Abstract
In the current study, we planned to predict the optimal gene functions for osteosarcoma (OS) by integrating network-based method with guilt by association (GBA) principle (called as network-based gene function inference approach) based on gene ontology (GO) data and gene expression profile. To begin with, differentially expressed genes (DEGs) were extracted using linear models for microarray data (LIMMA) package. Then, construction of differential co-expression network (DCN) relying on DEGs was implemented, and sub-DCN was identified using Spearman correlation coefficient (SCC). Subsequently, GO annotations for OS were collected according to known confirmed database and DEGs. Ultimately, gene functions were predicted by means of GBA principle based on the area under the curve (AUC) for GO terms, and we determined GO terms with AUC >0.7 as the optimal gene functions for OS. Totally, 123 DEGs and 137 GO terms were obtained for further analysis. A DCN was constructed, which included 123 DEGs and 7503 interactions. A total of 105 GO terms were identified when the threshold was set as AUC >0.5, which had a good classification performance. Among these 105 GO terms, 2 functions had the AUC >0.7 and were determined as the optimal gene functions including angiogenesis (AUC =0.767) and regulation of immune system process (AUC =0.710). These gene functions appear to have potential for early detection and clinical treatment of OS in the future.
Keywords: Osteosarcoma, Spearman correlation coefficient, Gene ontology, Area under the curve, Differential co-expression network, Guilt by association
1. Introduction
Osteosarcoma (OS) is the most commonly diagnosed histological form of primary bone tumor with high morbidity, and it is mainly prevalent in teenagers and young people [1]. Pain is the most common early symptom of OS and can cause fracture of the affected bone. Currently, multiple therapeutic strategies for OS, for example, surgical resection, chemotherapy and radiotherapy, have significantly improved the prognosis of patients with OS [2]. However, the overall survival rates rarely exceed 60–65% [3]. Moreover, a significant portion of OS patients develop metastasis even after curative resection of the primary tumor. There is still a long way to go for the management of OS [4]. Therefore, with an attempt to continue to make progress in the diagnosis and management of OS, identification of sensitive and specific minimally invasive signatures is one of the most important challenges.
Genetic aberration has been demonstrated as an important factor that may play significant roles in OS pathogenesis. For instance, Zhu and colleagues [5] have reported that SOX9 is over-expressed in OS tissues compared with controls. Moreover, it is indicated that FOXM1 is up-regulated, which suggests FOXM1 might be an valuable bio-signature for OS [6]. Nevertheless, these genes do not treat the OS efficiently or selectively, because molecules frequently don’t work individually, yet co-operate with the other genes. Additionally, genetic factors can disturb the protein levels, thereby in turn perturb the molecule interactions. Network is characterized by the complicated interactions and the complex interwoven relationships that control cellular functions [7]. Hence, understanding the networks will be beneficial to provide new insights to explore the molecular pathogenesis of OS. The concept of differential co-expression network (DCN) has been employed to the studies of OS, due to statistical confidence of single connections, overlap with protein interaction, and mathematical convenience [8]. In addition, improving our knowledge of gene function in uncharacterized genes is a major task [9]. Remarkably, gene interactions can be applied to deduce functional relationships based on a principle known as “guilt by association” (GBA) [10]. GBA has been indicated to predict gene function in various types of biological networks, for example, gene co-expression network [11].
Thus, in our work, we integrated network-based method with GBA principle (called as network-based gene function inference approach) to further identify the optimal gene functions for OS using gene ontology (GO) data and gene expression profile. To begin with, differentially expressed genes (DEGs) were extracted using linear models for microarray data (LIMMA) package. Then, construction of DCN based on DEGs was implemented, and sub-DCN was identified using Spearman correlation coefficient (SCC). Subsequently, GO annotations for OS were collected according to known confirmed database and DEGs. Ultimately, gene functions were predicted by means of GBA principle based on the area under the curve (AUC) for GO terms, and we determined GO terms with AUC >0.7 as the optimal gene functions for OS. These gene functions appear to have potential for early detection and clinical treatment of OS in the future.
2. Materials and methods
2.1. Gene expression profile and data pre-treatment
OS-related microarray data (accession number E-GEOD-36001) [12] were downloaded from ArrayExpress database based on the platform of A-MEXP-930-Illumina Human-6 v2 Expression BeadChip. There were 19 OS samples and 6 normal samples in E-GEOD-36001. Prior to analysis, we firstly pre-processed the microarray profile of E-GEOD-36001. Specifically, robust multi-array average (RMA) was used to perform background correction [13]. Quartile algorithm was utilized to implement quartile normalization [14], following by perfect match (PM)/mismatch (MM) correction using microarray suite (MAS) 5.0 package [15]. Ultimately, the data on probe levels were converted into gene symbols relying on annotate package [16]. Overall, 19,027 genes were reserved for further exploitation.
2.2. Identification of DEGs
The LIMMA package [17] in R language and an empirical Bayes framework were applied in our analysis to achieve DEGs between OS and normal samples. A t-test was conducted, and the multiple test was applied to correct the raw P values using the Benjamini & Hochberg [18] method based on false discovery rate (FDR). DEGs were extracted on the basis of FDR <0.05 and |log fold change (FC)| ≥2.
2.3. Generation of DCN
Cytoscape (http://cytoscape.org/) is an open source software which integrates bio-molecular interactions with high-throughput expression data as well as other molecular states into a unified conceptual network [19]. Therefore, we inputted DEGs into the Cytoscape software to visualize the DCN. Furthermore, with the goal of assessing the co-expressed strength of each interaction in the DCN, SCC was used in our study. As we all know, SCC is used to measure the co-expression probability of two variables by assessing the strength of association of two co-expressed variables and it ranges from −1 to 1 inclusive [20], [21]. The SCC absolute value of one interaction was determined as the weight value of the corresponding edge, and the higher the weight value was, the more relevant the interaction was related to the disease. Thus, we selected the edges with weight values higher than 0.8 to construct the sub-DCN. In order to display the sub-DCN more vividly, Cytoscape software was applied.
2.4. GO annotation for DEGs
GO annotation has been broadly utilized as functional enrichment studies for large-scale genes [22]. In our study, human GO annotations comprised of 19,003 GO terms covering 18,402 genes were retrieved from the GO consortium (http://geneontology.org/). In an attempt to obtain stable performance, we filtered for the GO annotations on gene size such that each remaining GO term had associated genes >20, and a total of 1313 GO terms were left to be used in our analysis. Next, DEGs identified above were mapped to the 1313 GO annotations. Finally, the GO slim set was obtained, consisting of 123 DEGs and 137 GO terms.
2.5. Identifying gene functions based on “GBA” prediction
Gene networks have been widely used in gene function prediction algorithms, many based on complex extensions of the “GBA” principle. As demonstrated here, GBA prediction approach was utilized to predict significant gene functions in OS progression. For GBA method, we used three-fold cross-validation to extract a ranked list scoring genes in DCN as to how they belonged in the known GO terms. The sum of co-expression values between the training set (co-expression) and the candidate gene was divided by the sum of co-expression values between the genes outside the training set and the candidate gene to analyze degree of candidacy. In detail, for each gene i in the DCN, all other neighbored genes of gene i were mapped to each GO category K, and the multifunctionality (MF) score for each gene i within the K-GO term based on the following equation:
In this formula, stood for the number of genes in GO term i, and denoted the count of genes outside GO category i.
Then, based on support vector machine (SVM), AUC for each GO group K was calculated, and the mean AUC across all GO terms was determined. Thus, the AUC scores were ordered from the highest to the lowest, the ranks of GO terms were ordered oppositely. The AUC of 0.5 stands for classification at chance levels, while the AUC of 1.0 denotes a perfect classification. In the literature about the gene function prediction, the AUC greater than 0.7 are regarded good [23]. In our study, GO terms with AUC >0.7 were identified and regarded as the optimal gene functions.
3. Results
3.1. DCN construction
Before DCN generation, we firstly identified DEGs between OS and normal samples using t-test. Based on FDR <0.05 and |log FC=≥ 2, a total of 123 DEGs were identified. The top 20 DEGs were shown in Table 1. The most significant DEGs were HOXB7, RHPN2, SRGN, FOXF2, and PLVAP.
Table 1.
List of the top 20 differentially expressed genes (DEGs).
| Genes | |log (fold change)| | False discovery rate (FDR) |
|---|---|---|
| HOXB7 | 2.085898 | 1.33E−07 |
| RHPN2 | 2.147406 | 6.32E−07 |
| SRGN | 5.174341 | 3.34E−06 |
| FOXF2 | 2.318869 | 4.51E−06 |
| PLVAP | 3.404251 | 1.98E−05 |
| COX7A1 | 2.988735 | 5.41E−05 |
| APOE | 3.399852 | 6.21E−05 |
| SPINT2 | 2.301193 | 6.65E−05 |
| LXN | 2.523381 | 8.93E−05 |
| TNFRSF1B | 2.427194 | 1.10E−04 |
| VAMP8 | 3.494650 | 1.61E−04 |
| CBS | 2.868625 | 1.76E−04 |
| HCLS1 | 2.686705 | 1.81E−04 |
| PHGDH | 2.362045 | 2.15E−04 |
| GIMAP7 | 2.258177 | 2.74E−04 |
| C1QC | 2.885076 | 2.76E−04 |
| HBB | 5.191561 | 2.86E−04 |
| C1QA | 3.521570 | 2.93E−04 |
| TYROBP | 3.511873 | 3.17E−04 |
| C1QB | 3.184835 | 3.17E−04 |
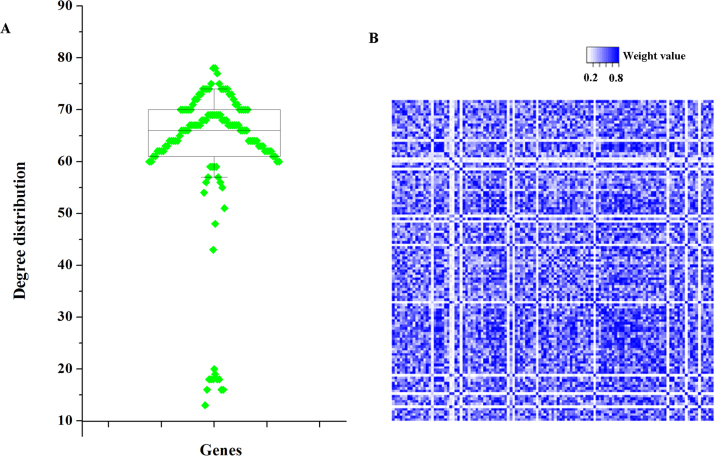
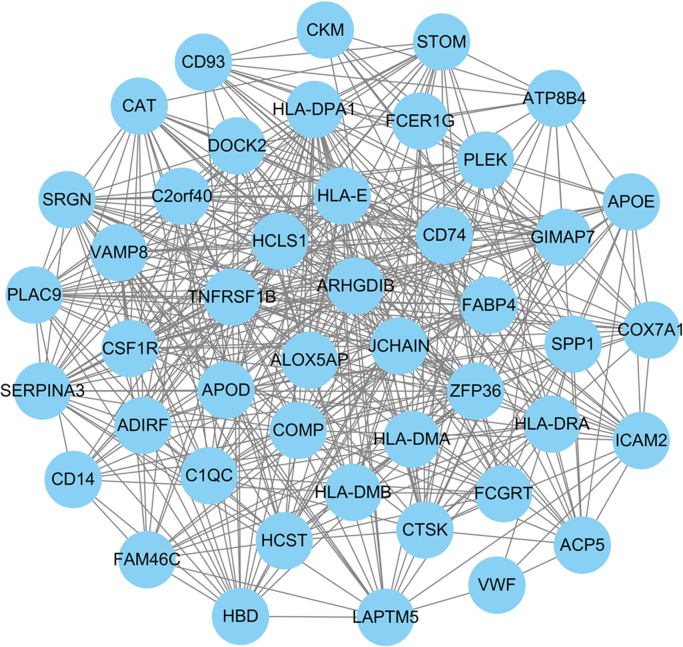
In order to further explore the biological activities of DEGs, we constructed a DCN using the above-identified 123 DEGs. In this DCN, there were 123 nodes and 7503 interactions. Under the DCN, node degree was more than just an statistic about a gene, and significantly, degree could explain the structures of the network. Then, topological degree for each node was computed through summing up the nodes it linked directly, and the degree distribution for each node was shown in Fig. 1A. We found that the ranges of degrees for a large number of DEGs (about 79%) was from 59 to 74, and genes (TNFRSF1B and HLA-E) had the highest degrees of 78. In addition to degree, the interacted strength was an index to estimate the interactions in the DCN. Thus, we used SCC to assign a weight value to each edge, and the specific information were demonstrated in Fig. 1B where a square stood for an edge in the DCN. The deeper the color was, the larger the weight value was. Significantly, among the interactions, there was a good liner correlation which indicated that the DCN had a good network scale property. Then, we selected the interactions with weight values >0.8 to establish the sub-DCN which was revealed in Fig. 2. In the sub-DCN, there were 46 nodes and 438 interactions. The degree distribution was shown in Table 2. Interestingly, in this sub-DCN, TNFRSF1B also had the highest degree (degree=33).
Fig. 1.
Differentially co-expressed network (DCN) construction for osteosarcoma (OS) based on differentially expressed genes (DEGs). A. Degree distribution of genes in the DCN. B. Weights distribution of edges in the DCN. Heatmap clarified weight distribution for each interaction.
Fig. 2.
Sub-DCN using the cut-off threshold of weight value >0.8. In the sub-DCN, there were 46 nodes and 438 interactions.
Table 2.
Degree distribution of 46 genes in the sub-differential co-expression network (DCN).
| Genes | Degree | Genes | Degree |
|---|---|---|---|
| TNFRSF1B | 33 | HBD | 20 |
| ZFP36 | 32 | STOM | 19 |
| HLA-E | 31 | DOCK2 | 18 |
| ARHGDIB | 28 | HLA-DMA | 17 |
| HLA-DPA1 | 28 | APOE | 17 |
| VAMP8 | 26 | LAPTM5 | 17 |
| FABP4 | 24 | ICAM2 | 17 |
| GIMAP7 | 24 | FCGRT | 16 |
| CD74 | 24 | ACP5 | 15 |
| PLEK | 23 | APOD | 15 |
| COMP | 23 | COX7A1 | 15 |
| HCLS1 | 23 | HLA-DMB | 15 |
| PLAC9 | 23 | CD93 | 14 |
| JCHAIN | 23 | ATP8B4 | 14 |
| SERPINA3 | 23 | FAM46C | 14 |
| C2orf40 | 22 | CD14 | 14 |
| SRGN | 22 | SPP1 | 13 |
| CTSK | 21 | HLA-DRA | 12 |
| CSF1R | 21 | FCER1G | 12 |
| CAT | 21 | ALOX5AP | 12 |
| ADIRF | 21 | CKM | 8 |
| HCST | 21 | VWF | 4 |
| C1QC | 21 |
3.2. Identifying gene functions using GBA prediction
In our study, the MF score was first calculated for each gene in a GO term, which influenced the counting membership in a given GO term by how much the gene contributed to the corresponding GO function. The higher the MF score of a gene was, the greater extent to which it should be a good candidate for owing any biological process. Consequently, a single ranked list of genes that best captured candidacy across all functions was equivalent to a list of genes sorted by MF score. Intuitively, if one was forced to choose a single ranking, the gene with the most GO annotations could be predicted as being in all GO categories. This was because if one gene was enriched in 100 GO terms (high MF score), and another was in only one (low MF score), by placing the former gene ahead of the latter gene in a fixed ranking, we made a correct prediction more often across all GO categories. Thus, with the goal of classifying OS patients and normal controls, 3-fold cross-validation was utilized to compute the AUC for GO functions based on MF scores.
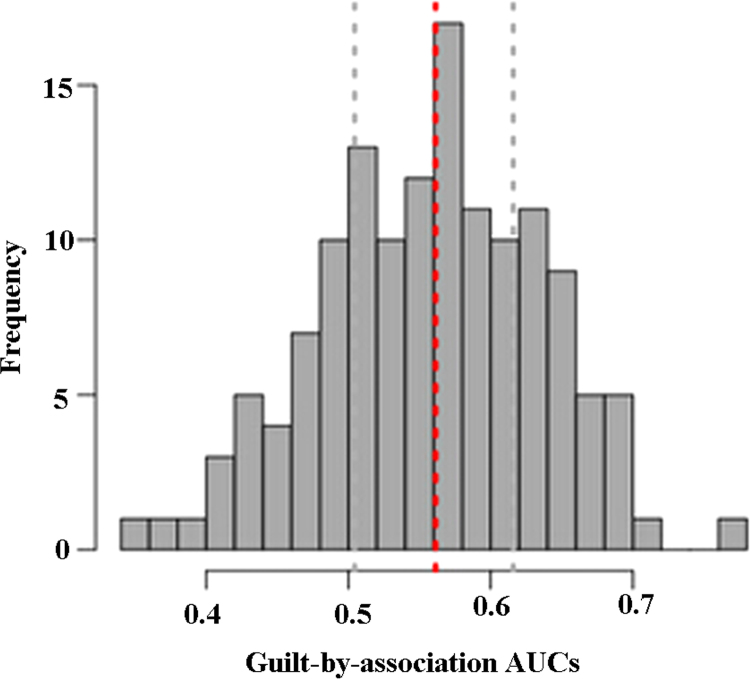
The AUC distribution for GO terms was shown in Fig. 3. The AUC for the majority of functional terms ranged from 0.4 to 0.7, especially from 0.5 to 0.65. AUC was used as a predictor to select significant GO terms, so, a total of 105 GO terms were identified based on AUC >0.5. Moreover, among these 105 GO terms, 2 GO terms had the AUC >0.7 and were determined as the optimal gene functions including angiogenesis (AUC =0.767) and regulation of immune system process (AUC =0.710).
Fig. 3.
Gene function prediction performance using guilt by association (GBA). The histogram of AUCs across all GO terms which can be obtained using a single list constructed from number of coexpression partners.
4. Discussion
Compared to gene focused researches, gene set and gene signature focused functional investigations appear much rewarding in understanding functional insights [24], and integrative functional genomic analysis of tumors promotes the understanding of the molecular mechanisms of cancers [25]. Network modeling on the basis of co-expression pattern analysis has been widely applied in a variety of cancers to understand the biology processes of cancers, and to obtain clinical insights [26]. Recently, a large amount of techniques have been created to extend GBA to indirect connections to predict gene functions [27], [28]. Gene interactions can be applied to deduce functional relationships based on a principle known as GBA [10]. The GBA principle forms the basis for most gene function prediction methods, which typically use relational information (e.g. interactions) to predict new gene membership in gene function categories [29]. However, combination of gene function prediction and network analysis are sparse. Generally speaking, network based-GBA analysis method can make exhaustively examining issues faster and easier than the simple GBA approach. Thus, in this work, we combined GBA principle with DCN-based analysis to further explore both direct and indirect optimal gene functions for OS on the basis of GO information as well as gene expression data. Finally, a total of 105 GO terms were identified based on AUC >0.5, which had a good classification ability. Moreover, 2 out of 105 GO terms had the AUC >0.7 and were determined as the optimal gene functions including angiogenesis and regulation of immune system process.
The ability of tumors to progress to more malignant phenotypes depends on tumor microenvironment. Blood capillaries, made up of endothelial cells, are usually static under physiological conditions but are induced to proliferate by direct exposure to angiogenic factors, for instance, vascular endothelial growth factor (VEGF) [30]. The proliferation and migration of endothelial cells play an important role in tumor angiogenesis [31]. As we all know, angiogenesis, development of new blood vessels from preexisting vascular bed, plays important roles during tumor growth and metastasis [32]. Significantly, the concept that malignant neoplasm onset, growth, and invasion rely on angiogenesis is broadly recognized and accepted [33], [34]. Theoretically, cancer cells, like normal cells, also need the delivery of oxygen as well as nutrients through blood vessels to survive and grow. In situ carcinoma, there seems to be a prolonged dormant period during which the cancer is not angiogenic, and is restrained in growth [35]. When sufficient cancer cells have transformed to the angiogenic phenotype from the quiescent phenotype, neovascularization may initiate, and thus rapid tumor growth can proceed. Through the literatures, we found that gene function of ‘angiogenesis’ has already been established to be important for OS. For example, previous studies have implicated that high recurrence rate and metastatic potential of OS are connected with high levels of angiogenesis [36], [37]. Moreover, Wang et al. [38] have used differential co-expression network to find that angiogenesis is closely related to OS progression and metastasis. Most importantly, suppression of cancer angiogenesis has been regarded as an attractive strategy for tumor therapy [39]. Thus, anti-angiogenic therapy might be a consideration for OS patients, with the majority of OS showing high vascularization [40].
During the tumor occurrence and metastasis, cancer cells can modulate the immune system of the host to find strategies that enable them to survive from the immune surveillance [41]. Cancer immunoediting is one of the two major strategies to get away from immune surveillance [42]. Innate and adaptive immunity appear to contribute to cancer immunoediting [43]. Previously, lymphocytes and IFNγ have indicated to prevent tumor immunoediting, thereby preventing the selection of less immunogenic tumor cells [44]. Indeed, immune cells (for example, macrophages cells) release soluble agents like chemokines and cytokines promoting the migration and infiltration of leukocytes that exert important functions in tumor development [45]. OS is frequently infiltrated by immune cells including macrophages and T cells [46]. Moreover, macrophage migration inhibitory factor is an proinflammatory cytokine which exerts an crucial function in the immune system [47]. Macrophage migration inhibitory factor contributes to cell proliferation, survival, and tumor-related angiogenesis [48], [49]. Moreover, Han et al. [50] have suggested that macrophage migration inhibitory factor can serve as a prognostic marker and a potential therapeutic target for OS. Interestingly, a link between p53 and IFN system was recently discovered in regulating tumor suppression and immunity [51]. Abnormalities of p53 gene in OS occur with a high incidences approaching 50% of all cases [52]. Currently, growing evidence implicates that the immune system is a fundamental player in cancer and a key determinant of prognosis and response to therapy [53], [54]. While, understanding the crosstalk between OS cells and the immune system, and how they drive tumorigenesis is still in its infancy. Hence, it is urgently needed to develop novel drugs that would potentiate the immune system in OS patients to act against this disease by means of immunomodulatory methods.
The current study had several limitations. First, there were limited clinical samples which might lead to biased estimates. Second, our analysis was conducted using bioinformatics methods but the conclusions have not been verified by means of any lab technique. Last but not least, we did not compare the obtained findings using other approaches. Despite these limitations, our findings had important implications for the molecular mechanisms underlying OS, yet further experimental study is still needed to validate our study.
In a nutshell, using network-based method with GBA based on GO and microarray profile of OS, we extracted 2 optimal gene functions during OS progression including angiogenesis and regulation of immune system process which might contribute to the successful identification of therapeutic targets for OS. Further studies might shed novel insights into the role of the gene functions in the pathophysiology of OS.
Conflicts of Interest
We declare that there are no conflicts of interest.
References
- 1.Jones K.B., Salah Z., Sara D.M., Galasso M., Gaudio E., Nuovo G.J. MicroRNA signatures associate with pathogenesis and progression of osteosarcoma. Cancer Res. 2012;72:1865–1877. doi: 10.1158/0008-5472.CAN-11-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang J., Zhang W. New molecular insights into osteosarcoma targeted therapy. Curr. Opin. Oncol. 2013;25:398–406. doi: 10.1097/CCO.0b013e3283622c1b. [DOI] [PubMed] [Google Scholar]
- 3.Folio C., Zalacain M., Zandueta C., Ormazábal C., Sierrasesúmaga L., San J.M. Cortactin (CTTN) overexpression in osteosarcoma correlates with advanced stage and reduced survival. Cancer Biomark. 2011;10:35. doi: 10.3233/CBM-2012-0227. [DOI] [PubMed] [Google Scholar]
- 4.Hameed M., Dorfman H. Elsevier; Amsterdam, Holland: 2011. Primary Malignant Bone Tumors—Recent Developments. in Seminars in Diagnostic Pathology. [DOI] [PubMed] [Google Scholar]
- 5.Zhu H., Jie T., Tang M., Cai H. Upregulation of SOX9 in osteosarcoma and its association with tumor progression and patients' prognosis. Diagn. Pathol. 2013;8 doi: 10.1186/1746-1596-8-183. (183-183) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fan C.L., Jiang J., Liu H.C., Yang D. Forkhead box protein M1 predicts outcome in human osteosarcoma. Int. J. Clin. Exp. Med. 2015;8:15563–15568. [PMC free article] [PubMed] [Google Scholar]
- 7.Sun S.-Y., Liu Z.-P., Zeng T., Wang Y., Chen L. Spatio-temporal analysis of type 2 diabetes mellitus based on differential expression networks. Sci. Rep. 2013:3. doi: 10.1038/srep02268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma S., Shah S., Bohnert H.J., Snyder M., Dineshkumar S.P. Incorporating motif analysis into gene co-expression networks reveals novel modular expression pattern and new signaling pathways. PLoS Genet. 2013;9:2184–2196. doi: 10.1371/journal.pgen.1003840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eisenberg D., Marcotte E.M., Xenarios I., Yeates T.O. Protein function in the post-genomic era. Nature. 2000;405:823–826. doi: 10.1038/35015694. [DOI] [PubMed] [Google Scholar]
- 10.Oliver S. Proteomics: guilt-by-association goes global. Nature. 2000;403:601–603. doi: 10.1038/35001165. [DOI] [PubMed] [Google Scholar]
- 11.Peña-Castillo L., Tasan M., Myers C.L., Lee H., Joshi T., Zhang C. A critical assessment of Mus musculus gene function prediction using integrated genomic evidence. Genome Biol. 2008;9(Suppl. 1):S2. doi: 10.1186/gb-2008-9-s1-s2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kresse S.H., Rydbeck H., Skårn M., Namløs H.M., Barraganpolania A.H., Cletonjansen A.M. Integrative analysis reveals relationships of genetic and epigenetic alterations in osteosarcoma. PLoS One. 2012;7 doi: 10.1371/journal.pone.0048262. (e48262-e48262) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Irizarry R.A., Hobbs B., Collin F., Beazer‐Barclay Y.D., Antonellis K.J., Scherf U. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 14.Bolstad B.M., Irizarry R.A., strand M., Speed T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 15.Pepper S.D., Saunders E.K., Edwards L.E., Wilson C.L., Miller C.J. The utility of MAS5 expression summary and detection call algorithms. BMC Bioinforma. 2007;8:1. doi: 10.1186/1471-2105-8-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu L.J., Gazin C., Lawson N.D., Pagès H., Lin S.M., Lapointe D.S. ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinforma. 2010;11:1–10. doi: 10.1186/1471-2105-11-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smyth G.K. Springer; Berlin, Germany: 2005. Limma: Linear Models for Microarray Data, in Bioinformatics and Computational Biology Solutions Using R and Bioconductor; pp. 397–420. [Google Scholar]
- 18.Benjamini Y., Drai D., Elmer G., Kafkafi N., Golani I. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 19.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hauke J., Kossowski T. Comparison of values of Pearson's and Spearman's correlation coefficients on the same sets of data. Quaest. Geogr. 2011;30:87–93. [Google Scholar]
- 21.Mukaka M.M. Statistics corner: a guide to appropriate use of correlation coefficient in medical research. Malawi Med. J.: J. Med. Assoc. Malawi. 2012;24:69–71. [PMC free article] [PubMed] [Google Scholar]
- 22.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gillis J., Pavlidis P. The role of indirect connections in gene networks in predicting function. Bioinformatics. 2011;27:1860–1866. doi: 10.1093/bioinformatics/btr288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ponomarev I., Wang S., Zhang L., Harris R.A., Mayfield R.D. Gene co-expression networks in human brain identify epigenetic modifications in alcohol dependence. J. Neurosci. Off. J. Soc. Neurosci. 2012;32:1884–1897. doi: 10.1523/JNEUROSCI.3136-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gerlinger M., Rowan A.J., Horswell S., Larkin J., Endesfelder D., Gronroos E. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doig T.N., Hume D.A., Theocharidis T., Goodlad J.R., Gregory C.D., Freeman T.C. Coexpression analysis of large cancer datasets provides insight into the cellular phenotypes of the tumour microenvironment. BMC Genom. 2013;14:469. doi: 10.1186/1471-2164-14-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chua H.N., Sung W.K., Wong L. Exploiting indirect neighbours and topological weight to predict protein function from protein-protein interactions. Bioinformatics. 2008;22:1623–1630. doi: 10.1093/bioinformatics/btl145. [DOI] [PubMed] [Google Scholar]
- 28.Yip A.M., Horvath S. Gene network interconnectedness and the generalized topological overlap measure. BMC Bioinforma. 2007;8:22. doi: 10.1186/1471-2105-8-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mostafavi S., Ray D., Warde-Farley D., Grouios C., Morris Q. GeneMANIA: a real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 2008;9(Suppl. 1):S4. doi: 10.1186/gb-2008-9-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bussolino F., Mantovani A., Persico G. Molecular mechanisms of blood vessel formation. Trends Biochem. Sci. 1997;22:251–256. doi: 10.1016/s0968-0004(97)01074-8. [DOI] [PubMed] [Google Scholar]
- 31.Leung D.W., Cachianes G., Kuang W.J., Goeddel D.V., Ferrara N. Vascular endothelial growth factor is a secreted angiogenic mitogen. Science. 1989;246:1306–1309. doi: 10.1126/science.2479986. [DOI] [PubMed] [Google Scholar]
- 32.Folkman J. Angiogenesis in cancer, vascular, rheumatoid and other disease. Nat. Med. 1995;1:27–31. doi: 10.1038/nm0195-27. [DOI] [PubMed] [Google Scholar]
- 33.Folkman J. The role of angiogenesis in tumor growth. Semin. Cancer Biol. 1992;3:65–71. [PubMed] [Google Scholar]
- 34.Thompson W.D., Shiach K.J., Fraser R.A., Mcintosh L.C., Simpson J.G. Tumours acquire their vasculature by vessel incorporation, not vessel ingrowth. J. Pathol. 1987;151:323–332. doi: 10.1002/path.1711510413. [DOI] [PubMed] [Google Scholar]
- 35.Folkman J. Tumor angiogenesis: therapeutic implications. N. Engl. J. Med. 1971;285:1182. doi: 10.1056/NEJM197111182852108. [DOI] [PubMed] [Google Scholar]
- 36.Yang J., Yang D., Sun Y., Sun B., Wang G., Trent J.C. Genetic amplification of the vascular endothelial growth factor (VEGF) pathway genes, including VEGFA, in human osteosarcoma. Cancer. 2011;117:4925–4938. doi: 10.1002/cncr.26116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mcmanus M.M., Weiss K.R., Hughes D.P.M. Understanding the role of Notch in osteosarcoma. Adv. Exp. Med. Biol. 2014;804:67–92. doi: 10.1007/978-3-319-04843-7_4. [DOI] [PubMed] [Google Scholar]
- 38.Wang Q. Identification of biomarkers for metastatic osteosarcoma based on DNA microarray data. Neoplasma. 2015;62:365. doi: 10.4149/neo_2015_044. [DOI] [PubMed] [Google Scholar]
- 39.Cooney M.M., Heeckeren W.V., Bhakta S., Remick O.S.C. Drug insight: vascular disrupting agents and angiogenesis--novel approaches for drug delivery. Nat. Clin. Pract. Oncol. 2006;3:682–692. doi: 10.1038/ncponc0663. [DOI] [PubMed] [Google Scholar]
- 40.Kunz P., Fellenberg J., Moskovszky L., Sápi Z., Krenacs T., Machado I. Improved survival in osteosarcoma patients with atypical low vascularization. Ann. Surg. Oncol. 2015;22:489. doi: 10.1245/s10434-014-4001-2. [DOI] [PubMed] [Google Scholar]
- 41.Zitvogel L., Tesniere A., Kroemer G. Cancer despite immunosurveillance: immunoselection and immunosubversion. Nat. Rev. Immunol. 2006;6:715–727. doi: 10.1038/nri1936. [DOI] [PubMed] [Google Scholar]
- 42.Dunn G.P., Koebel C.M., Schreiber R.D. Interferons, immunity and cancer immunoediting. Nat. Rev. Immunol. 2006;6:836–848. doi: 10.1038/nri1961. [DOI] [PubMed] [Google Scholar]
- 43.Smyth M.J., Dunn G.P., Schreiber R.D. Cancer immunosurveillance and immunoediting: the roles of immunity in suppressing tumor development and shaping tumor immunogenicity. Adv. Immunol. 2006;90:1. doi: 10.1016/S0065-2776(06)90001-7. [DOI] [PubMed] [Google Scholar]
- 44.Shankaran V., Ikeda H., Bruce A.T., White J.M., Swanson P.E., Old L.J. IFN|[gamma]| and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature. 2001;410:1107. doi: 10.1038/35074122. [DOI] [PubMed] [Google Scholar]
- 45.Visser K.E.D., Eichten A., Coussens. De Visser L.M., Eichten K.E., Coussens LMParadoxical A. roles of the immune system during cancer development. Nat. Rev. Cancer. 2006;6:24–37. doi: 10.1038/nrc1782. [DOI] [PubMed] [Google Scholar]
- 46.Buddingh E.P., Kuijjer M.L., Duim R.A., Bürger H., Agelopoulos K., Myklebost O. Tumor-infiltrating macrophages are associated with metastasis suppression in high-grade osteosarcoma: a rationale for treatment with macrophage activating agents. Clin. Cancer Res.: Off. J. Am. Assoc. Cancer Res. 2011;17:2110–2119. doi: 10.1158/1078-0432.CCR-10-2047. [DOI] [PubMed] [Google Scholar]
- 47.Mitchell R.A. Mechanisms and effectors of MIF-dependent promotion of tumourigenesis. Cell. Signal. 2004;16:13–19. doi: 10.1016/j.cellsig.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 48.Es W., Flaherty K.R., Carskadon S., Brant A., Iannettoni M.D., Yee J. Macrophage migration inhibitory factor and CXC chemokine expression in non-small cell lung cancer: role in angiogenesis and prognosis. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2003;9:853–860. [PubMed] [Google Scholar]
- 49.EHMD, Takashi O.M.D., Dipok K.D.M.D., Osama N.E.-aM.D., Yoshitaka H.M.D., Yamanoi A. Overexpression of macrophage migration inhibitory factor induces angiogenesis and deteriorates prognosis after radical resection for hepatocellular carcinoma. Cancer. 2005;103:588–598. doi: 10.1002/cncr.20818. [DOI] [PubMed] [Google Scholar]
- 50.Han I., Mi R.L., Nam K.W., Oh J.H., Moon K.C., Kim H.S. Expression of macrophage migration inhibitory factor relates to survival in high-grade osteosarcoma. Clin. Orthop. Relat. Res.®. 2008;466:2107. doi: 10.1007/s11999-008-0333-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ternovoi V.V., Curiel D.T., Smith B.F., Siegal G.P. Adenovirus-mediated p53 tumor suppressor gene therapy of osteosarcoma. Lab. Investig. 2006;86:748–766. doi: 10.1038/labinvest.3700444. [DOI] [PubMed] [Google Scholar]
- 52.Ragland B.D., Bell W.C., Lopez R.R., Siegal G.P. Cytogenetics and molecular biology of osteosarcoma. Lab. Investig. 2002;82:365–373. doi: 10.1038/labinvest.3780431. [DOI] [PubMed] [Google Scholar]
- 53.M.D. Vesely, M.H. Kershaw, R.D. Schreiber, M.J. Smyth. Natural Innate and Adaptive Immunity to Cancer, Annu. Rev. Immunol., 29(1):235. [DOI] [PubMed]
- 54.Schreiber R.D., Old L.J., Smyth M.J. Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion. Science. 2011;331:1565. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]