Abstract
As part of the effort to create a high resolution representative sequence database of the medieval Hungarian conquerors we have resequenced the entire mtDNA genome of 24 published ancient samples with Next Generation Sequencing, whose haplotypes had been previously determined with traditional PCR based methods. We show that PCR based methods are prone to erroneous haplotype or haplogroup determination due to ambiguous sequence reads, and many of the resequenced samples had been classified inaccurately. The SNaPshot method applied with published ancient DNA authenticity criteria is the most straightforward and cheapest PCR based approach for testing a large number of coding region SNP-s, which greatly facilitates correct haplogroup determination.
Introduction
Comparing ancient DNA (aDNA) sequences extracted from well dated archaeological remains from different periods and locations provide crucial information about past human population history (reviewed in [1]). Phylogeographic inferences are drawn from phylogenetic and population genetic analyses of sequence variations, the quality of which can be biased by data quantity and quality. Nowadays Next Generation Sequencing technology (NGS) provides a growing number of high quality aDNA sequence data, but until recently the majority of aDNA studies have been restricted to short fragments from the hypervariable region-1 (HVR-I) of the mitochondrial DNA (mtDNA) genome, using PCR based methods. PCR based methods are very sensitive for contamination, as low amounts of exogenous DNA can easily dominate PCR products resulting in the recovery of irrelevant sequences [2–5]. As a result, in spite of the applied authenticity criteria [6], many of the published databases may contain unreliable sequences, which distort statistical analyses. This problem is especially relevant for many of the ancient populations, from which only PCR based HVR data are available.
Recently several aDNA studies were published aiming to shed light on the origin of ancient Hungarians, two of these [7,8] applied restriction fragment length polymorphism (RFLP) to identify 11 or 14 haplogroup (Hg) specific coding region SNP-s in addition to HVR sequencing, while another study [9] tested 22 coding region SNP-s with multiplex PCR and GenoCoRe22 assay described in [10].
Using the NGS method combined with hybridization enrichment, we have sequenced the entire mtDNA genome of 9 samples from the Tömöry et al. 2007 [7] study, and 15 samples from the Neparáczki et al. 2016 [9] study, so we could compare the reliability of two different traditional approaches.
Materials and methods
Archaeological samples
Bone samples from the Hungarian conquest period used in the study of [7] are carefully maintained in the anthropological collection at the Department of Biological Anthropology, University of Szeged, Hungary, so we could unambiguously identify and resample these remains. Bone powder remains of samples from the study of [9], were saved in the Department of Genetics, University of Szeged, and were reused to build NGS sequencing libraries.
DNA extraction
Ancient DNA work was performed in the specialized ancient DNA (aDNA) facilities of the Department of Genetics, University of Szeged, Hungary with strict clean-room conditions. 100 mg bone powder from tooth roots, femurs or metatarsus was predigested in 1 ml 0,5 M EDTA 100 μg/ml Proteinase K for 30 minutes at 48°C, to increase the proportion of endogenous DNA [11], then DNA solubilisation was done overnight, in 1 ml extraction buffer containing 0.45 M EDTA, 250 μg/ml Proteinase K, 1% Triton X-100, and 50 mM DTT. DNA was bound to silica [12] adding 6 ml binding buffer (5,83 M GuHCl, 105 mM NaOAc, 46,8% isopropanol, 0,06% Tween-20 and 150 μl silica suspension to the 1 ml extract, and the pH was adjusted between 4–6 with HCl. After 3 hours binding at room temperature silica was pelleted, and washed twice with 80% ethanol, then DNA was eluted in 100 μl TE buffer.
NGS library construction
First 50 μl DNA extract was subjected to partial uracil-DNA-glycosylase (UDG) treatment followed by blunt end repair, as described in [13]. DNA was then purified on MinElute column (Qiagen), and double stranded library was made as described in [14], except that all purifications were done with MinElute columns, and after adapter fill-in libraries were preamplified in 2 x 50 μl reactions containing 800 nM each of IS7 and IS8 primers, 200 μM dNTP mix, 2 mM MgCl2, 0,02 U/μl GoTaq G2 Hot Start Polymerase (Promega) and 1X GoTaq buffer, followed by MinElute purification. PCR conditions were 96°C 6 min, 16 cycles of 94°C 30 sec, 58°C 30 sec, 72°C 30 sec, followed by a final extension of 64°C 10 min. Libraries were eluted from the column in 50 μl 55°C EB buffer (Qiagen), and concentration was measured with Qubit (Termo Fisher Scientific). Libraries below 5 ng/μl concentration were reamplified in the same reaction for additional 5–12 cycles, depending on concentration, in order to obtain 50 μl preamplified library with a concentration between 10–50 ng/μl.
50 ng preamplified libraries were double indexed according to [15] in a 50 μl PCR reaction containing 1 x KAPA HiFi HotStart ReadyMix (Kapa Biosystems) and 1000 nM each of P5 and P7 indexing primers. PCR conditions were 98°C 3 min, 6 cycles of 98°C 20 sec, 66°C 10 sec, 72°C 15 sec followed by a final extension of 72°C 30sec. Indexed libraries were MinElute purified and their concentration was measured with Qubit, and size distribution was checked on Agilent 2200 TapeStation Genomic DNA ScreenTape.
Control libraries without UDG treatment were also made for assessing the presence of aDNA specific damages in the extract, as well as DNA free negative control libraries, to detect possible contamination during handling or present in materials.
Mitochondrial DNA capture and sequencing
Biotinilated mtDNA baits were prepared from three overlapping long-range PCR products as described in [16], but using the following primer pairs, L14759-H06378, L10870-H14799, L06363-H10888, described in [10].
Capture was done according to [16] with the following modifications: Just four blocking oligos, given below were used in 3 μM (each) final concentration:
BO1.P5.part1F: AATGATACGGCGACCACCGAGATCTACAC-Phosphate,
BO2.P5.part2F ACACTCTTTCCCTACACGACGCTCTTCCGATCT-Phosphate,
BO4.P7.part1 R GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT-Phosphate,
BO6.P7.part2 R CAAGCAGAAGACGGCATACGAGAT-Phosphate.
For one capture 300 ng biotinilated bait was used with 30 μl Dynabeads MyOne Streptavidin C1 magnetic beads (Thermo Fisher Scientific). Double indexed libraries of 20 samples (300 ng each) were mixed and concentrated on MinElute columns, then captured together in a 64 μl hybridization reaction. When fewer samples were enriched, we used proportionally smaller amounts of baits. After washing, bead-bound enriched libraries were resuspended in 20 μl water and released from the beads in a 60 μl PCR reaction containing 1 X KAPA HiFi HotStart ReadyMix and 2000 nM each of IS5- IS6 library primers. PCR conditions were: 98°C 1 min, 10 cycles of 98°C 20 sec, 60°C 30 sec, 72°C 30 sec, followed by a final extension of 72°C 30 sec. The captured and amplified library mix was purified on MinElute column and eluted in 15 μl EB.
Before sequencing, libraries were quantified with Qubit, and quality checked and Agilent 2200 TapeStation Genomic DNA ScreenTape. Sequencing was done at the SeqOmics Biotechnology Ltd., using MiSeq sequencer with MiSeq Reagent Kit v3 (Illumina, MS-102-3003) generating 2x150bp paired-end sequences.
Data analysis
The adapters of paired-end reads were trimmed with the cutadapt software [17] in paired end mode. Read quality was assessed with FastQC [18]. Sequences shorter than 25 nucleotide were removed from this dataset. The resulting analysis-ready reads were mapped to the GRCh37.75 human genome reference sequence using the Burrows Wheeler Aligner (BWA) v0.7.9 software [19] with the BWA mem algorithm in paired mode and default parameters. Aligning to the GRCh37.75 human reference genome that also contains the mtDNA revised Cambridge Reference Sequence (rCRS, NC_012920.1) [20] helped to avoid the forced false alignment of homologous nuclear mitochondrial sequences (NumtS) to rCRS, though the proportion of NumtS, derived from low copy nuclear genome, is expexted to be orders of magnitudes lower than mtDNA in aDNA libraries. Samtools v1.1 [21] was used for sorting and indexing BAM files. PCR duplicates were removed with Picard Tools v 1.113 [22]. Ancient DNA damage patterns were assessed using MapDamage 2.0 [23], and read quality scores were modified with the rescale option to account for post-mortem damage. Freebayes v1.02 [24] was used to identify variants and generate variant call format (VCF) files with the parameters -q 10 (exclude nucleotids with <10 phred quality) and -P 0.5 (exclude very low probability variants). Each variant call was also inspected manually. From VCF files FASTA format was generated with the Genom Analysis Tool Kit (GATK v3.5) FastaAlternateReferenceMaker walker [25].
Results
NGS sequencing
We have sequenced 24 complete mtDNA genomes of the ancient Hungarians with multiple coverage (Table 1) without gaps and determined the haplotypes of the individuals (Table 2 and S1 Table). For two samples (Table 1) we have replicated the experiments from two independent extracts, one from bone another from tooth derived from the same individual, and in each case received identical sequence reads. UDG treated and non UDG treated libraries derived from the same extract also gave the same sequence reads. MapDamage profile of our partial UDG treated and control non treated library molecules displayed typical aDNA damage distribution (S1 Fig), as described in [13]. MapDamage computed proportions of sequence reads with aDNA specific C-to-T and G-to-A transitions at the ends of molecules which remained after partial UDG treatment are shown in Table 1. The average length of the obtained mtDNA fragments ranged from 56 to 85 bp (Table 1), an expected size range for aDNA [26]. These data indicated that the majority of sequences were derived from endogenous DNA molecules. Then we have estimated the percentage of possible contaminating molecules (Table 1) with a similar logic as in [27], by calculating the proportion of reads which did not correspond with the diagnostic positions of the consensus sequence given in S1 Table, which revealed very low contamination levels. Phylogenetic analyses (HaploGrep 2, [28]) of all consensus sequences resulted unambigous classifications without contradictory positions. Consensus sequences were submitted to NCBI GenBank under Accession No: KY083702-KY083725.
Table 1. Details of NGS data for each sample.
| cemetery/grave no. /sample name | sample source | total no. of reads | no. of reads mapped on rCRS | no. of unique mapped reads | average fragment length | Average coverage | (%) of nucleotides above 5x coverage | estimated contamination (%) | (%) G to A misincorp. at 3' end (MapDamage) | (%) C to T misincorp. at 5' end (MapDamage) |
|---|---|---|---|---|---|---|---|---|---|---|
| Magyarhomoróg/120/anc2 | tooth | 26152 | 12519 | 2994 | 55.97 | 10.2 | 89.62 | 0.00 | 6.11 | 7.81 |
| Orosháza-Görbics tanya/2/ anc3 | femur | 85178 | 50555 | 5516 | 79.37 | 24.8 | 99.89 | 0.43 | 8.13 | 8.89 |
| Szabadkígyós-Pálliget/7/anc4 | tooth | 53176 | 27002 | 5006 | 63.85 | 19.3 | 97.46 | 0.45 | 8.19 | 9.22 |
| Szegvár-Oromdülő/412/anc5 | tooth femur | 242925 | 139136 | 74712 | 68.45 | 302.9 | 100.00 | 1.47 | 15.11 | 16.15 |
| Szegvár-Oromdülő/593/anc6 | tooth | 66798 | 35260 | 6488 | 58.09 | 22.8 | 98.64 | 2.08 | 6.47 | 8.70 |
| Sárrétudvari-Hízóföld/5/anc10 | tooth | 17632 | 6664 | 4284 | 69.94 | 17.9 | 95.11 | 0.00 | 7.40 | 10.29 |
| Sárrétudvari-Hízóföld/118/anc12 | tooth | 36214 | 14708 | 6218 | 63.96 | 24.0 | 98.81 | 0.36 | 8.75 | 11.53 |
| Sárrétudvari-Hízóföld/213/anc13 | tooth | 42326 | 20383 | 9424 | 62.44 | 34.2 | 95.01 | 0.83 | 9.01 | 10.22 |
| Harta-Freifelt/10/anc25 | tooth | 135472 | 66169 | 7938 | 80.75 | 35.8 | 99.93 | 1.75 | 6.00 | 7.10 |
| Karos-III/1 | femur | 30830 | 5968 | 4738 | 81.44 | 22.4 | 98.07 | 0.00 | 13.09 | 11.48 |
| Karos-III/3 | femur | 58982 | 12858 | 7414 | 76.84 | 32.0 | 98.91 | 1.68 | 12.51 | 10.66 |
| Karos-III/4 | femur | 82194 | 22930 | 9316 | 68.71 | 38.3 | 99.98 | 0.33 | 12.93 | 11.98 |
| Karos-III/5 | metatarsus | 60797 | 25043 | 20236 | 85.13 | 100.2 | 100.00 | 3.76 | 7.99 | 6.65 |
| Karos-III/6 | femur | 41054 | 3863 | 1886 | 70.68 | 7.9 | 87.24 | 0.00 | 6.71 | 6.75 |
| Karos-III/8 | femur | 75724 | 26747 | 11426 | 64.26 | 43.9 | 99.67 | 1.79 | 11.37 | 11.08 |
| Karos-III/10 | femur | 67416 | 6371 | 3438 | 68.22 | 14.1 | 89.61 | 0.00 | 9.89 | 9.04 |
| Karos-III/11 | femur tooth | 203927 | 75860 | 57508 | 69.95 | 240.7 | 100.00 | 1.43 | 13.48 | 15.45 |
| Karos-III/12 | femur | 52738 | 5843 | 4742 | 70.76 | 20.0 | 95.80 | 3.96 | 12.40 | 11.19 |
| Karos-III/14 | femur | 61346 | 16134 | 8778 | 69.29 | 35.7 | 99.69 | 1.82 | 14.53 | 14.13 |
| Karos-III/15 | femur | 142977 | 83486 | 24702 | 81.12 | 115.4 | 100.00 | 0.22 | 9.78 | 9.39 |
| Karos-III/16 | femur | 90950 | 23233 | 5334 | 75.65 | 23.6 | 99.14 | 1.34 | 11.34 | 11.12 |
| Karos-III/17 | femur | 43330 | 2626 | 2382 | 79.64 | 11.2 | 87.55 | 0.00 | 10.31 | 9.91 |
| Karos-III/18 | femur | 9184 | 3208 | 3154 | 68.48 | 12.9 | 90.49 | 0.00 | 15.42 | 12.11 |
| Karos-III/19 | tooth | 59102 | 30135 | 5948 | 69.07 | 24.6 | 98.78 | 0.00 | 6.39 | 6.55 |
Data refer to paired-end sequences from UDG treated libraries. The Szegvár-Oromdülő/412/anc5 and Karos-III/11 samples were sequenced twice from tooth and femur with identical results, then these sequence reads were merged, and statistics are given for the merged reads.
Table 2. Comparison of Haplogroups identified with different PCR based methods and NGS.
| cemetery/grave no. / sample name | HVR-I muations found (position -16000) | HVR-II and coding region mutations studied / method | HVR-II and coding region mutations found | Hg described in the sudy (Hg with Haplogrep) | Haplotype identified by NGS in the present study | unnoticed SNP-s, or erroneously identified SNP-s in the region studied | |
|---|---|---|---|---|---|---|---|
| Tömöry et al. 2007 samples | Magyarhomoróg/120/ anc2 | CRS | 73 7028 14766 / RFLP | - | H (H2a2a1) | H84 | none |
| Orosháza-Görbics tanya/2/ anc3 | 147A 172C 183C 189C 223T 320T 355T | 10238 / RFLP | 10238C | N1a (L3e2b) | N1a1a1a1a | none | |
| Szabadkígyós-Pálliget/7/ anc4 | 223T 356C | 10400 12308 12705 / sequencing | 12308G | U4 (U4a2b) | N1a1a1a1 | 16147A 16172C 16248T 16320T 16355T 16356C 10398G 12308G 12705T | |
| Szegvár-Oromdülő/412/ anc5 | CRS | 73 7028 14766 / RFLP | 14766T | H (R0) | K1c1d | 16224C 16311C 73G 7028T | |
| Szegvár-Oromdülő/593/ anc6 | 114A 192T 256T 270T 294T | 12308 / sequencing | 12308G | U5a1 (U5a2a) | U5a2a1b | none | |
| Sárrétudvari-Hízóföld/5/ anc10 | 129A 148T 223T | 10238 / RFLP 12705 / sequencing | 10238C 12705T | I (N1) | I5a1a | 16391A | |
| Sárrétudvari-Hízóföld/118/ anc12 | 126C 182C 183C 189C 294T 296T 298C | 9 bp del / elecrtophoresis | 9 bp del* | T (T2f1a) | T2f1a1 | none | |
| Sárrétudvari-Hízóföld/213/ anc13 | 311C | 73 14766 / RFLP 11719 12308 12705 / sequencing | 73G 11719A 14766T | R (R1) | J1c3g | 16069T 16126C 16311C | |
| Harta-Freifelt/10/ anc25 | 294T 304C | 73 7028 14766 / RFLP 10310 / sequencing | - | H (H5a4) | H5e1a | none | |
| Neparáczki et al. 2016 samples | Karos-III/1 | 183C 189C 217C | HVR-II: nt.190-309 sequenced and coding region 22 SNP-s of the GenoCoRe22 assay determined in all cases | 263G 7028T 9bp del 11719A 14766T | B4 | B4d1 | 207A |
| Karos-III/3 | 362C | 239C 263G | H6 | H6a1b | none | ||
| Karos-III/4 | 069T 092C 126C 261T | 228A 263G 295T 7028T 11719A 12612G 14766T | J1c7 | J1c7a | none | ||
| Karos-III/5 | 183C 189C 217C | 263G 7028T 9bp del 11719A 14766T | B4 | B4d1 | none | ||
| Karos-III/6 | 189C | 263G 7028T 9bp del 11719A 14766T | B4’5 | B4d1 | 16183C, 16217C | ||
| Karos-III/8 | 051G 189C 362C | 263G 7028T 11467G 11719A 14766T | U2e | U2e1b | 217C, 16129C, 16256T, | ||
| Karos-III/10 | 304C | 263G | H5 | H5e1 | 16189C 16294T | ||
| Karos-III/11 | 189C 223T 278T | 195C 257G 263G 6371T 7028T 11719A 12705T 14766T | X2f | X2f | 16093C | ||
| Karos-III/12 | 183C 189C 223T 290T 319A | 235G 263G 4248C 7028T 11719A 12705T 14766T | A | A12 | none | ||
| Karos-III/14 | 126C 163G 186T 189C 294T | 195G 263G 7028T 11719A 13368A 14766T | T1a | T1a1b | none | ||
| Karos-III/15 | 069T 126C 362C | 263G 295T 7028T 11719A 12612G 14766T | J | J2a1 | 16263 del | ||
| Karos-III/16 | 256T 270T | 263G 7028T 11467G 11719A 14766T | U5a | U5a1a2a | 16399G | ||
| Karos-III/17 | 362C | 239C 263G | H6 | H6a1a | none | ||
| Karos-III/18 | 126C 163G 186T 189C 294T | 214G 263G 7028T 11719A 13368A 14766T | T1a10a | T1a10a | none | ||
| Karos-III/19 | 126C 163G 186T 189C 294T | 214G 263G 7028T 11719A 13368A 14766T | T1a10a | T1a10a | none |
Hg-s determined incorrectly with PCR methods are hightlighted with pink background, while yellow bakground hightlights correct Hg-s with incorrect haplotypes. Erroneously identified SNP-s are labelled with bold italic and lined through. Haplogroups and haplotypes were determined with the HaploGrep 2 version 2.1.0 [28] based on Phylotree 17 [39] from the available SNP-s. For the [7] samples HaploGrep assignment, based on their identified SNP positions is given in parenthesis.
*data from Ph.D thesis of Tömöry 2008.
In NGS sequence reads typical aDNA sequence alterations, present in individual molecules, are disclosed and excluded by averaging multiple reads. Moreover aDNA specific sequence alterations, primarily C-T and G-A transitions accumulating at the end of molecules, serve as markers to distinguish ancient molecules from contaminating modern DNA. Therefore NGS eliminates most sequencing uncertainties inherent in PCR based methods (reviewed in [29]), resulting in very reliable sequence reads. So we could use our NGS data to reevaluate and compare previous haplotyping strategies used in [7–9]. For this end, from our NGS data, we collected all SNP-s within the HVR stretches and coding region positions, which had been examined in [7] and [9], then contrasted these with the original dataset (Table 2).
Contrasting NGS and PCR based sequence data
We found that in [7] haplotypes of 5 out of 9 samples were determined correctly, while in one sample haplogroup was correct with inaccurate haplotype, and in 3 samples NGS detected entirely different haplogroups. In the 15 samples of [9] the same haplogroups were assigned from NGS data in all cases, however only 8 haplotypes proved to be correct. In both studies the majority of deviations originated from undetected SNP-s in sequencing reactions of PCR fragments, but [7] also identified 3 SNP-s erroneously (lined through nucleotide positions in Table 2). These results indicate that haplotypes from both studies were rather unreliable, but haplogroup classification with the approach of [9] is more trustworthy than with approach used in [7].
Discussion
As multicopy mtDNA is best preserved in archaeological remains than low copy nuclear DNA, most ancient sequences are derived from mitochondria [30]. Within mtDNA, the most polymorphic HVR control region contains outstanding phylogenetic information, therefore HVR sequencing has been the primary method of choice for mtDNA hapolotyping. However HVR polymorphisms have a limited reliability for haplogroup determination, therefore in addition several informative coding region SNP-s (CR-SNP) were selected to unambiguously define haplogroups [31]. At the beginning individual CR-SNP-s were determined with RFLP [32] or direct sequencing of PCR clones, but soon multiplex PCR combined with the SNaPshot technique [33] offered a more straightforward solution for identifying multiple SNP-s simultaneously. Latter method was soon adapted in the ancient DNA field [34] [10].
Determining individual CR-SNP-s separately is very time consuming and expensive, so it is tempting to test just those CR-SNP-s which are in line with HVR-I data. This is exactly what we read in [7]: “In cases when haplogroup categorization was not possible on the basis of HVSI motifs alone, analysis of the diagnostic polymorphic sites in the HVSII region and mtDNA coding region was also performed.” A major problem with this approach is the ambiguity of sequence reads derived from aDNA PCR clones, as amplification typically starts from a mixture of endogenous and contaminating human DNA molecules [3]. Erroneous HVR reading will lead to inappropriate CR-SNP selection, and in case of dubious CR-SNP results, false Hg classification. This is the most probable explanation of the 3 incorrectly defined haplogroups in [7] (Table 2). A major advantage of the GonoCore22 SNaPshot assay is that all Hg specific CR-SNP-s are examined irrespectively of HVR reads. The 22 CR-SNP alleles independently define a certain Hg, which must correspond with that based on HVR sequence. As both HVR and CR-SNP reads may give ambiguous results, this approach provides a double control for correct Hg designation, but is not immune against incorrect HVR haplotype reads. This is the explanation of correct Hg-s and erroneous haplotypes in [9] (Table 2).
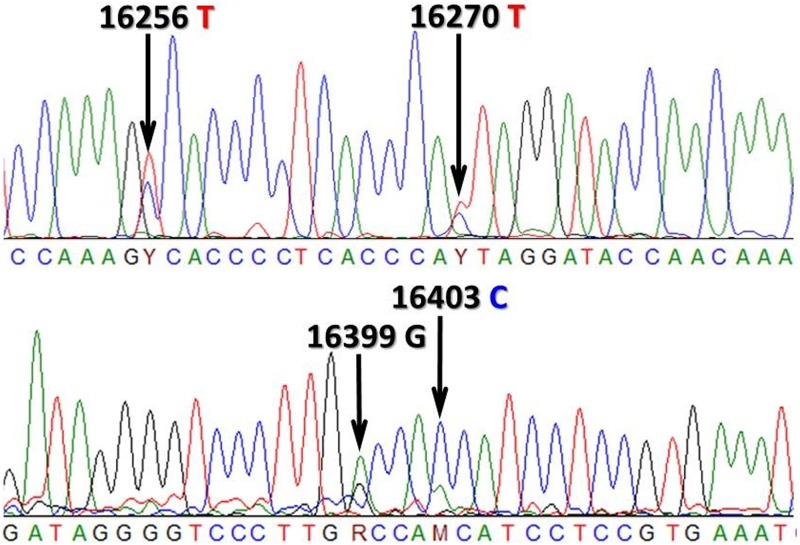
The problem of ambiguous aDNA sequence reads is demonstrated on Fig 1. In [9] consequently the higher peaks were taken into account, which also matched with the GenoCoRe22 data. However in position 16399 the correct nucleotide is defined by the neglected lower peak (G instead of A, see Table 2), which resulted in incorrect haplotyping. In contrast in the neighboring double peak (16403 in Fig 1), the correct nucleotide is defined by the selected higher peak.
Fig 1. Chromatogram of two HVR-I sequence fragments of the Karos-III/16 sample from [9].
Arrows label double peaks, correct reads according to NGS data are listed above the arrows.
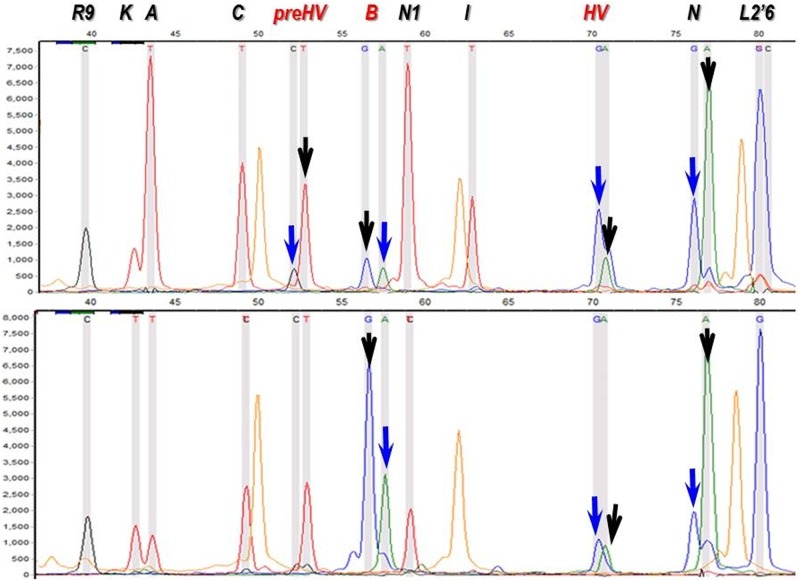
Coding region SNP testing with either RFLP, sequencing or SNaPshot method also suffers from the same problem as demonstrated on Fig 2. After multiplex PCR amplification of 22 mtDNA fragments two separate Single Base Extension (SBE) reactions are performed, and each reveals 11 Hg defining alleles. Both independent SBE reactions shown in Fig 2 contain several double peaks, and one of each must have derived from contamination. Some of these can be excluded from repeated SNaPshot reactions, for example the lower electropherogram excludes the ancestral preHV allele, since it has a single peak (T) in this position. If such exclusion is not possible, the higher peaks are preferably chosen, as the blue peak (G) for Hg B and the green (A) for Hg N on Fig 2. These decisions however must be handled with caution, therefore the presence of the B Hg defining 9 bp deletion also had been confirmed in [9], with singleplex PCR and agarose gelelectrophoresis. In other cases phylogenetic relations are taken into account [35], for example if the preHV allele is derived the HV allele must also be derived, this is why we have considered the lower peak (A) for HV in Fig 2 [9]. The summary of repeated SNaPshot reactions considered together with multiple HVR sequence reads warrants trustable Hg classification.
Fig 2. Electropherograms of two SNaPshot SBE-II reactions from two extracts of the same Karos-III/6 sample [9].
Characters at the top indicate Hg-s defined by the corresponding peaks. Black characters indicate peaks defining the ancestral allele, read characters indicate peaks defining the derived allele. Arrows point at double peaks. As each dye has a different influence on DNA mobility, positions of identical fragments with different dyes are not the same. Black arrows point at peaks taken into account, while blue arrows indicate neglected peaks, considered to have been derived from contamination. Orange peaks are size standards (GeneScan-120 LIZ, Applied Biosystems).
The studied conqueror samples were excavated between the 1930-90s, and had been handled by a large number of researchers, many with untraceable identity. It follows that these samples were inevitably contaminated during sample collection and storage. Tömöry et al. 2007 [7] collected samples from a large number of cemeteries, and published the ones with best DNA preservation. In spite of careful sampling their available method was error prone. Neparáczki et al. 2016 [9] aimed at characterizing an entire cemetery which limited the ability of sample selection, so in spite of the more reliable method their haplotype determination proved error prone. The lesson from this study is that PCR based haplotypes need to be handled cautiously, which has been well known in the aDNA field [2] [36–38]. It also follows that incorrect haplotypes particularly distort sequence based statistical analysis, like Fst statistics or shared haplotype analysis applied in [7,8]. The accumulation of authentical NGS ancient DNA sequence data in databases will greatly facilitate reliable population genetic studies.
Supporting information
SNPs are provided against rCRS. Following the recommendations in [40], we excluded common indels (hotspots) at nucleotide positions: 309.1C(C), 315.1C, 523-524del (or 522-523del), 3106del, 16182C, 16183C, 16193.1C(C), 16519C.
(XLSX)
a. non UDG treated library shownig C to T (and complementary G to A) misincorporations at the 5’ and 3’ termini of the last 25 nucleotides. b. Damage pattern of partial UDG treated library derived from the same extract. As expected the nontreated library contains much higher rate of transitions, most of which was removed by partial UDG treatment. Only data from one extract are shown, as all libraries displayed similar pattern.
(TIF)
Acknowledgments
The generous support of Avicenna Foundation grant no. GF/JSZF/814/9/2015 to I.R. and encouragement of professor Miklos Maroth is highly appreciated. This research was also supported in part by OTKA NN 78696. IN was supported by the János Bolyai Research Scholarship of the Hungarian Academy of Sciences.
Data Availability
Sequences were submitted to NCBI GenBank under the accession numbers: KY083702-KY083725.
Funding Statement
The generous support of Avicenna Foundation grant no. GF/JSZF/814/9/2015 to I.R. and encouragement of professor Miklos Maroth is highly appreciated. This research was also supported in part by OTKA NN 78696. SeqOmics Biotechnology Ltd. IN was supported by the János Bolyai Research Scholarship of the Hungarian Academy of Sciences. Hereby we certify that this company did not provide any funding for our study, they simply performed NGS sequencing services, and had consulting roles. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Pickrell JK, Reich D. Toward a new history and geography of human genes informed by ancient DNA. Trends in Genetics. 2014. pp. 377–389. 10.1016/j.tig.2014.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Richards MB, Sykes BC, Hedges REM. Authenticating DNA Extracted From Ancient Skeletal Remains. J Archaeol Sci. 1995;22: 291–299. [Google Scholar]
- 3.Malmström H, Storå J, Dalén L, Holmlund G, Götherström A. Extensive human DNA contamination in extracts from ancient dog bones and teeth. Mol Biol Evol. 2005;22: 2040–2047. 10.1093/molbev/msi195 [DOI] [PubMed] [Google Scholar]
- 4.Pilli E, Modi A, Serpico C, Achilli A, Lancioni H, Lippi B, et al. Monitoring DNA Contamination in Handled vs. Directly Excavated Ancient Human Skeletal Remains. PLoS One. 2013;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heupink TH, Subramanian S, Wright JL, Endicott P, Westaway MC, Huynen L, et al. Ancient mtDNA sequences from the First Australians revisited. Proc Natl Acad Sci. 2016; 201521066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knapp M, Clarke AC, Horsburgh KA, Matisoo-Smith EA. Setting the stage—building and working in an ancient DNA laboratory. Ann Anat. 2012;194: 3–6. Available: 10.1016/j.aanat.2011.03.008 [DOI] [PubMed] [Google Scholar]
- 7.Tömöry G, Csányi B, Bogácsi-Szabó E, Kalmár T, Czibula Á, Csősz A, et al. Comparison of maternal lineage and biogeographic analyses of ancient and modern Hungarian populations. Am J Phys Anthropol. 2007;134: 354–368. 10.1002/ajpa.20677 [DOI] [PubMed] [Google Scholar]
- 8.Csősz A, Szécsényi-Nagy A, Csákyová V, Langó P, Bódis V, Köhler K, et al. Maternal Genetic Ancestry and Legacy of 10th Century AD Hungarians. Sci Rep. 2016;6: 33446 10.1038/srep33446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neparáczki E, Juhász Z, Pamjav H, Fehér T, Csányi B, Zink A, et al. Genetic structure of the early Hungarian conquerors inferred from mtDNA haplotypes and Y-chromosome haplogroups in a small cemetery. Molecular Genetics and Genomics. 2016: 1–14. [DOI] [PubMed] [Google Scholar]
- 10.Haak W, Balanovsky O, Sanchez JJ, Koshel S, Zaporozhchenko V, Adler CJ, et al. Ancient DNA from European early Neolithic farmers reveals their near eastern affinities. PLoS Biol. 2010;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Damgaard PB, Margaryan A, Schroeder H, Orlando L, Willerslev E, Allentoft ME. Improving access to endogenous DNA in ancient bones and teeth. Sci Rep. 2015;5: 11184 10.1038/srep11184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rohland N, Hofreiter M. Ancient DNA extraction from bones and teeth. Nat Protoc. 2007;2: 1756–1762. Available: http://www.nature.com/nprot/journal/v2/n7/full/nprot.2007.247.html%5Cn http://www.nature.com/nprot/journal/v2/n7/pdf/nprot.2007.247.pdf 10.1038/nprot.2007.247 [DOI] [PubMed] [Google Scholar]
- 13.Rohland N, Harney E, Mallick S, Nordenfelt S, Reich D. Partial uracil-DNA-glycosylase treatment for screening of ancient DNA. Philos Trans R Soc Lond B Biol Sci. 2015;370: 20130624 10.1098/rstb.2013.0624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meyer M, Kircher M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb Protoc. 2010;5. [DOI] [PubMed] [Google Scholar]
- 15.Kircher M, Sawyer S, Meyer M. Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res. 2012;40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maricic T, Whitten M, Pääbo S. Multiplexed DNA Sequence Capture of Mitochondrial Genomes Using PCR Products. PLoS One. 2010;5: e14004 10.1371/journal.pone.0014004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17: 10–12. [Google Scholar]
- 18.Andrews S. FastQC: A quality control tool for high throughput sequence data [Internet]. babraham bioinformatics. 2016.
- 19.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25: 1754–60. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23: 147 10.1038/13779 [DOI] [PubMed] [Google Scholar]
- 21.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25: 2078–9. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Broad Institute. Picard tools. https://broadinstitute.github.io/picard/. 2016; http://broadinstitute.github.io/picard/
- 23.Jónsson H, Ginolhac A, Schubert M, Johnson PLF, Orlando L. MapDamage2.0: Fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics. 2013. pp. 1682–1684. 10.1093/bioinformatics/btt193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. arXiv Prepr arXiv12073907. 2012; 9.
- 25.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20: 1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sawyer S, Krause J, Guschanski K, Savolainen V, Pääbo S. Temporal patterns of nucleotide misincorporations and DNA fragmentation in ancient DNA. PLoS One. 2012;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fu Q, Mittnik A, Johnson PLF, Bos K, Lari M, Bollongino R, et al. A revised timescale for human evolution based on ancient mitochondrial genomes. Curr Biol. 2013;23: 553–559. 10.1016/j.cub.2013.02.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weissensteiner H, Pacher D, Kloss-Brandstätter A, Forer L, Specht G, Bandelt H-J, et al. HaploGrep 2: mitochondrial haplogroup classification in the era of high-throughput sequencing. Nucleic Acids Res. 2016;44: W58–63. 10.1093/nar/gkw233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rizzi E, Lari M, Gigli E, De Bellis G, Caramelli D. Ancient DNA studies: new perspectives on old samples. Genet Sel Evol. 2012;44: 21 10.1186/1297-9686-44-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hofreiter M, Serre D, Poinar HN, Kuch M, Pääbo S. Ancient DNA. Nat Rev Genet. 2001;2: 353–9. 10.1038/35072071 [DOI] [PubMed] [Google Scholar]
- 31.Behar DM, Rosset S, Blue-Smith J, Balanovsky O, Tzur S, Comas D, et al. The genographic project public participation mitochondrial DNA database. PLoS Genet. 2007;3: 1083–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown WM. Polymorphism in mitochondrial DNA of humans as revealed by restriction endonuclease analysis. Proc Natl Acad Sci U S A. 1980;77: 3605–3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salas A, Quintáns B, Álvarez-iglesias V. SNaPshot Typing of Mitochondrial DNA Coding Region Variants. Forensic DNA Typing Protocols. 2005. pp. 197–208. [DOI] [PubMed] [Google Scholar]
- 34.Bouakaze C, Keyser C, Amory S, Crubézy E, Ludes B. First successful assay of Y-SNP typing by SNaPshot minisequencing on ancient DNA. Int J Legal Med. 2007;121: 493–499. 10.1007/s00414-007-0177-3 [DOI] [PubMed] [Google Scholar]
- 35.Cooper A, Poinar HN. Ancient DNA: do it right or not at all. Science (New York, N.Y.). 2000. p. 1139. [DOI] [PubMed] [Google Scholar]
- 36.Handt O, Höss M, Krings M, Pääbo S. Ancient DNA: Methodological challenges. Experientia. 1994. pp. 524–529. [DOI] [PubMed] [Google Scholar]
- 37.Gilbert MTP, Rudbeck L, Willerslev E, Hansen AJ, Smith C, Penkman KEH, et al. Biochemical and physical correlates of DNA contamination in archaeological human bones and teeth excavated at Matera, Italy. J Archaeol Sci. 2005;32: 785–793. [Google Scholar]
- 38.Sampietro ML, Gilbert MTP, Lao O, Caramelli D, Lari M, Bertranpetit J, et al. Tracking down human contamination in ancient human teeth. Mol Biol Evol. 2006;23: 1801–1807. 10.1093/molbev/msl047 [DOI] [PubMed] [Google Scholar]
- 39.van Oven M. PhyloTree Build 17: Growing the human mitochondrial DNA tree. Forensic Sci Int Genet Suppl Ser. 2015;5: 9–11. [Google Scholar]
- 40.van Oven M. Revision of the mtDNA tree and corresponding haplogroup nomenclature. Proc Natl Acad Sci U S A. 2010;107: E38-NaN-e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SNPs are provided against rCRS. Following the recommendations in [40], we excluded common indels (hotspots) at nucleotide positions: 309.1C(C), 315.1C, 523-524del (or 522-523del), 3106del, 16182C, 16183C, 16193.1C(C), 16519C.
(XLSX)
a. non UDG treated library shownig C to T (and complementary G to A) misincorporations at the 5’ and 3’ termini of the last 25 nucleotides. b. Damage pattern of partial UDG treated library derived from the same extract. As expected the nontreated library contains much higher rate of transitions, most of which was removed by partial UDG treatment. Only data from one extract are shown, as all libraries displayed similar pattern.
(TIF)
Data Availability Statement
Sequences were submitted to NCBI GenBank under the accession numbers: KY083702-KY083725.