Abstract
For drug delivery, characterization of liposomes regarding size, particle number concentrations, occurrence of low-sized liposome artefacts and drug encapsulation are of importance to understand their pharmacodynamic properties. In our study, we aimed to demonstrate the applicability of nano Electrospray Gas-Phase Electrophoretic Mobility Molecular Analyser (nES GEMMA) as a suitable technique for analyzing these parameters. We measured number-based particle concentrations, identified differences in size between nominally identical liposomal samples, and detected the presence of low-diameter material which yielded bimodal particle size distributions. Subsequently, we compared these findings to dynamic light scattering (DLS) data and results from light scattering experiments coupled to Asymmetric Flow-Field Flow Fractionation (AF4), the latter improving the detectability of smaller particles in polydisperse samples due to a size separation step prior detection. However, the bimodal size distribution could not be detected due to method inherent limitations. In contrast, cryo transmission electron microscopy corroborated nES GEMMA results. Hence, gas-phase electrophoresis proved to be a versatile tool for liposome characterization as it could analyze both vesicle size and size distribution. Finally, a correlation of nES GEMMA results with cell viability experiments was carried out to demonstrate the importance of liposome batch-to-batch control as low-sized sample components possibly impact cell viability.
Keywords: Liposome, Gas-phase electrophoretic mobility, molecular analysis, Transmission electron microscopy, Dynamic light scattering, Cytotoxicity, Flow field flow fractionation
1. Introduction
Liposomes are artificial vesicles formed by one or more concentric phospholipid bilayers that are separated by aqueous water compartments (Munin and Edwards-Levy, 2011). Within the aqueous compartments and the lipid membrane they have the unique ability of transporting hydrophilic, hydrophobic, and amphiphilic compounds (Nii and Ishii, 2005). Liposomes emerged in the 60s’ (Bangham et al., 1962) and have gained widespread use during the last decades as versatile drug carriers, cellular transfection agents and carriers of diverse contrast agents among other things (Allen and Cullis, 2013; Burdinski et al., 2010; Ramamoorth and Narvekar, 2015; Terreno et al., 2009). Liposomes may be unilamellar or multilamellar with spatial dimensions (sizes) ranging from tens of nanometers to tens of micrometers.
For drug delivery purposes, liposome size, size distribution and vesicle numbers per sample volume are of great importance. These factors affect passive targeting and accumulation around tumor areas, the so-called Enhanced Permeability and Retention (EPR) effect, sedimentation and biodistribution, but also the amount of drug that can be encapsulated within the liposome (Drummond et al., 1999). For optimal drug delivery, liposomes should have a size between 70 up to 300 nm and are usually protected by a steric shield, like PEGylation, to avoid vesicle detection by the reticuloendothelial system (Hupfeld et al., 2006; Nag and Awasthi, 2013). To date, several liposomal drugs have been approved by the US Food and Drug Administration (FDA) like Marqibo (liposomal vinCRIStine), Myocet and DOXIL/Caelyx (liposomal doxorubicin) (Allen and Cullis, 2013). The liposomes used in this study are of a similar lipid composition to that of DOXIL/Caelyx, differing only in lipid molar ratios.
Characterization of liposomes has always been challenging and still, to date no technique offers a full range analysis without any drawbacks or the need of complementary data to make a full characterization of the lipid vesicles. Among the common techniques used for characterization of liposomes are (cryo) Transmission Electron Microscopy (TEM), negative stain and freeze-fracture Electron Microscopy, Atomic Force Microscopy (AFM), Size Exclusion Chromatography (SEC) and Dynamic/Static Light Scattering (DLS/SLS).
For instance, electron microscopy techniques are suitable for surface and morphological studies of analytes but have several drawbacks. Apart from cost-related issues, sample preparation and sample analysis being carried out in vacuo might affect vesicle shape (Almgren et al., 2000; Bibi et al., 2011). AFM is also suitable for characterization of liposome shape, morphology and surface properties. In contrast to Electron Microscopy techniques, imaging is carried out at ambient pressure with little sample preparation needed at a solid/gas interphase (or in a liquid). However, deformation or rupture of liposomes might be induced after deposition onto the AFM sample support due to interaction either with the carrier material or the cantilever during contact mode (Laouini et al., 2012; Muraji et al., 2013; Pignataro et al., 2000; Ruozi et al., 2011). A limitation common for both AFM and Electron Microscopy is the small sampling area while imaging as a high number of particles has to be considered to achieve a representative (i.e. statistically valid) and reliable size characterization.
For size determination and distribution, light scattering techniques are usually preferred. DLS is nowadays one of the most common techniques for the determination of liposome and particle sizes in the nano-range. It gives a fast and reliable reading with little to no sample preparation (Nickel et al., 2014). DLS is based on a particle’s light scattering intensity, which gives a reading of its mean hydrodynamic radius depending on the particle Brownian motion and is ideal for homogeneous, monodisperse samples. However, for more heterogeneous samples, when small particles are measured in the presence of a few larger particles, the mean particle diameter will be strongly biased towards larger constituent sizes (Hupfeld et al., 2006, 2010). Another limitation seldom taken into account when using DLS is the PEGylation (steric shield) of stealth liposomes used for drug delivery. For PEGylated liposomes, the hydrodynamic radii might give a larger size reading than the actual size of the vesicle.
A strategy to overcome the strong bias of light scattering data to larger sample components can be found in a preceding size fractionation technique, such as Field Flow Fractionation (FFF). In the FFF channel, particles are fractionated based on their size and then analyzed with light scattering or other detectors (fluorescence, UV–vis, ICP-MS, etc.) (Dubascoux et al., 2010; Kammer et al., 2005). Although ease of handling for light scattering as a stand-alone method is lost, its combination with FFF helps to overcome the problem of small particle masking in the presence of larger particles (Ranville and Montano, 2015).
Nano electrospray Gas-phase Electrophoretic Mobility Molecular Analysis (nES GEMMA) as an alternative method to analyze liposomal vesicles separates single charged analytes. These are obtained from a nES process with subsequent drying of droplets and charge conditioning in a 210Po induced bipolar atmosphere. Separation occurs in a high laminar flow of compressed air and an orthogonal, tunable electric field. By variation of the field strength during a voltage scan, only particles of a given size (electrophoretic mobility, EM, diameter) are able to pass the differential mobility analyzer (DMA) and are subsequently counted (Bacher et al., 2001; Kaufman et al., 1996). nES GEMMA allows for single-particle, number-concentration based detection of analytes and is therefore in perfect accordance with recommendations of the EU for nanoparticle characterization (2011/696/EU from October 18th, 2011). In contrast to FFF and DLS, nES GEMMA relates surface-dry particle diameters. Therefore, the application of volatile electrolyte solutions for the nES process is a necessary prerequisite for sample analysis, as else unspecified analyte/salt aggregates would interfere with vesicle size detection (Weiss et al., 2014).
In a previous study (Weiss et al., 2016), based on work by Epstein and the group of Biswas (Chadha et al., 2012; Chattopadhyay et al., 2010; Epstein et al., 2006) we took nES GEMMA to the field of liposome characterization and analysis of sample composition to allow for vesicle batch control. The aim of the present study is to investigate the use of nES GEMMA as a stand-alone or complementary technique for determining particle size and size distributions of liposomal drug carriers. This will be done by comparing nES GEMMA readings to DLS data and SLS coupled to Asymmetric Flow-FFF (AF4). Additionally, electron microscopy will be carried out. Correlation of nES GEMMA results to cell viability experiments demonstrates the importance of liposome characterization prior application as drug delivery vesicles.
2. Materials and methods
2.1. Materials
l-α-phosphatidylcholine, hydrogenated (Soy) (HSPC), (Chol) and 1,2-distearoyl-sn-glycero-3-phosphoethanol-amine-N-[methoxy(polyethylene glycol)-2000 (DSPE-mPEG2000) were purchased from Avanti Polar Lipids (Alabaster, AL, USA). Polystyrene size standard suspensions (20, 60, 100, and 150 nm) from polymer microspheres (Duke Scientific Corporation, Palo Alto, CA, USA) used for calibrating the AF4 were prepared in MilliQ water (18.2 M Ω cm resistivity at 25 °C). All other chemicals and solutions used were the same as previously described (Weiss et al., 2016).
2.2. Liposome production
Liposomes of HSPC:Chol:DSPE-mPEG2000 (55:40:5 molar ratio) were made by the thin lipid film hydration technique. Briefly, phospholipids were dissolved in a round bottomed flask using a chloroform/methanol (3:1, v/v) mixture. Organic solvents were evaporated under nitrogen flux until a thin and homogenous lipid film was formed. Traces of organic solvents were removed by placing the films in a desiccator connected to a vacuum pump (<8 mbar, from Ilmvac, Ilmenau, Germany) for at least one hour at room temperature. Lipid films were hydrated with a 40 mM ammonium acetate solution, pH 8.4 (NH4OAc), including 10 μM ATTO633 fluorophore (NH4OAc was filtered through surfactant-free cellulose acetate membrane syringe filters, 0.2 μm pore size; Sartorius, Göttingen, Germany), by vortex-mixing until lipid films were completely dissolved to achieve a 10 mM lipid concentration in solution. Multilamellar liposomes were submitted to 5 cycles of freezing in liquid nitrogen and thawing in a water bath set above transition temperature of the main lipid (approx. 70 °C). Finally, liposome solutions were serially extruded 21 times each through 400, 200 and 100 nm stacked polycarbonate filters, respectively, to yield stock solutions. The liposome preparation process was repeated for in total n = 5 times to yield nominally identical vesicle batches.
2.3. nES GEMMA measurements
The TSI Inc (Shoreview, MN, USA) nES GEMMA instrumentation employed consisted of (i) a nES unit including a 210Po source (model 3480), (ii) a nano differential mobility analyzer (nDMA, model 3080) and (iii) an ultrafine particle counter (CPC, series 3025A). Compressed, particle-free sheath air for the nES process (1.0 L/m) was additionally dried (Donaldson Variodry Membrane Dryer Superplus obtained through R. Ludvik Industriegeräte, Vienna, Austria). 0.1 Lpm CO2 were likewise included in the nES sheath flow. Samples were introduced into the nES unit via a cone-tipped fused silica capillary with 25 μm inner diameter (TSI Inc.) by application of approx. 28 kPa to the sample vial. The nES was operated with a positive 2.2 kV voltage resulting in approx. −340 nA current. Scan ranges were set between 4.8–184.3 nm EM diameter by employment of 2.5 L pm sheath flow in the nDMA. Scan times were set to 150 s for voltage variation with 30 s retrace time to allow for a reset of the employed voltage. Between samples, the capillary was flushed with NH4OAc including 0.0005 [v/v%] Tween20 followed by blank NH4OAc for capillary equilibration. Samples were loaded for 5 min before recording of spectra. Liposomal stock solutions were diluted 1:25 in NH4OAc to obtain samples (0.4 mM total lipid concentration per sample). Each sample was measured by four scans. The resulting GEMMA spectra obtained are medians of these four complete scans, respectively. Fitting of symmetric, Gauss-shaped peaks to spectra via Origin software (OriginPro 9.1.0) allowed for EM diameter determination at the peak apex. If not indicated otherwise, raw count particle values per detection channel were taken for data analysis. It is of note that due to analyte heterogeneity resulting in possible calculation artifacts, we decided to reduce data processing to a minimum. In a case of raw particle count data evaluation, additional data correction e.g. for particle charging probabilities are not considered. However, the correction for size dependent analyte charging probability would neither influence the qualitative findings (detection of smaller sized sample compounds and determination of dry liposome size values) nor the quantitative outcome (comparison of liposome encapsulation capacities) of our study and was therefore omitted.
2.4. Particle size determination by DLS and zeta potential
Mean sizes expressed as Z-averages from cumulant fit analyses of liposomes were evaluated by dynamic light-scattering using a Zetasizer (size range: 1 nm–1 μm) (Malvern Instruments, Spring Lane South, Worcestershire, UK), a photo-correlation spectroscopy apparatus. The intensity-weighted average hydrodynamic diameter of the suspensions was determined using incident light (λ = 633 nm), detected at a scattering angle of 173°.
The material refractive index was set to 1.45, the medium refractive index was set to 1.33 and the viscosity to 0.887 cP at 20 °C. Samples were diluted with NH4OAc. Dilutions at 1:100 and 1:1000 were prepared in a quartz cuvette immediately before measurement (dilutions had been previously empirically tested until attenuator optimal position was achieved). Every sample and dilution was analyzed three times, each containing 5 sequences with an integration time of 10 s. All experiments were carried out at 20 °C temperature. Results are given as Z-average. Polydispersity indices (PDI) were determined with the cumulant analysis method.
2.5. Asymmetric flow-field flow fractionation (AF4)
The AF4 equipment used was an Eclipse Dualtech Asymmetric Field Flow Fractionation system from Wyatt Technology (Dernbach, Germany), equipped with a polyether ether ketone spacer of 0.35 mm height and a 10 kDa nominal cutoff regenerated cellulose membrane (Postnova Analytics, Landsberg, Germany). Liposomal stock solutions were diluted 1:37 in NH4OAc to obtain samples. Injection volume was 50 μL for separation runs and 10 mL for recovery runs. In general, two and three replicates were performed for separation and recovery runs, respectively. Tip to tip channel length was 26.5 cm, the detector flow rate was set to 1 mL/min, and the focus flow rate to 0.8 mL/min with 12 min focusing time. A shallow cross-flow gradient profile was programmed: the initial crossflow rate was set to 0.8 mL/min and gradually decreased to 0.31 mL/min within 30 min, resulting in a cross-flow reduction rate of 0.0163 mL/min, which prevents unwanted effects of secondary relaxation and release of non-fractionated particles from the membrane due to reduction of the cross-flow to zero during the analytical run. After 30 min, cross-flow was reduced to 0.15 mL/min within 5 min, held stable for 5 min at 0.15 mL/min, and finally decreased to zero within 3 min. This was done to check if parts of the sample would elute under very low cross-flow field strengths, although no size information would be obtained from this part of the analysis. No such parts were observed and fractograms only originated from the region of the shallow gradient. Due to non-linearities, questionable applicability of FFF theory and missing calibration points, no data from the later part of the field-program were used. The carrier solution was 10 mM NaNO3 (Hupfeld et al., 2006) and was delivered with a 1200 series quaternary pump (Agilent Technologies, Waldbronn, Germany) equipped with a micro-vacuum degasser. Detection was achieved by means of a Dawn EOS multi angle light scattering detector (Wyatt Technology, Santa Barbara, CA, USA) and a fluorescence detector with excitation wavelength set to 629 nm, emission wavelength to 657 nm (1200 series FLD, Agilent Technologies, Santa Clara, CA, USA).
Two methods were used to calculate particle size. First, a second order calibration curve of particle size with retention time was acquired, using polystyrene standards with 20, 60, 100, and 150 nm diameter; liposome size was then calculated by converting retention time to diameter. Second, using the ASTRA software (Version 5), the spherical model was applied on MALLS data. This model was used since liposomes were expected to be of spherical shape.
2.6. Cryogenic transmission electron microscopy (TEM) measurements
Specimens for electron microscopy were prepared in a controlled environment vitrification system (CEVS) to ensure stable temperature and to avoid loss of solution during sample preparation. The specimens were prepared as thin liquid films, <300 nm thick, on lacey carbon filmed copper grids and plunged into liquid ethane at −180 °C. This leads to vitrified specimens, avoiding component segmentation and rearrangement, and water crystallization, thereby preserving original microstructures. The vitrified specimens were stored under liquid nitrogen until measured. An Oxford CT3500 cryoholder and its workstation were used to transfer the specimen into the electron microscope (Philips CM120 BioTWIN Cryo) equipped with a post-column energy filter (Gatan GIF100). The acceleration voltage was 120 kV. The images were recorded digitally with a CCD camera under low electron dose conditions.
Analysis of cryo-TEM images was performed using ImageJ 1.50i software (National Institutes of Health, Bethesda, Maryland). For each sample, more than 3000 liposomes from over 10 images were measured and size determined.
2.7. Cell culture
The human pancreatic adenocarcinoma cell line BxPc-3 was purchased from ATCC-LGC Standards (Manassas, VA, USA). The cells were maintained in RPMI60 (Gibco, Life Technologies, Grand Island, NY, USA) supplemented with 10% fetal bovine serum and antibiotics (100 U/ml penicillin and 100 μg/ml streptomycin). The cells were grown in T-75 culturing flasks (Sarstedt, Nümbrecht, Germany) and kept in a humidified 5% CO2 atmosphere at 37 °C. Culturing cell media were changed twice a week and the cells passaged before reaching confluence. Before experiments, cells were washed with Dulbecco’s phosphate-buffered saline without Ca+ or Mg2+ (PBS; Gibco), detached using trypsin-EDTA (Gibco) for 5 min, harvested and pelleted at 1.2 × 103 rpm for 4 min. After pellet dissociation by gentle pipetting, cell concentration and viability were determined using 0.06% trypan blue (Thermo Fisher Scientific, Waltham, MA, USA).
2.8. Cell proliferation assays
To assess cellular toxicity, MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) tests were carried out. For the experiments, cells were seeded (5 × 103 cells/well) in 96-well plates in standard culturing medium for 72 h at 37 °C, thus allowing cell adhesion and confluence before changing to serum-free media (SFM) for 24 h. After 24 h, SFM was removed and replaced with fresh SFM containing liposomes at different phospholipid concentrations. Untreated cells with only SFM at every plate were used as controls. Following incubation for 24 and 48 h, cell proliferation was assessed by MTT according to the manufacturer’s instructions (Roche Life Science, Indianapolis, Indiana, USA). Briefly, 10 μL of MTT reagent 1 (MTT labeling reagent) was added to each well and incubated for 4 h at 37 °C before adding 100 μL of MTT reagent 2 (solubilization solution, 10% SDS in 0.01 M HCl) and incubating at 37 °C before reading plates. The samples were measured on a Multiskan GO plate reader (test wavelength 595 nm, reference wavelength 660 nm) using the SkanIt Sotware 3.2 (both Thermo Fisher Scientific, Waltham, MA, USA).
2.9. Statistical analysis
Proliferation data are expressed as means ± SD of four replicate wells per plate. Statistical analyses were performed by one-way ANOVA and Student t-test using GraphPad Prism software (GraphPad Software Inc., La Jolla, CA, USA). A P-value of <0.05 was considered statistically significant.
For statistical analysis and creation of size distribution histograms, size distribution data from cryo-TEM images was exported from ImageJ software to Graphpad Prism software.
3. Results and discussion
Liposomes were prepared by the thin lipid film hydration technique, followed by repeated freezing and thawing cycles and serial extrusions through 400, 200 and 100 nm filters to ensure (i) an even liposomal size distribution around 100 nm, as well as (ii) employment of unilamellar vesicles for subsequent analyses.
3.1. nES GEMMA, DLS, AF4 and cryo-TEM measurements
We conducted size and size distribution studies for our nominally identical liposome batches by means of nES GEMMA, DLS, AF4 and cryo-TEM measurements.
-
(i)
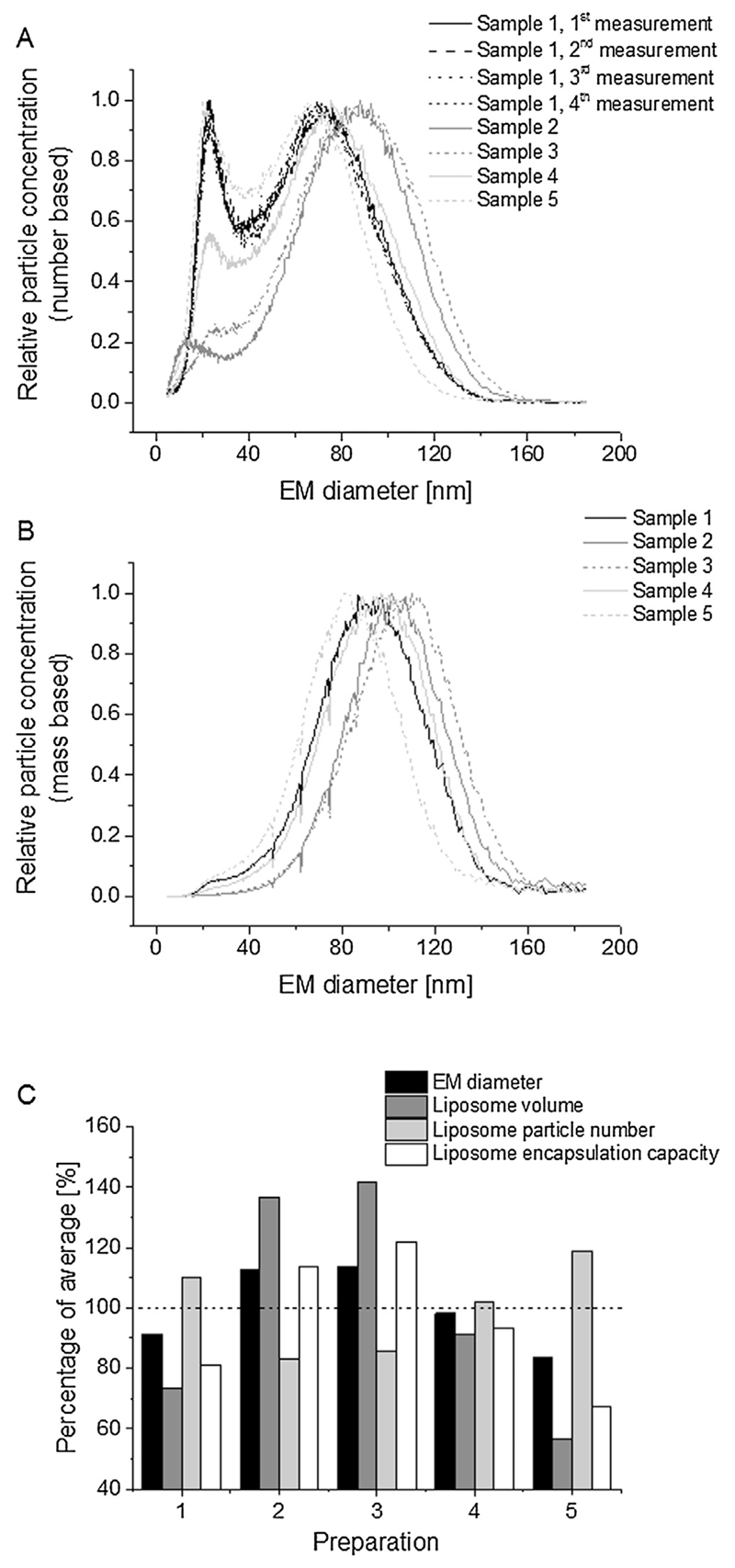
As already described in the introduction part, nES GEMMA yields diameters of surface-dry particles. Additionally, data can either be interpreted on the basis of number- or mass-concentrations. For number-based analysis, each nanoparticle enclosed in a droplet will be counted disregarding their size or mass. For the latter, information on smaller sized sample components (present to a varying degree in vesicle preparations) is lost, as preferentially particles with larger EM diameter values are displayed. However, as shown in Fig. 1A applying number-concentration based data analysis on nES GEMMA derived data, the five investigated, nominally identical liposome preparations varied significantly in the amount of low EM diameter sample components (around 20 nm EM diameter). The loss of information on smaller EM diameter sample compounds becomes evident in Fig. 1B plotting the same data as Fig. 1A but mass-concentration based. At the same time (upon mass-concentration based data evaluation), the EM diameter of the main vesicle peak apex is shifted to higher EM diameter values as presented in Table 1 as the mean of at least five independent readings for each sample for number- and mass-concentration based data evaluation. Finally, peak apex values for the main liposome peak of the five nominally identical liposome preparations showed a significant variation with EM diameter values between 63.2 and 87.3 nm EM diameter (number-concentration based data). The measurement method itself exhibited a good repeatability as demonstrated in Fig. 1A for one representative preparation batch. (For all batches, intrabatch relative standard deviation (RStD) values below 1.6%, n ≥ 5, were recorded, respectively.) Hence, the cause of this variation can be traced back to PEGylated lipid inclusion in liposomes as shown in a previous study (Weiss et al., 2016). Based on peak apex values as well as on particle numbers the putative encapsulation capacity of vesicles can be calculated from obtained particle numbers and EM diameters, the latter allowing the calculation of vesicle volumes (Fig. 1C).
-
(ii)
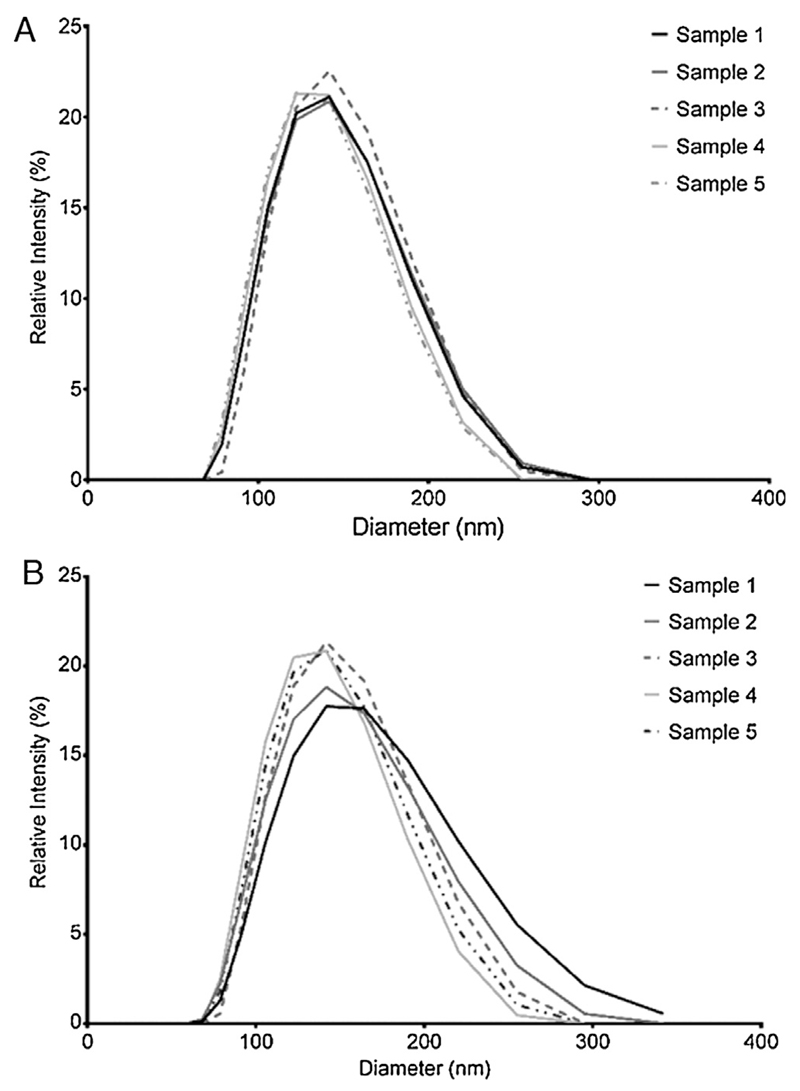
DLS size characterization yields readings of particle hydrodynamic diameter (Fig. 2). Data in Table 1 is presented as the mean ± standard deviation of three independent analyses at two different dilutions for the five nominally identical liposome preparation batches. DLS readings only showed one peak for every sample. The mean hydrodynamic diameter for both dilutions were 132.8 ± 3.5 and 138.5 ± 5.5 nm, respectively. PDI values (relative PDI for scattering = (peak width/mean)2) were below 0.1 for all liposome preparations and no larger species were detectable up to 1 μm particle diameter. Likewise, no smaller sized sample components (as detected with nES GEMMA upon number-based data evaluation) were detected. This latter finding was in accordance with mass-concentration based nES GEMMA data as expected from a light scattering technique exhibiting a preferential detection of larger sample compounds.
-
(iii)
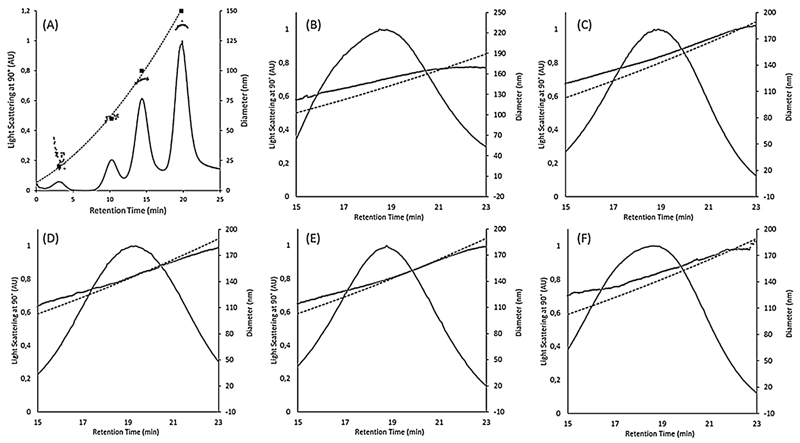
We used AF4 for the possibility to separate sample components, according to their size, prior to light scattering analysis (Dudkiewicz et al., 2015). However, it has to be kept in mind that even though particle separation occurs, which in principle simplifies the detection of multimodal size distributions in static light scattering as a series of measurements is performed on nearly monodisperse sample fractions, the sensitivity of the system decreases with decreasing particle size, as demonstrated in Fig. 3A for polystyrene size standards of comparable concentrations (5–10 ppm). Overall, for the setup used in this study, particles smaller than approximately 50 nm would pre-elute and may be influenced by the void peak (Kammer et al., 2005).
Fig. 1.
nES GEMMA analysis of n = 5 preparations of HSPC:Chol:DSPE-mPEG2000 (55:40:5 molar ratio) liposomes. To demonstrate the repeatability of measurements four standard spectra of sample 1 are shown (A). (B) Upon mass-concentration based data analysis, information on smaller-sized sample components is lost. (C) Calculation of liposome encapsulation capacities based on number-concentration based nES GEMMA data (percentage of average).
Table 1.
nES GEMMA data showing the EM diameter [nm] for recorded peaks in samples 1–5. Samples were diluted 1:25 in NH4OAc. Average and standard deviation values from at least n = 5 measurements are shown. Number- and mass-based data evaluation is compared. DLS data showing the Z-average in nm for each sample and two dilutions. Data presented as ‘mean diameter ± SD’ or ‘mean PDI ± SD’ from n = 3 measurements. No statistical difference could be found for DLS data between dilutions within the same sample (calculations not shown here).
| Sample | nES GEMMA |
DLS |
||||
|---|---|---|---|---|---|---|
| Number-based |
Mass-based | Dilution [v/v] | Z-average (diameter, nm) | Mean PDI | ||
| Peak 1 (EM diamter, nm) | Peak 2 (EM diameter, nm) | EM diameter (nm) | ||||
| 1 | 24.0 ± 0.4 | 69.5 ± 0.6 | 92.3 ± 0.5 | 1–100 1–1000 |
133.8 ± 0.9 147.2 ± 3.5 |
0.02 ± 0.02 0.09 ± 0.02 |
| 2 | 16.8 ± 0.9 | 85.3 ± 0.4 | 102.3 ± 0.6 | 1–100 1–1000 |
133.8 ± 0.3 138.9 ± 1.7 |
0.03 ± 0.01 0.06 ± 0.02 |
| 3 | 21.3 ± 2.8 | 87.3 ± 0.7 | 106.6 ± 0.6 | 1–100 1–1000 |
138.1 ± 1.4 140.4 + 1.1 |
0.01 ± 0.00 0.03 ± 0.01 |
| 4 | 25.2 ± 0.9 | 75.7 ± 1.2 | 95.0 ± 0.7 | 1–100 1–1000 |
129.5 ± 0.9 131.0 ± 1.4 |
0.02 ± 0.03 0.03 ± 0.03 |
| 5 | 23.5 ± 0.8 | 63.2 ± 0.6 | 84.1 ± 1.1 | 1–100 1–1000 |
128.5 ± 1.7 135.0 ± 3.4 |
0.02 ± 0.01 0.03 ± 0.02 |
Fig. 2.
DLS data presented for (A) 1–100 dilution and (B) 1–1000 dilution of n = 5 liposome preparations batches in NH4OAc.
Fig. 3.
MALLS signal (detector at 90°) over retention time for AF4 fractograms (solid black lines) of PS standards and five liposome preparation batches. (A) Fractogram of PS standard mix of 20, 60, 100, and 150 nm diameter (shown as solid squares). The detectability of PS particles decreases with their size. (B), (C), (D), (E), (F): Fractograms of samples 1, 2, 3, 4, and 5, respectively. Also shown are geometric diameters calculated from MALLS data using the spherical model (grey dots) and the hydrodynamic diameter calibration curve calculated from the elution time of the PS standards (dashed black line).
Nominally identical liposome preparation batches demonstrated similar fractograms (Fig. 3B–F). Hydrodynamic diameters were obtained by means of a calibration curve with polystyrene standards; in addition, geometric diameter was obtained by means of the multi-angle laser light scattering (MALLS). Overall, there was good agreement between the hydrodynamic diameter and the geometric diameter, which verifies that the liposomes are of spherical shape and fractionation occurs according to FFF theory in normal mode. Only for samples 1 and 5 and to a lesser extent sample 2, could small differences be seen between the geometric and hydrodynamic radius, indicating a possible deviation from the spherical shape, interactions of the particles with the membrane or multimodal particle size distributions.
Concerning the size differences for vesicles detected with AF4 when compared to nES GEMMA, it should be noted that very small particles (in the order of 20 nm) may not have been in large enough numbers to produce a readable intensity of scattered light in the MALLS detector of the AF4 system. An incontrovertible evidence for smaller-sized particle components on AF4 measurements alone is therefore not possible. However, smaller sized sample components in the order of 20 nm were indeed detected via nES GEMMA upon number-based data evaluation. Furthermore, samples 1 and 5 exhibited peak shapes that are not completely Gaussian, showing a shoulder towards the smaller size range (compare Fig. 3B and F). The same tendency can be seen for sample 1 and 5 in the DLS readings where they have the highest difference between dilutions of all samples as well as the highest standard deviation. This is evidence that these two samples contained a larger number of particles below the mean as compared to samples 2, 3, and 4 and explains differences in geometric and hydrodynamic radii for these samples.
-
(iv)
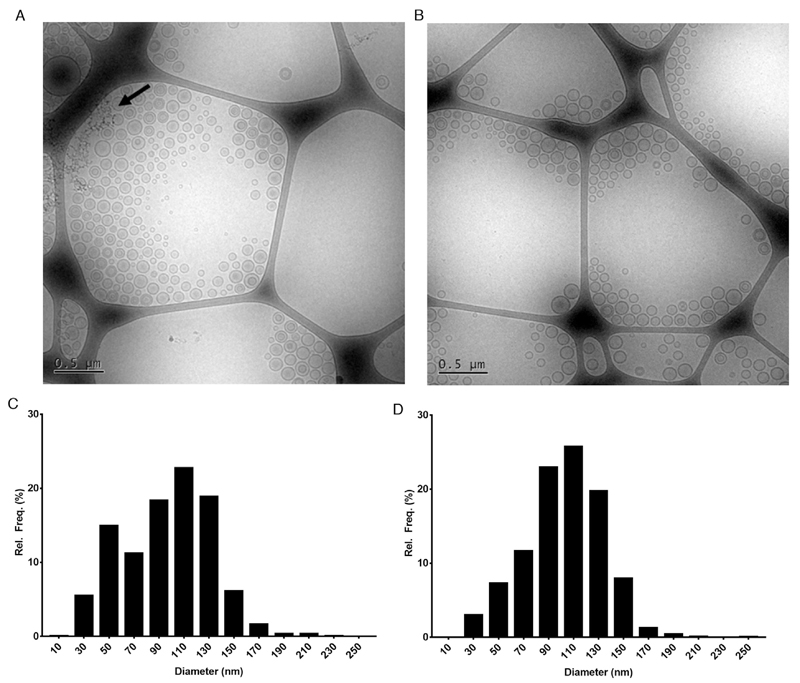
Representative cryo-TEM images for liposome preparation batches 1 and 3 are shown in Fig. 4 as well as their respective size distribution histograms. Sample 1 and 3 were selected for imaging as they show again the most and least pronounced, respectively, peaks for low diameter populations in nES GEMMA analysis. Sample 1 clearly shows the presence of many small diameter liposomes. In contrast, sample 3, although still showing the presence of small diameter populations, the frequency at which these small-diameter liposomes were observed was clearly less than for sample 1. These observations were corroborated by creating size distribution histograms by measuring more than 3000 particles from over 10 images for each sample. For sample 1, the smallest recorded liposome was at 16 nm in diameter and 20.42% of the total observed liposome population below 60 nm in diameter. For sample 3 on the other hand, the smallest recorded liposome was at 22 nm diameter and 10.22% of the total observed liposome population below 60 nm in diameter confirming that there indeed was a larger presence of low diameter liposomes in sample 1. Finally, the size distribution histograms clearly show a bimodal size distribution for sample 1 and a unimodal size distribution for sample 3, hence, corroborating nES GEMMA number-concentration based data.
Fig. 4.
Cryo-Transmission Electron Microscopy (cryo-TEM) images of liposomal samples (A) 1 and (B) 3 and their respective size distribution histograms in (C) and (D). In sample 1 there is a larger presence of small diameter liposomal populations compared to sample 3 (in accordance with number-based nES GEMMA data). Dark spots pointed out by an arrow in (A) are due to frost artifacts. Size distribution histograms of samples 1 (C) and 3 (D) confirmed the same size distributions as seen in nES GEMMA data. For the histograms, more than 3000 particles from several TEM images were counted and measured.
3.2. Interpretation of results obtained for analysis of liposome preparation batches
Analyte characterization via nES GEMMA yields surface-dry particle diameters and enables the calculation of particle volumes and thereof encapsulation capacities. DLS and AF4-MALLS in contrast yield hydrodynamic and geometric diameter values, which differ significantly from dry particle diameters; not only but also due to PEG-chains on the surface of vesicles being solvated and moving freely when in contact with a liquid but collapsing on a dry particle. Nevertheless, hydrodynamic values are necessary to estimate the behavior of vesicles in suspension, which is especially of importance when vesicles are administered in that form. Furthermore, AF4 with SLS and standalone DLS enables the detection of analytes and vesicle aggregates, whose size exceeds by far the detectable EM diameter range of the current nES GEMMA setup (mind that other GEMMA setups allow detection of particles up to 1 μm EM diameter and above). However, DLS detection alone is not capable to characterize bi- or multimodal analyte size distributions, even though various methods exist to transform the correlation curve to a size distribution, due to the limited size-resolution offered by the technique and the preferential detection of larger components. This effect can likewise be demonstrated by number- and mass-based evaluation of the same set of data obtained from nES GEMMA measurements. Upon mass-based data evaluation, information on smaller sized sample components is lost. These findings are supportive of the EU recommendations concerning nanoparticle characterization based on number concentrations (2011/696/EU from October 18th, 2011).
Occurrence of low EM diameter material influences (i) the overall size distribution in preparations as well as (ii) the number of larger liposomes due to inclusion of lipids in low EM diameter material instead of larger vesicles. Hence, the detection of low EM diameter material is of importance. The problem of bias in the intensity-based analyte detection via DLS is partially circumvented upon application of AF4 in combination with MALLS, a SLS detection system. Particle size separation prior to detection allows for separation of particles in a multimodal distribution and improves the detectors capabilities. However, the dependence of signal intensities on particle size in MALLS reduces the sensitivity for small particles with diameters <~50 nm. In principle, the intensity signal from any of the MALLS angles reflects a similar intensity-weighted characteristic as DLS shows. On top of the intensity-weighted signals from MALLS the detection principle relies on the mass of the particles in the detector. If the number-based distribution of nES GEMMA is transformed into a mass-based distribution, there are clear similarities between both techniques. However, AF4/MALLS results alone are not sufficient to demonstrate unambiguously the occurrence of a multimodal particle-number size distribution when such particles are present and information from additional methods is necessary.
Cryo-TEM reflects the multimodal size distribution of analytes besides giving a notion on overall vesicle shape and lamellarity. However, the large number of vesicle images necessary to obtain statistically valid information on liposomes renders this technique time- and cost-consuming. Nevertheless, cryo-TEM data supports our nES GEMMA findings, whereas AF4/MALLS and DLS failed to detect the presence of smaller-sized sample components. To conclude, besides differences in the determined liposome size (surface-dry particle diameters vs. hydrodynamic diameter values) employed analytical methods for vesicle characterization show differences especially in the detection of lower sized sample material. If this material indeed has an influence on the toxicological behavior of liposomes upon application as carrier vesicles will be targeted in the following section.
3.3. Does smaller-sized material influence cytotoxicity of liposome carriers?
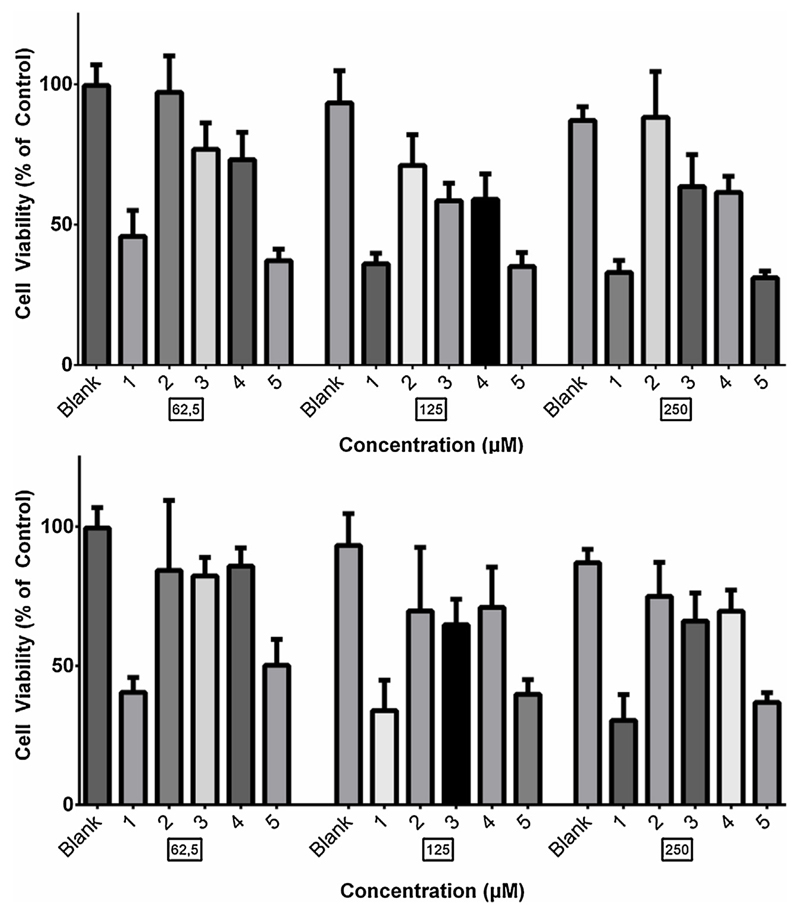
To assess whether the low diameter populations detected, especially in samples 1 and 5, during nES GEMMA measurements had a cytotoxic effect for bare vesicles, the cytotoxic profiles of the liposomal samples were acquired on BxPC-3 Pancreatic Cancer cells using the MTT assay (Fig. 5). Liposomes were diluted to match lipid concentrations similar to the ones used in studies where liposomal drug delivery was tested (Papa et al., 2012). For each sample and concentration, four replicates were made. Results in Fig. 5 are presented as “% of control” where control cells were left untreated, adding only SFM during incubation periods.
Fig. 5.
In-vitro dose- and time-dependent cytotoxic effect induced by liposomal samples on BxPC-3 cells. Analysis was performed by a standard MTT assay. Data are reported as percent of control (untreated cells). Results are means ± standard deviation, n = 4 from two different experiments.
The assays showed a clear correlation between increased cytotoxic effect and the presence of low diameter material. Samples 1 and 5 which showed the largest presence of small diameter particles in nES GEMMA measurements, induced a significantly higher cytotoxic effect compared to samples 4 and especially, 2 and 3 which showed the lowest presence of small diameter particles. Liposomal sample 2 showed the least cytotoxic effect and a significant difference compared to samples 1 and 5 could be seen for both 24 and 48 h. However, compared to samples 3 and 4, a significant difference in cytotoxic effect could only be seen at 24 h. Blank cells were treated with SFM and liposomal suspension medium at same dilution as liposomal samples. No significant cytotoxic effect could be seen from suspension medium compared to control cells. This indicates that even a small presence of low diameter material might have an undesired cytotoxic effect on cells. A summary of comparison between liposomal samples can be found in Table 2 underscoring the above mentioned differences between liposome preparations.
Table 2.
P-value and statistical significance of sample 2 and 3 in comparison to each other and liposomal samples 1, 4 and 5.
| (a) 24 h | ||||||
|---|---|---|---|---|---|---|
| Concentration | Sample | 1 | 2 | 3 | 4 | 5 |
| 62,5 | 2 | 0.0002 *** |
– – |
0.007 ** |
0.0011 ** |
0.0002 *** |
| 3 | 0.003 *** |
0.007 ** |
– – |
0.7768 n.s. |
0.0002 *** |
|
| 125 | 2 | 0.002 *** |
– – |
0.0148 * |
0.0648 n.s. |
0.0002 *** |
| 3 | 0.0002 *** |
0.0148 * |
– – |
0.854 n.s. |
0.0002 *** |
|
| 250 | 2 | 0.0002 *** |
– – |
0.006 *** |
0.0002 *** |
0.0002 *** |
| 3 | 0.0002 *** |
0.006 *** |
– – |
0.7768 n.s. |
0.0003 *** |
|
| (b) 48 h | ||||||
| Concentration | Sample | 1 | 2 | 3 | 4 | 5 |
| 62,5 | 2 | 0.0002 *** |
– – |
>0.99 – |
>0.99 n.s. |
0.003 ** |
| 3 | 0.0002 *** |
>0.99 – |
– – |
0.3754 n.s. |
0.0002 *** |
|
| 125 | 2 | 0.0011 ** |
– – |
0.7768 – |
0.7768 n.s. |
0.0006 *** |
| 3 | 0.0002 *** |
0.7768 n.s. |
– – |
0.4331 n.s. |
0.0002 *** |
|
| 250 | 2 | 0.0002 *** |
– – |
0.1296 n.s. |
0.2317 n.s. |
0.0002 *** |
| 3 | 0.0002 *** |
0.1296 n.s. |
– – |
0.7768 n.s. |
0.0002 *** |
|
n.s. stands for p > 0.05.
p ≤ 0.05.
p ≤ 0.01.
p ≤ 0.001.
4. Conclusions
The aim of our current work was to compare several independent, i.e. based on different physical principles, analysis methods in the characterization of liposomal vesicles applicable for drug delivery. In doing so we were able to highlight the ability of nES GEMMA to target surface-dry nanoparticles with number-concentration based analyte detection. Hence, differences between nominally identical liposome preparations concerning the size of liposomes and the presence of lower-sized sample material were demonstrated. These differences cannot be detected by DLS alone as the measurement is biased by the presence of preferentially detected larger particles. Additionally, it has to be considered that DLS yields overall higher diameter values (hydrodynamic radii vs. dry particles). Coupling of SLS detection with a preceding separation step (AF4) leading to separation of smaller and larger sample components did not allow detection of the multimodal size distribution, likely due to decreased detection sensitivity in the lower particle size range. In contrast, this multimodal particle size distribution of vesicles is demonstrated by cryo-TEM, a method that additionally allows the assessment of liposome lamellarity. Overall, it has to be stressed that especially the combination of independent analysis techniques allows an as complete as possible characterization of carrier vesicle preparations concerning (i) analyte size, (ii) lamellarity and (iii) the occurrence of lower-sized sample components. The latter information can be deduced from number-concentration based nES GEMMA data. From our data, it appears that lower-sized sample components play a major role in in vitro cytotoxicity as preparation batches with larger amounts of lower-sized particles exhibited a larger cytotoxic effect in cell culture. However, more experiments in the in vitro field are necessary in order to further strengthen our findings.
Acknowledgments
This project was supported by the Swedish Society for Medical Research (to CU) and the Austrian Science Fund (FWF), grant P25749-B20 (to VUW).
We thank Gunnel Karlsson at the Biomicroscopy Unit, Polymer and Materials Chemistry, Chemical Centre, Lund University, Lund, Sweden for the cryo-TEM work. The work was done at Biomicroscopy Unit, Polymer and Materials Chemistry, CAS, P.O. Box 124, S-221 00 LUND, Sweden.
We would also like to thank Erik Wisaeus at the Danish Technological Institute, Taastrup, Denmark for his assistance in cryo-TEM image analysis.
Abbreviations
- AF4
Asymmetric Flow-Field Flow Fractionation
- CPC
Condensation Particle Counter
- DLS
Dynamic Light Scattering
- EM
Electrophoretic Mobility
- GEMMA
Gas-phase Electrophoretic Mobility Molecular Analysis
- nDMA
nano Differential Mobility Analyzer
- nES
nano Electrospray
- PDI
Polydispersity Index
- PEG
Polyethylene Glycol
- SEC
Size Exclusion Chromatography
- SLS
Static Light Scattering
- TEM
ransmission Electron Microscopy
Footnotes
Conflict of interest
The authors of this paper have no conflict of interest to disclose.
References
- Allen TM, Cullis PR. Liposomal drug delivery systems: from concept to clinical applications. Adv Drug Deliv Rev. 2013;65:36–48. doi: 10.1016/j.addr.2012.09.037. [DOI] [PubMed] [Google Scholar]
- Almgren M, Edwards K, Karlsson G. Cryo transmission electron microscopy of liposomes and related structures. Colloids Surf A. 2000;174:3–21. [Google Scholar]
- Bacher G, Szymanski WW, Kaufman SL, Zollner P, Blaas D, Allmaier G. Charge-reduced nano electrospray ionization combined with differential mobility analysis of peptides, proteins, glycoproteins, noncovalent protein complexes and viruses. J Mass Spectrom. 2001;36:1038–1052. doi: 10.1002/jms.208. [DOI] [PubMed] [Google Scholar]
- Bangham AD, Glover JC, Hollingshead S, Pethica BA. The surface properties of some neoplastic cells. Biochem J. 1962;84:513–517. doi: 10.1042/bj0840513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibi S, Kaur R, Henriksen-Lacey M, McNeil SE, Wilkhu J, Lattmann E, Christensen D, Mohammed AR, Perrie Y. Microscopy imaging of liposomes: from coverslips to environmental sem. Int J Pharm. 2011;417:138–150. doi: 10.1016/j.ijpharm.2010.12.021. [DOI] [PubMed] [Google Scholar]
- Burdinski D, Pikkemaat JA, Emrullahoglu M, Costantini F, Verboom W, Langereis S, Grull H, Huskens J. Targeted lipocest contrast agents for magnetic resonance imaging: alignment of aspherical liposomes on a capillary surface. Angew Chem Int Ed Engl. 2010;49:2227–2229. doi: 10.1002/anie.200905731. [DOI] [PubMed] [Google Scholar]
- Chadha TS, Chattopadhyay S, Venkataraman C, Biswas P. Study of the charge distribution on liposome particles aerosolized by air-jet atomization. J Aerosol Med Pulm Drug Deliv. 2012;25:355–364. doi: 10.1089/jamp.2011.0967. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay S, Modesto-Lopez LB, Venkataraman C, Biswas P. Size distribution and morphology of liposome aerosols generated by two methodologies. AS&T. 2010;44:972–982. [Google Scholar]
- Drummond DC, Meyer O, Hong K, Kirpotin DB, Papahadjopoulos D. Optimizing liposomes for delivery of chemotherapeutic agents to solid tumors. Pharmacol Rev. 1999;51:691–743. [PubMed] [Google Scholar]
- Dubascoux S, Le Hecho I, Hassellov M, Von der Kammer F, Gautier MP, Lespes G. Field-flow fractionation and inductively coupled plasma mass spectrometer coupling: history, development and applications. J Anal Atom Spectrom. 2010;25:613–623. [Google Scholar]
- Dudkiewicz A, Wagner S, Lehner A, Chaudhry Q, Pietravalle S, Tiede K, Boxall AB, Allmaier G, Tiede D, Grombe R, von der Kammer F, et al. A uniform measurement expression for cross method comparison of nanoparticle aggregate size distributions. Analyst. 2015;140:5257–5267. doi: 10.1039/c5an00561b. [DOI] [PubMed] [Google Scholar]
- Epstein H, Afergan E, Moise T, Richter Y, Rudich Y, Golomb G. Number-concentration of nanoparticles in liposomal and polymeric multiparticulate preparations: empirical and calculation methods. Biomaterials. 2006;27:651–659. doi: 10.1016/j.biomaterials.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Hupfeld S, Holsaeter AM, Skar M, Frantzen CB, Brandl M. Liposome size analysis by dynamic/static light scattering upon size exclusion-/field flow-fractionation. J Nanosci Nanotechnol. 2006;6:3025–3031. doi: 10.1166/jnn.2006.454. [DOI] [PubMed] [Google Scholar]
- Hupfeld S, Moen HH, Ausbacher D, Haas H, Brandl M. Liposome fractionation and size analysis by asymmetrical flow field-flow fractionation/multi-angle light scattering: influence of ionic strength and osmotic pressure of the carrier liquid. Chem Phys Lipids. 2010;163:141–147. doi: 10.1016/j.chemphyslip.2009.10.009. [DOI] [PubMed] [Google Scholar]
- Kammer FVD, Baborowski M, Friese K. Field-flow fractionation coupled to multi-angle laser light scattering detectors: applicability and analytical benefits for the analysis of environmental colloids. Anal Chim Acta. 2005;552:166–174. [Google Scholar]
- Kaufman SL, Skogen JW, Dorman FD, Zarrin F, Lewis KC. Macromolecule analysis based on electrophoretic mobility in air: globular proteins. Anal Chem. 1996;68:1895–1904. doi: 10.1021/ac951128f. [DOI] [PubMed] [Google Scholar]
- Laouini AJ-MC, Limayem-Blouza I, Sfar S, Charcosset C, Fessi H. Preparation, characterization and applications of liposomes: state of the art. J Colloid Sci Biotechnol. 2012;1:22. [Google Scholar]
- Munin A, Edwards-Levy F. Encapsulation of natural polyphenolic compounds: a review. Pharmaceutics. 2011;3:793–829. doi: 10.3390/pharmaceutics3040793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraji Y, Fujita T, Itoh H, Fujita D. Preparation procedure of liposome-absorbed substrate and tip shape correction of diameters of liposome measured by afm. Microsc Res. 2013;1:4. [Google Scholar]
- Nag OK, Awasthi V. Surface engineering of liposomes for stealth behavior. Pharmaceutics. 2013;5:542–569. doi: 10.3390/pharmaceutics5040542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickel C, Angelstorf J, Bienert R, Burkart C, Gabsch S, Giebner S, Haase A, Hellack B, Hollert H, Hund-Rinke K, Jungmann D, et al. Dynamic light-scattering measurement comparability of nanomaterial suspensions. J Nanopart Res. 2014;16 [Google Scholar]
- Nii T, Ishii F. Encapsulation efficiency of water-soluble and insoluble drugs in liposomes prepared by the microencapsulation vesicle method. Int J Pharm. 2005;298:198–205. doi: 10.1016/j.ijpharm.2005.04.029. [DOI] [PubMed] [Google Scholar]
- Papa AL, Basu S, Sengupta P, Banerjee D, Sengupta S, Harfouche R. Mechanistic studies of gemcitabine-loaded nanoplatforms in resistant pancreatic cancer cells. BMC Cancer. 2012;12:419. doi: 10.1186/1471-2407-12-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignataro B, Steinem C, Galla HJ, Fuchs H, Janshoff A. Specific adhesion of vesicles monitored by scanning force microscopy and quartz crystal microbalance. Biophys J. 2000;78:487–498. doi: 10.1016/S0006-3495(00)76611-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamoorth M, Narvekar A. Non viral vectors in gene therapy—an overview. J Clin Diagn Res. 2015;9:GE01–06. doi: 10.7860/JCDR/2015/10443.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranville J, Montano MD. Size distributions. In: Mohammed B, Jamie RL, editors. Frontiers of Nanoscience. Chapter 3. Elsevier; 2015. pp. 91–1212. [Google Scholar]
- Ruozi B, Belletti D, Tombesi A, Tosi G, Bondioli L, Forni F, Vandelli MA. Afm, Esem, Tem, and Clsm in liposomal characterization: a comparative study. Int J Nanomed. 2011;6:557–563. doi: 10.2147/IJN.S14615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terreno E, Delli Castelli D, Violante E, Sanders HM, Sommerdijk NA, Aime S. Osmotically shrunken lipocest agents: an innovative class of magnetic resonance imaging contrast media based on chemical exchange saturation transfer. Chemistry. 2009;15:1440–1448. doi: 10.1002/chem.200801766. [DOI] [PubMed] [Google Scholar]
- Weiss VU, Kerul L, Kallinger P, Szymanski WW, Marchetti-Deschmann M, Allmaier G. Liquid phase separation of proteins based on electrophoretic effects in an electrospray setup during sample introduction into a gas-phase electrophoretic mobility molecular analyzer (CE-GEMMA/CE-ES-DMA) Anal Chim Acta. 2014;841:91–98. doi: 10.1016/j.aca.2014.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss VU, Urey C, Gondikas A, Golesne M, Friedbacher G, von der Kammer F, Hofmann T, Andersson R, Marko-Varga G, Marchetti-Deschmann M, Allmaier G. Nano electrospray gas-Phase electrophoretic mobility molecular analysis (Nes gemma) of liposomes: applicability of the technique for nano vesicle batch control. Analyst. 2016 doi: 10.1039/c6an00687f. [DOI] [PMC free article] [PubMed] [Google Scholar]