches M, Gazit E. Science. 2003;300:625. doi: 10.1126/science.1082387. [DOI] [PubMed] [Google Scholar]; 7b Yan X, Zhu P, Li J. Chem Soc Rev. 2010;39:1877. doi: 10.1039/b915765b. [DOI] [PubMed] [Google Scholar]; 7c Adamcik J, Castelletto V, Bolisetty S, Hamley IW, Mezzenga R. Angew Chem Int Ed. 2011;50:5495. doi: 10.1002/anie.201100807. [DOI] [PubMed] [Google Scholar]; Angew Chem. 2011;123:5609. [Google Scholar]; 7d Castelletto V, Hamley IW, Hule RA, Pochan D. Angew Chem Int Ed. 2009;48:2317. doi: 10.1002/anie.200805500. [DOI] [PubMed] [Google Scholar]; Angew Chem. 2009;121:2353. [Google Scholar]
- 8.Hartgerink JD, Beniash E, Stupp SI. Science. 2001;294:1684. doi: 10.1126/science.1063187. [DOI] [PubMed] [Google Scholar]
- 9a.Gazit E. FASEB J. 2002;16:77. doi: 10.1096/fj.01-0442hyp. [DOI] [PubMed] [Google Scholar]; 9b Bowerman AJ, Liyanage W, Federation AJ, Nilsson BL. Biomacromolecules. 2011;12:2735. doi: 10.1021/bm200510k. [DOI] [PubMed] [Google Scholar]
- 10.Liu P, Ni R, Mehta AK, Childers WS, Lakdawala A, Pingali SV, Thiyagarajan P, Lynn DG. J Am Chem Soc. 2008;130:16867. doi: 10.1021/ja807425h. [DOI] [PubMed] [Google Scholar]
- 11.Petty JT, Zheng J, Hud NV, Dickson RM. J Am Chem Soc. 2004;126:5207. doi: 10.1021/ja031931o. [DOI] [PubMed] [Google Scholar]
- 12.Chen C-L, Zhang P, Rosi NL. J Am Chem Soc. 2008;130:13555. doi: 10.1021/ja805683r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H, Rothberg L. Proc Natl Acad Sci USA. 2004;101:14036. doi: 10.1073/pnas.0406115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kimura-Suda H, Petrovykh DY, Tarlov MJ, Whitman LJ. J Am Chem Soc. 2003;125:9014. doi: 10.1021/ja035756n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.