Abstract
Peanut is a major oilseed crop worldwide. In the Brazilian peanut production, silvering thrips and red necked peanut worm are the most threatening pests. Resistant varieties are considered an alternative to pest control. Many wild diploid Arachis species have shown resistance to these pests, and these can be used in peanut breeding by obtaining hybrid of A and B genomes and subsequent polyploidization with colchicine, resulting in an AABB amphidiploid. This amphidiploid can be crossed with cultivated peanut (AABB) to provide genes of interest to the cultivar. In this study, the sterile diploid hybrids from A. magna V 13751 and A. kempff-mercadoi V 13250 were treated with colchicine for polyploidization, and the amphidiploids were crossed with A. hypogaea cv. IAC OL 4 to initiate the introgression of the wild genes into the cultivated peanut. The confirmation of the hybridity of the progenies was obtained by: (1) reproductive characterization through viability of pollen, (2) molecular characterization using microsatellite markers and (3) morphological characterization using 61 morphological traits with principal component analysis. The diploid hybrid individual was polyploidized, generating the amphidiploid An 13 (A. magna V 13751 x A. kempff-mercadoi V 13250)4x. Four F1 hybrid plants were obtained from IAC OL 4 × An 13, and 51 F2 seeds were obtained from these F1 plants. Using reproductive, molecular and morphological characterizations, it was possible to distinguish hybrid plants from selfed plants. In the cross between A. hypogaea and the amphidiploid, as the two parents are polyploid, the hybrid progeny and selves had the viability of the pollen grains as high as the parents. This fact turns the use of reproductive characteristics impossible for discriminating, in this case, the hybrid individuals from selfing. The hybrids between A. hypogaea and An 13 will be used in breeding programs seeking pest resistance, being subjected to successive backcrosses until recovering all traits of interest of A. hypogaea, keeping the pest resistance.
1. Introduction
Peanut (Arachis hypogaea L.) is considered as the fifth largest oilseed crop in the world, after soybean, rapeseed, cotton and sunflower. World production during 2014–2015 was 39.83 million tons [1]. The five major countries producing peanut 2014–2015 were China, India, Nigeria, United States, and Burma. Brazil ranked 17th in production, where approximately 90% of peanut production comes from the state of São Paulo [2, 3].
Pests and foliar diseases are among the factors that mostly limit the economically sustainable production of peanuts in Brazil. The silvering thrips (Enneothrips flavens Moulton) and red necked peanut worm (Stegasta bosquella Chambers) are considered key pests [4, 5].
Thrips are tiny sucking insects of the order Thysanoptera and usually have between 0.5 and 5.0 mm in length, which when feed, destroy plant cells. The red necked peanut worm is an insect of the order Lepidoptera and usually has between 6.0 and 7.0 mm in length, which feeds on the peanut leaflet while still closed [5, 6].
Peanut crop must be chemically protected from pests to achieve satisfactory yields. Infestations of these insects have an important and peculiar aspect: silvering thrips and red necked peanut worm lodge in the buds (tips) of the branches, causing damage more or less severe to the vegetative growth of plants. This mode of attack requires the use of systemic insecticides for the control of these insects, which are more efficient, but more expensive [5, 6]. The use of resistant varieties is one of the best alternatives for pest control, because they do not harm the environment, keep pests at low levels and reduce costs with pesticides and crop treatment [7].
In the case of peanuts, it is known that many wild species of Arachis have resistance to pests, which can be introgressed into cultivars [8]. Obtaining cultivars resistant to pests through plant breeding is an alternative to reduce the cost of production for this crop. Efficient use of exotic peanut germplasm favors research programs aimed at the production of new improved cultivars from germplasm adapted to potential types of resistance to diseases and pests [9].
The main difficulty in the use of wild species in peanut breeding is that the majority of the species of Section Arachis are diploids and have genomes A, B, D, F, G or K, while the cultivated species A. hypogaea is allotetraploid and has genomic formula AABB [10, 11, 12, 13, 14, 15, 16, 17].
To overcome the ploidy barrier between the wild and cultivated peanuts, Simpson [18] and Simpson and Starr [19] showed three forms of gene introgression in A. hypogaea. The first is the cross between the wild diploid species (2n = 20) with A. hypogaea AABB (4n = 40), generating a triploid hybrid (3n = 30) which would be treated with colchicine for doubling of chromosomes, making it hexaploid (6n = 60) and fertile. This hexaploid would be backcrossed with A. hypogaea several times until there is loss of chromosomes, and the progeny again has 40 chromosomes. The second introgression process would be the doubling of chromosomes from wild species with genome A and B (2n = 20) making them tetraploid AAAA and BBBB (4n = 40), with subsequent cross between them, producing a hybrid AABB (4n = 40), which would be crossed with A. hypogaea AABB (4n = 40). The third method would be the cross of a species with genome A (2n = 20) with a species with genome B (2n = 20), generating a sterile hybrid AB (2n = 20), which would be tetraploidized with colchicine, becoming a fertile amphidiploid AABB (4n = 40) that would be crossed with A. hypogaea AABB (4n = 40) and backcrossed several times until all the traits of interest in A. hypogaea are recovered. The third way showed the most promising results by producing an amphidiploid (AABB) and crossing it with cultivated peanut A. hypogaea AABB (4 n = 40), as was done in this study. The peanut cultivar COAN showing high resistance to root-knot nematodes (Meloidogyne arenaria and M. javanica), was obtained from crosses between A. batizocoi x (A. cardenasii x A. diogoi) by Simpson and Starr [19]. The hybrid of this cross was sterile and was treated with colchicine for chromosome doubling. This amphidiploid was crossed with A. hypogaea cv. Florunner, generating a hybrid registered as TxAG-6. After five backcrosses and successive selection for agronomic traits and resistance to nematodes, it was released as the cultivar COAN.
Accessions of present study, A. magna V 13751 and A. kempff-mercadoi V 13250 showed tolerance to thrips and rednecked peanutworm [8]. Crosses between the accessions of A. magna V 13751 (female parent, genome B) and A. kempff-mercadoi V 13250 (male parent, genome A) were performed and individuals of progenies were analyzed by reproductive and morphological characterizations. Two diploid hybrid plants were obtained (Paula et al., unpublished data).
In this context, this study aimed to produce a new amphidiploid from wild species of Arachis with distinct genomes, cross the new amphidiploid with an elite cultivar of A. hypogaea, and perform morphological, molecular and reproductive characterization of the progenies.
Material and methods
Development of amphidiploid
The crosses between A. magna V 13751 (female parent) and A. kempff-mercadoi V 13250 (male parent) were performed manually in a greenhouse at Embrapa Southeast Livestock (São Carlos, state of São Paulo, Brazil) from December to March, 2010/2011. Flower buds of the female parents were emasculated in the late afternoon and flowers pollinated in the morning the next day. To calculate the percentage of success (PS) of hybridization, the following formula was used: PS = (number of hybrids/number of pollinations) x 100.
During the 2011–2012 growing season, fifteen cuttings of approximately 20 cm were taken from the hybrid A. magna V 13751 x A. kempff-mercadoi V 13250 (Ferreira et al., unpublished data), with the aid of scissors. Only the apical leaves still closed were kept and the apexes of cuttings were immersed into test tubes containing 0.2% colchicine. The tubes were closed and placed in BOD incubator (Biochemical Oxygen Demand), using fluorescent white light and 28°C for eight hours. After eight hours, the cuttings were washed in running water for about 20 minutes [20, 21, 22, 23]. Thus, with the aid of a scalpel, the cuttings were cut in a bevel (obliquely) over the last node and planted in plastic cups (180 mL) with the same substrate of pots to develop. At this stage, the cuttings were covered with plastic bags to minimize water loss. When cuttings rooted and grew, the plants were transplanted in pots.
During the growing season of 2012–2013, concurrently to the second crossing season, we analyzed the polyploidization of A. magna V 13751 x A. kempff-mercadoi V 13250. AB genome diploid plants usually do not produce seeds, sometimes nor flowers. So, they were considered sterile. Those sterile diploid hybrids with the colchicine treatment, showing presence of “peg” and the appearance of seeds were confirmed as successful amphidiploid. The progeny was evaluated by means of reproductive and morphological characterizations.
Crosses between A. hypogaea and amphidiploid
Cultivar IAC OL4 (A. hypogaea subsp. hypogaea) as female parent was crossed with the amphidiploid An 13 (A. magna V 13751 x A. kempff-mercadoi V 13250)4x as male parent during 2012/2013 growing season. Reproductive, morphological and molecular characterization was carried out on resultant progenies during 2013–2014 growing season.
Seed germination
All seeds were treated with Thiram + Distilled water (1:2) for two minutes and placed on germitest paper soaked in 0.65% Ethrel for germination. Germination conditions were 16 h at 20°C in the dark and 08 h at 35°C under fluorescent light. Disposable glasses of 180 mL were prepared with substrate and perforated bottom to receive the newly germinated seeds. Plants remained in the cups to achieve size and vigor to be transplanted to pots with volume of 25x40x40 cm.
Reproductive characterization
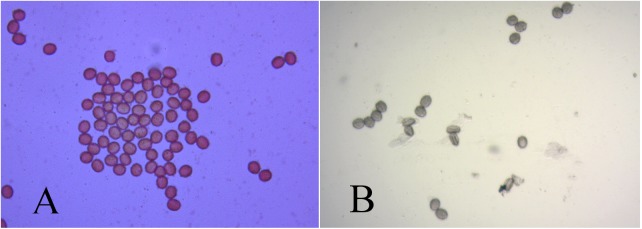
Four flowers of each individual used in the research were randomly collected. With the aid of tweezers, pollen grains were taken from the anthers, placed on slides and stained with 2% carmine acetic acid with glycerin (CA) or 0.25% tetrazolium (TZ) for three minutes. Slides were analyzed for viable and non-viable grains under a microscope. Pollen grains were considered as viable when they showed proper development and complete staining (Fig 1). Two hundred grains in each sample (repetition) derived from a single flower, totaling 800 grains per individual were counted. The percentages of stained pollen grains were calculated for each sample of all individuals involved. Analyses of variance and Tukey’s test for means comparison were performed using the software Statistical Analysis System (SAS).
Fig 1. Pollen grains of the hybrid IAC OL4 x An 13.
A) Pollen grains stained with 2% carmine acetic acid with glycerin. B) Pollen grains stained with 0.25% tetrazolium. Arrows indicate inviable grains. 100 X magnification under an optical microscope.
Morphological characterization
Four leaves of the side branch, a leaf of the main axis and four flowers of all individuals were randomly collected. The leaf collected was always the last leaf expanded of both lateral branches and of the main axis.
Sixty-one morphological traits were analyzed (Table 1), which have been previously evaluated in other studies [21, 22, 23]. According to the trait, the material was measured by ruler or caliper, or observed under a stereomicroscope.
Table 1. Descriptors evaluated in genotypes of Arachis, respective codes, unit of measurement and in which structure of the plant it was evaluated.
| Descriptors | Codes | Unit | MA2 | LB3 | Fl4 |
|---|---|---|---|---|---|
| Proximal leaflet length | Pll | Millimeter | X | X | - |
| Proximal leaflet width | Plw | Millimeter | X | X | - |
| Distal leaflet length | Dll | Millimeter | X | X | - |
| Distal leaflet width | Dlw | Millimeter | X | X | - |
| Petiole length | Pl | Millimeter | X | X | - |
| Petiolule length | Pol | Millimeter | X | X | - |
| Lenght of free part of stipule | Lfps | Millimeter | X | X | - |
| Width of free part of stipule | Wfps | Millimeter | X | X | - |
| Lenght of adnate part of stipule | Laps | Millimeter | X | X | - |
| Trichomes on the abaxial leaflet border | Tablb | Scale 1 to 31 | X | X | - |
| Trichomes on the abaxial leaflet center | Tablc | Scale 1 to 31 | X | X | - |
| Trichomes on the abaxial leaflet midvein | Tablm | Scale 1 to 31 | X | X | - |
| Trichomes on the adaxial leaflet border | Tadlb | Scale 1 to 31 | X | X | - |
| Trichomes on the adaxial leaflet center | Tadlc | Scale 1 to 31 | X | X | - |
| Trichomes on the adaxial leaflet midvein | Tadlm | Scale 1 to 31 | X | X | - |
| Bristles on the leaflet border | Blm | Scale 1 to 31 | X | X | - |
| Trichomes on the petiole | Tp | Scale 1 to 31 | X | X | - |
| Trichomes on the petiolule | Tpo | Scale 1 to 31 | X | X | - |
| Bristles on the petiole | Bp | Scale 1 to 31 | X | X | - |
| Bristles on the petiolule | Bpo | Scale 1 to 31 | X | X | - |
| Trichomes on the stipule (free part) center | Tfpsc | Scale 1 to 31 | X | X | - |
| Trichomes on the stipule (free part) border | Tfpsb | Scale 1 to 31 | X | X | - |
| Trichomes on the stipule (adnate part) center | Tadpsc | Scale 1 to 31 | X | X | - |
| Trichomes on the stipule (adnate part) border | Tadpsb | Scale 1 to 31 | X | X | - |
| Bristles on the stipule (free part) | Bfps | Scale 1 to 31 | X | X | - |
| Bristles on the stipule (adnate part) | Badps | Scale 1 to 31 | X | X | - |
| Anthocyanin in the stipule | As | Absence or presence | X | X | - |
| Standard length | Sl | Millimeter | - | - | X |
| Standard width | Sw | Millimeter | - | - | X |
| Wing length | Wl | Millimeter | - | - | X |
| Wing width | Ww | Millimeter | - | - | X |
| Lower lip length | Lll | Millimeter | - | - | X |
| Upper lip length | Ull | Millimeter | - | - | X |
| Hypanthium length | Hl | Millimeter | - | - | X |
1(1) Absence, (2) Few, (3) Many
2MA: Main axis
3LB: lateral branch
4FL: flower.
Data were analyzed using Principal Component Analysis (PCA) generated by SAS software. The results of components 1 and 2 were multiplied by the mean values of each trait for each individual and the resulting values were used to construct a Biplot graph using software Microsoft Excel.
Molecular characterization
Total genomic DNA was isolated from young leaves using the protocol based on CTAB (Cationic Hexadecyl Trimethyl Ammonium Bromide) described by Grattapaglia and Sederoff [24], with the inclusion of an additional precipitation with 1.2 M NaCl, immediately after CTAB buffer. Quantification of total DNA was performed with a spectrophotometer (NanoDrop ND-1000).
Three microsatellite markers were pre-selected from a larger set of markers according to the polymorphism in the genitors and to its amplification profile, and were evaluated in this study (Table 2), as follows: Seq3D09, IPAHM406 and RI2A06 [25, 26, 27, 28].
Table 2. Microsatellite markers used in this study.
| SSR | Amplification temperature °C | Size (pb) | References |
|---|---|---|---|
| IPAHM-406 | 59 | 202 | Cuc et al., 2008 |
| seq3D9 | 58 | 168 | Ferguson et al., 2004 |
| RI2A06 | 52 | 159 | Moretzsohn et al., 2004 |
Amplification conditions of markers were established from testing under different annealing temperature of primers in the polymerase chain reaction (PCR). PCR reactions were performed in a thermocycler (BioRad T100), with a final volume of 15 μl, as follows: 120ng of genomic DNA, 0.65U Taq DNA polymerase, 1x PCR buffer (200 mM Tris pH 8.4, 500 mM KCl), 1.5 mM MgCl2, 0.2 μM dNTP, and 0.165 μM of each primer. The protocol used for amplification consisted of 95°C for 5 min, 30 cycles (94°C for 45 sec; X°C for 45 sec.; 72°C for 45 sec.), and 72°C for 10 min, where X°C is the specific annealing temperature of the primers. Fragments were visualyzed on 2.5% agarose gel and markers that succeeded in amplification were subjected to electrophoresis in 6% polyacrylamide gels and stained with silver nitrate [29] for visualization of the fragments and genotyping using the 10 bp ladder (Invitrogen). Hybrids were considered those individuals F1 who had an allele from the male parent which was not present in the female parent in the evaluated polymorphic loci. As peanut is able to perform self-fertilization, the individuals F1 who had only alleles from the female parent and did not have alleles from the male parent were considered as self-plants.
Results and discussion
Development of amphidiploid
A total of 105 pollinations were performed between the A. magna V 13751 x A. kempff-mercadoi V 13250, which produced seven pegs and two seeds. The two seeds were germinated and were transplanted into pots. Ten cuttings were collected from each hybrid of A. magna V 13751 x A. kempff-mercadoi V 13250 (15), totaling 20 stakes treated with colchicine.
The 20 cuttings of the hybrid A. magna V 13751 x A. kempff-mercadoi V 13250 (15) treated with colchicine have developed up and just one seed was obtained that produced a plant and was called as An 13 (A. magna V 13751 x A. kempff-mercadoi V 13250)4x. The presence of seed indicates successful production of amphidiploid. This amphidiploid, An 13 produced a few amount of seeds. For this reason, this amphidiploid was not evaluated in field for pest resistance, as it was done for its progenitors (A. magna V 13751 and A. kempff-mercadoi V 13250).
Development of hybrid between the cultivar IAC OL4 and an amphidiploid An 13
Hybridization between cultivar IAC OL4 and an amphidiploid An 13 was undertaken with 98 pollinations which resulted in 10 pegs in turn 10 seeds. All these 10 seeds were germinated and were planted in cups and transplanted into pots. Among the 10 plants, four were characterized as hybrids and they produced 51 F2 seeds with percentage of success of 4.08%. The techniques of emasculation and pollination are among the factors that can influence the percentage of success [30]. Besides these techniques, according to Tallury et al. [31], fertilization itself is difficult and may not occur after pollination or occur late (delayed development of the pollen tube). Problems, such as inability of proembryo to grow after the peg reaches the ground or very slow growth of proembryo may be present.
In a peanut breeding program, for the production of amphidiploid, it is necessary to perform crosses between wild species of Arachis which results in sterile interspecific hybrids AB (2n = 20). It is common in these crosses the presence of abortions of “peg” that did not develop seeds, developed seeds that did not germinate and germinated seeds that did not had the vigor to survive in pots [21, 22, 23]. Further, greater number of abortions was due to interspecific hybrids with difference between genomes. In this study, the pollination difficulties cited by Nigam et al. [30] and Tallury et al. [31] are still present, but the problem of the difference between genomes was solved to obtain the amphidiploid. Thus, when performing crosses between A. hypogaea and amphidiploid with AABB genome, the number of abortions recorded was smaller in comparison with crosses between wild species. This also influences the observed percentage of success. Fávero [21] selected 14 combinations with species with genome A and B in order to obtain interspecific hybrids AB and 2,234 pollinations were performed, which generated 21 plants confirmed as hybrid, reaching a percentage of success of 0.9%. In the same study, when crosses between A. hypogaea and amphidiploid were conducted, there were 1,359 pollinations, which resulted in 107 hybrid plants, and the success percentage was 7.8%.
Reproductive characterization
ANOVA evidenced significant differences between individuals, but there were no differences between repetitions in the two tested stainings (CA and TZ) (Table 3, S1 Table).
Table 3. Viability of pollen grains of 22 individuals analyzed by staining.
| Identification | Individual | Mean percentage viability of pollen grains according to the staining1 | |
|---|---|---|---|
| CA2 | TZ3 | ||
| 14 | V 13250 | 98.00 a | 89.00 ab |
| 6 | OL4 x AN13 | 97.50 a | 88.00 ab |
| 10 | OL4 x AN13 | 96.75 a | 94.00 a |
| 13 | V 13751 | 96.75 a | 77.75 b |
| 8 | OL4 x AN13 | 96.25 a | 94.75 a |
| 11 | IAC OL4 | 95.75 a | 93.75 a |
| 7 | OL4 x AN13 | 95.50 a | 95.75 a |
| 9 | OL4 x AN13 | 94.00 a | 96.50 a |
| 5 | OL4 x AN13 | 92.50 a | 96.00 a |
| 4 | OL4 x AN13 | 91.25 a | 87.25 ab |
| 2 | OL4 x AN13 | 84.75 b | 80.75 b |
| 3 | OL4 x AN13 | 82.50 bc | 89.50 ab |
| 1 | OL4 x AN13 | 80.75 bc | 86.50 ab |
| 12 | An 13 | 77.00 c | 88.25 ab |
| 15 | V 13751 x V 13250 | 1.25 d | 0.75 c |
| CV % | 2.79 | 4.96 | |
1Means followed by same letters are significantly equal at 5% probability by Tukey’s test
2 2% carmine acetic acid with glycerin
30.25% Tetrazolium solution.
The diploid hybrid A. magna V 13751 x A. kempff-mercadoi V 13250 (15) and the progenitors were included in the reproductive characterization to compare them with amphidiploids and the F1 hybrids with A. hypogaea. The parents had a high percentage of pollen grains stained, the female parent A. kempff-mercadoi V 13250 (plant 14) showed 98.00% pollen grains stained with CA and 89.00% with TZ; the male parent A. magna V 13751 (13) showed 96.75% with CA and 77.75% with TZ. The hybrid A. magna V 13751 x A. kempff-mercadoi V 13250 (15), different from parents, showed low values of pollen grains stained, 1.25% with CA and 0.75% with TZ.
On the other hand, the amphidiploid An 13 (12) presented high viability of pollen grains, both with CA (77.00%) and with TZ (88.25%) (Table 3).
During cell division, colchicine acts as an antimitotic agent, binding to tubulin dimers, preventing the formation of microtubules and consequently the formation of spindle fibers [32]. During mitosis, when the achromatic spindle is damaged or absent, no separation of the duplicated chromosomes in anaphase, and consequently, cell division does not occur and the cell starts a new cell cycle with amount of DNA duplicated [33]. Thus, colchicine induces polyploidy, cells start to have homologous chromosomes, and the problem of irregular meiosis in diploid interspecific hybrids is solved, leading the plant to produce viable pollen grains.
With the problem of irregular meiosis solved, when making the reproductive characterization by means of pollen grain staining, the amphidiploid shows high viability of pollen grains, being largely different from the sterile diploid hybrid, which has low viability of pollen grains. Thus, reproductive characterization is able to clearly identify this sterile hybrid diploid and an amphidiploid.
A high viability of pollen grains was observed due to CA (over 76%) and TZ (over 77%) staining in the hybrids obtained between IAC OL4 and an amphidiploid An 13 and parents (Table 3).
The pollen staining technique can be used to identify diploid interspecific hybrids, due to the high number of non-viable pollen grains. When the diploid interspecific hybrid plants are treated with colchicine, they become polyploid (amphidiploid). These amphidiploids have homologous chromosomes of the two distinct genomes (stable genetic material) and their pollen grains become viable and the plant fertile. In crossing a cultivar with an amphidiploid, the two individuals have stable genetic material with homologous chromosomes. Their progeny will also have a stable genetic material and there is no difference between the parents and the offspring with respect to the viability of pollen grains. Therefore, in this case, pollen staining technique as a method for identification of hybrids is not conclusive. Hence, it is necessary to use other methods such as morphological and molecular characterizations.
A cytogenetic study conducted by Stalker [10] reported the viability of pollen grains associated with meiosis in intra- and interspecific crosses. When the genetic material was stable, the viability showed 90% stained pollen grains with low presence of univalent (below 0.05 univalent) and high rate of bivalent (approximately 10 bivalent) during meiosis. When the genetic material was unstable, that is, without homologous chromosomes, there was low viability of pollen grains, high number of univalent and low amount of bivalent.
The diploid interspecific hybrid has two distinct genomes in the cells with occasional or total absence of chromosome pairing. Given the low genetic similarity between chromosomes of different genomes, the pairing at metaphase I will be compromised, thus causing an irregular meiosis and non-viable pollen grains. Arachis species with genome A and species with genome B have around 20% genetic similarity [28].
When considered together the work of Lüdke [23] and Moretzsohn et al. [28], it was possible to establish a relationship between genetic similarity and the viability of pollen grains. Lüdke [23] studied four rounds of intra and interspecific crosses between species of the Arachis section, always using accessions of genome B as female parent. They found that the hybrids with the highest average percentage viability of pollen grains were exactly those which had parents with the same genome, in this case, the genome B. The intraspecific hybrid with genome B having the greatest viability of pollen grains was A. valida V 13514 x A. valida V 15096 [B x B], which showed 98.40% pollen grains stained with genetic similarity of 58% between the parents. All other intraspecific hybrids had less than 39% genetic similarity between the parents and below 40% viability of pollen grains. As for interspecific hybrids, all combinations presented less than 20% genetic similarity between the parents and all hybrids presented below 4.6% viability of pollen grains. In this context, the lower the genetic similarity between the parents, the smaller the percentage of viability of pollen grains in the produced hybrid.
Morphological characterization
All individuals were morphologically characterized, including the diploid hybrid A. magna V 13751 x A. kempff-mercadoi V 13250 (15) and its progenitors in order to compare them with amphidiploids and the F1 hybrids with A. hypogaea (S2 Table). PCA showed that the three first principal components accounted for 98% of the total variation of morphological traits.
According to the principal component 1, among the 15 most important morphological descriptors explaining the observed variation, five were collected in the main axis of the plant, eight in the lateral branch and two in the flower (Table 4).
Table 4. Order of descriptors that contributed most to the morphological variation observed in the principal component 1 (Prin 1) of the Principal Component Analysis of all the individuals evaluated in this work.
| Order | Descriptors | Codes1 | Prin 1 | Individuals2 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 11 | 12 | 13 | 14 | 15 | ||||
| 1 | Petiolule length MA | PolMA | 0.487 | 39,260 | 17,630 | 40,090 | 14,260 | 15,550 | 17,420 | 13,220 | 12,620 | 11,470 |
| 2 | Petiolule length LB | PolLB | 0.487 | 11,870 | 6,590 | 7,980 | 11,378 | 11,350 | 10,850 | 11,048 | 10,770 | 8,355 |
| 3 | Bristles on the petiole LB | BpLB | 0.360 | 6,110 | 6,585 | 6,780 | * | 7,800 | 7,650 | 7,995 | 7,448 | 14,263 |
| 4 | Distal leaflet length MA | DllMA | 0.305 | 44,230 | 35,670 | 37,090 | 63,100 | 41,900 | 36,650 | 41,710 | 33,530 | 42,510 |
| 5 | Distal leaflet length LB | DllLB | 0.305 | 24,430 | 27,840 | 34,490 | 47,978 | 34,955 | 30,480 | 34,423 | 33,323 | 32,670 |
| 6 | Bristles on the petiolule LB | BpoLB | 0.230 | 1,000 | 1,000 | 1,000 | 1,000 | 1,000 | 1,000 | 1,000 | 1,000 | 1,000 |
| 7 | Width of free part of stipule MA | WfpsMA | 0.184 | 2,500 | 3,290 | 2,330 | 2,150 | 2,150 | 1,840 | 1,250 | 2,410 | 2,460 |
| 8 | Width of free part of stipule LB | WfpsLB | 0.184 | 2,950 | 3,150 | 3,190 | 2,510 | 2,865 | 2,718 | 3,160 | 3,373 | 4,145 |
| 9 | Distal leaflet width MA | DlwMA | 0.148 | 20,460 | 14,910 | 13,530 | 28,690 | 17,640 | 15,850 | 17,630 | 14,820 | 20,060 |
| 10 | Distal leaflet width LB | DlwLB | 0.148 | 17,440 | 17,620 | 19,830 | 25,248 | 21,613 | 18,345 | 20,498 | 20,275 | 23,055 |
| 11 | Bristles on the leaflet border LB | BlmLB | 0.115 | 1,000 | 1,000 | 1,000 | 1,000 | 1,750 | 1,250 | 1,500 | 1,250 | 1,500 |
| 12 | Lower lip length FL | BlmFL | 0.077 | 8,080 | 7,760 | 8,770 | * | 7,650 | 7,720 | 7,265 | 8,043 | 9,520 |
| 13 | Lenght of adnate part of stipule MA | LapsMA | 0.070 | 12,750 | 10,090 | 8,500 | 11,550 | 15,290 | 14,840 | 17,280 | 15,220 | 15,830 |
| 14 | Lenght of adnate part of stipule LB | LapsLB | 0.070 | 7,280 | 7,675 | 8,420 | 8,773 | 12,570 | 10,370 | 11,045 | 10,655 | 7,343 |
| 15 | Upper lip length FL | UllFL | 0.067 | 6,570 | 6,745 | 7,280 | * | 8,525 | 7,458 | 7,575 | 8,423 | 8,600 |
1 Codes ending with MA refer to the main axis, with LB, to the lateral branch and with FL, to flower.
21–4: F1 hybrids from IAC OL4 x An 13; 11: IAC OL4, 12: An 13; 13: V13751; 14: V13250; 15: V13751 x V13250
* Not evaluated
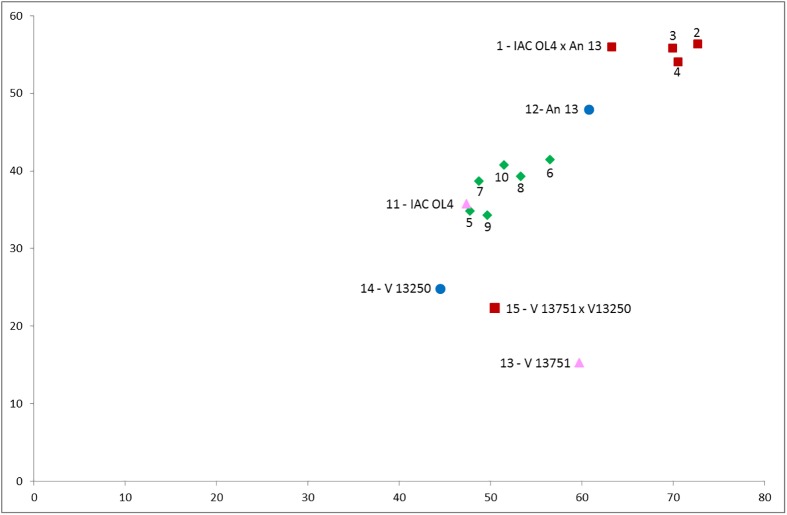
From the components 1 and 2, multiplied by the mean values of each trait for each individual, it was possible to construct a biplot graph (Fig 2). The male parent A. kempff-mercadoi V 13250 (14) was located at the center of the graph and the female parent A. magna V 13751 (13) was located at the bottom right of the graph, the plant resulting from the cross de A. magna V 13751 x A. kempff-mercadoi V 13250 (15) was located closer to the male parent than to the female parent, which allows to characterize this plant as a hybrid by means of PCA.
Fig 2. Biplot graph resulting from the cross between IAC OL4 x An 13, obtained by Principal Component Analysis considering the 61 descriptors for the principal components 1 and 2.
Triangle indicates female parents V 13751 (13) and IAC OL4 (11); circles, male parents V 13250 (14) and An 13 (12); and representing the progenies, square indicates the hybrid plants V 13751 x V 13250 (15) and IAC OL4 x An 13(1 to 4), and, diamonds, the plants derived from selfing IAC OL4 x An 13 (5 to 10).
Analyzing the most important descriptor, it can be observed that the female and male parents, respectively, presented 39.260 and 17.630 mm for the petiolule length of the main axis (PolMA), thus, the hybrid plant A. magna V 13751 x A. kempff-mercadoi V 13250 (15) presenting 27.240 mm, was located closest to the male parent. This proximity of the hybrid to the male parent can be also perceived in other descriptors in Table 4.
On the graph, the plant An 13 (amphidiploid) was located far from the diploid plant A. magna V 13751 x A. kempff-mercadoi V 13250 (15), indicating morphological variation between these plants (Fig 2). This variation may be explained by the giantism effects observed. Amphidiploid has twice the genetic material in relation to the sterile interspecific hybrid. So, this plant may have an increase in the cell size, which may result in the increasing of size of morphological structures.
With respect to the most important descriptor, it was observed that the hybrid A. magna V 13751 x A. kempff-mercadoi V 13250 (15) presented 27.240 mm, and An 13 (12) presented 11.470 mm for the petiolule length of the main axis (PolMA). The difference between the values observed in the descriptors led the hybrid to be located at the bottom right of the graph, and the amphidiploid, at the top right of the graph. Other descriptors and respective measures responsible for the location of these plants on the graph can be seen in Table 4.
It was obtained ten seeds from the cross between IAC OL4 x An 13. Plants of IAC OL4 x An 13 (1, 2, 3 and 4), characterized as a result of hybridization, were located in the top left graph (Fig 2) nearest to the male parent An 13 (12) than to the female parent OL4 IAC (11) on the PCA analysis. The plants IAC OL4 x An 13 (5, 6, 7, 8, 9 and 10) were identified as a result of selfing by molecular paternity test, which were located at the center of the biplot graph, near the female parent IAC OL4 (11). Analyzing the measures of the most important descriptors (Table 4), in most of them, the measurements of hybrids were more similar to the male parent than to the female parent. Thus, the hybrid plants IAC OL4 x An 13 (1, 2, 3 and 4) have measurements that were more similar to the male parent data than to the female parent results. So, they were located closer to the male parent in the graph.
During the evaluations, the descriptor “number of lateral branches” (nLB), which had no evaluation predicted in the methods, was interesting to identify hybrids between IAC OL4 and amphidiploid An 13. In the growth stage of plants still in disposable cups, four plants (hybrids) presented lower numbers of side branches, compared the other six plants (selfed), which had the same morphological profile of cultivar IAC OL4 (female parent), showing higher number of lateral branches (Fig 3). Thus, at this stage of the breeding program, the descriptor nLB proved efficient to distinguish the hybrid individuals and selfed.
Fig 3. Comparison in the number of lateral branches between plants from.
(A) hybridization and (B) selfing.
Molecular characterization of the F1 hybrids from A. hypogaea and An 13
Loci evaluated allowed the identification of hybridization in individuals 1, 2, 3 and 4 and selfing in individuals 5, 6, 7, 8, 9 and 10, all from the cross IAC OL4 x An 13.
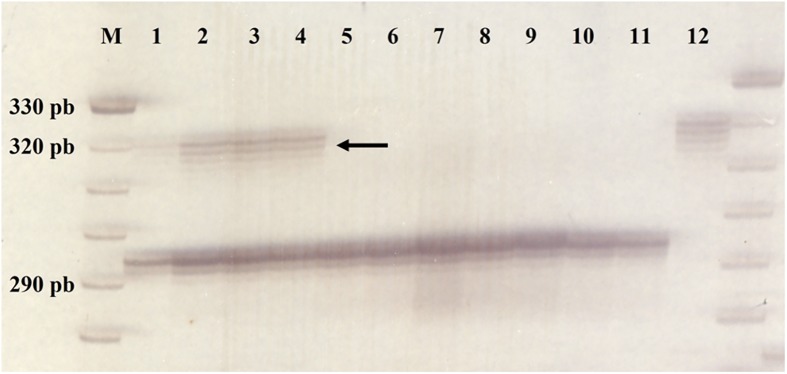
The Fig 4 shows the amplification profile of the marker IPAHM-406, in which the presence of the 342 bp band in the male parent An 13 (12), and its absence in the female parent IAC OL4 (11) allowed the identification of selfing or hybridization in the progenies. The markers RI2A06 and Seq3d9 showed the same results with respect to the identification of progenies as a result of selfing or hybridization.
Fig 4. Amplification profile of the marker IPAHM-406 for progenies of IAC OL4 x An 13.
Individuals: Hybrids (1 to 4), self-fertilized (5 to 10), female parent IAC OL4 (11) and male parent An 13 (12). Arrow indicates the polymorphic band identifying hybridization. M: 10 bp ladder (Invitrogen).
Molecular markers are very important tools in molecular characterization of genetic resources and in plant breeding and pre-breeding. In Arachis, several studies used molecular markers to characterize germplasm collections, to study genetic diversity [25, 26, 34, 35, 36] and to confirm hybridizations. Fávero [22] using the SSR Lec-1 on 1.2% agarose gel, was able to identify 17 hybrids derived from crosses between KG 30006 x V13710 e KG 30076 x V 12812. Moretzsohn et al. [28] studied the molecular genetic relations between cultivated peanut (Arachis hypogaea) and wild species. Thus, it was possible to obtain important information about the genomes and genetic similarity of wild species, which is important to explain the irregular meiosis that diploid interspecific hybrids have, and how this problem is solved by obtaining the amphidiploid. In this study, microsatellites were extremely efficient in identifying hybridization in controlled crosses between IAC OL4 and amphidiploid An 13.
The obtainment of F2 plants of an interspecific hybrid that include three very distinct species is the first step for the use of these germplasm in breeding programs. After that it will be necessary the confirmation of the introgression of pest resistance genes in the progenies, select the best ones and backcross these genotypes with A. hypogaea for at least eight times. Bioassays for pest resistance may be done before each backcrossing season.
Observation of mite resistance during evaluations
At the end of 2014–2015 growing season, parents and progenies plants were attacked by mites (Mononychellus planki (McGregor)) in greenhouse. Realizing that the plants An 13 (12) and the hybrids IAC OL4 x An 13 (1, 2, 3 and 4) were resisting the attack, all pots (parents and progenies) were no longer sprayed with acaricide. The female parent IAC OL4 (11) and plants of the progeny of the cross IAC OL4 x An 13 (5, 6, 7, 8, 9 and 10) considered as resulting from selfing suffered such a great pressure from mites that no longer developed and leaves began to yellow (Figs 5 and 6). While the female parent IAC OL4 (11) was attacked by mites, the male parent An 13 (12) and the plants IAC OL4 x An 13 (1, 2, 3 and 4), considered hybrids, continued their development normally (Fig 6). It was observed the presence of mites in hybrid plants, but at a much lower level compared to plants arising from selfing. By observations of plants in this attack, it was possible to identify resistance to mites in hybrids and amphidiploid An 13, and observe that hybrids have resistance that the IAC OL4 does not have.
Fig 5. Progeny of IAC OL4 x An 13 under attack by mites.
The blue arrow indicates the plants resistant to mite, result from hybridization. The red arrow indicates the plants susceptible to mite, a result from selfing.
Fig 6. Parents and progeny of IAC OL4 x An 13 under attack by mites.
The blue arrow represents the plants resistant to mite (An 13 and hybrid). The red arrow represents the plants susceptible to mite (IAC OL4 and selfed).
6. Conclusions
In the present study, sterile diploid hybrid was obtained from the cross between the accessions of A. magna V 13751 (genome B) and A. kempff-mercadoi V 13250 (genome A) showing resistance to thrips and rednecked peanutworm. The resultant hybrid was polyploidized to obtain an amphidiploid which was crossed with an elite cultivar of A. hypogaea and F2 seeds were generated.
Using the reproductive, molecular and morphological characterizations, it was possible to distinguish hybrid plant individuals from selfing.
In the cross between A. hypogaea cv. IAC OL4 and amphidiploid An 13, the hybrid progeny had a pollen grain viability as high as that of parents. This prevented the use of reproductive traits for discriminating, in this case, the hybrids of individuals from selfing.
The 51 F2 seeds produced by the hybrids IAC OL4 x An 13 will be used in breeding programs, and may confer resistance to thrips and rednecked peanutworm for the cultivated peanut.
Supporting information
* Not evaluated.
(XLSX)
1See Descriptors details in Table 1. * Not evaluated.
(XLSX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
APF received funds by Embrapa (no. 0211080060008) and National Council for Scientific and Technological Development (CNPq, grant number 471657/2011-5). AFP received a grant by Coordination for the Improvement of Higher Education Personnel (CAPES). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.USDA. Table 01 Major Oilseeds: World Supply and Distribution (Commodity View). 2015. Available: http://apps.fas.usda.gov/psdonline/psdReport.aspx?hidReportRetrievalName=Table+01%3a+Major+Oilseeds%3a+World+Supply+and+Distribution+(Commodity+View)&hidReportRetrievalID=531&hidReportRetrievalTemplateID=5. Accessed.
- 2.CONAB. Acompanhamento da safra brasileira. 2015. Available: http://www.conab.gov.br/OlalaCMS/uploads/arquivos/15_09_11_10_42_03_boletim_graos_setembro_2015.pdf. Accessed.
- 3.USDA. Table 13 Peanut Area, Yield, and Production. 2015. Available: http://www.fas.usda.gov/psdonline/psdreport.aspx?hidReportRetrievalName=BVS&hidReportRetrievalID=918&hidReportRetrievalTemplateID=1. Accessed.
- 4.Calcagnolo G, Leite FM, Gallo JR. Efeitos da infestação do tripes nos folíolos do amendoinzeiro Enneothrips (Enneothripiella) flavens Moulton, 1941, no desenvolvimento das plantas, na qualidade da produção de uma cultura “das águas”. O Biológico. 1974; 40: 239–242. [Google Scholar]
- 5.Gallo D, Nakano O, Silveira Neto S, Carvalho RPL, Batista GC, Berti Filho E, et al. Entomologia Agrícola. Piracicaba: FEALQ; 2002. [Google Scholar]
- 6.Borror DJ, De Long DM. Introdução ao Estudo dos Insetos. Rio de Janeiro: USAID; 1969. [Google Scholar]
- 7.Lara FM. Princípios de Resistência de Plantas a Insetos. São Paulo: Ícone; 1991. [Google Scholar]
- 8.Janini, JC: Resistência de germoplasma silvestre de amendoim (Arachis spp.) a Enneothrips flavens Moulton, 1941 (Thysanoptera:Thripidae) e Stegasta bosquella (Chambers, 1875) (Lepidoptera: Gelechiidae). M.Sc Thesis, Universidade Estadual Paulista. 2011. Available: http://repositorio.unesp.br/handle/11449/102283
- 9.Wynne JC, Halward TM. Germplasm enhancement in peanut In: Stalker HT, Chapman C, editors. Science Management of Germplasm: Characterization, Evaluation and Enhancement. Rome: International Board for Plant Genetic Resources; 1989. pp. 155–174. [Google Scholar]
- 10.Stalker HT. A New Species in Section Arachis of Peanuts with a D Genome. American Journal of Botany. 1991; 78: 630–637. [Google Scholar]
- 11.Fernández A, Krapovickas A. Cromosomas y evolución en Arachis (Leguminosae). Bonplandia. 1994; 8: 187–220. [Google Scholar]
- 12.Peñaloza APS, Valls JFM. Chromosome number and satellited chromosome morphology of eleven species of Arachis (Leguminosae). Bonplandia. 2005; 14: 65–72. [Google Scholar]
- 13.Lavia GI. Karyotypes of Arachis palustris and A. praecox (section Arachis), two species with basic chromosome number x = 9. Cytologia. 1998; 63: 177–181. [Google Scholar]
- 14.Robledo G, Seijo G. Characterization of the Arachis (Leguminosae) D genome using fluorescence in situ hybridization (FISH) chromosome markers and total genome DNA hybridization. Genetics and Molecular Biology. 2008; 31: 717–724. [Google Scholar]
- 15.Robledo G, Seijo G. Species relationships among the wild B genome of Arachis species (section Arachis) based on FISH mapping of rDNA loci and heterochromatin detection: a new proposal for genome arrangement. Theoretical and Applied Genetics. 2010; 121: 1033–1046. 10.1007/s00122-010-1369-7 [DOI] [PubMed] [Google Scholar]
- 16.Robledo G, Lavia GI, Seijo G. Species relations among wild Arachis species with the A genome as revealed by FISH mapping of rDNA loci and heterochromatin detection. Theorical Applied Genetic. 2009; 118: 1295–1307. [DOI] [PubMed] [Google Scholar]
- 17.Silvestri MC, Ortiz AM, Lavia GI. rDNA loci and heterochromatin positions support a distinct genome type for ‘x = 9 species’ of section Arachis (Arachis, Leguminosae). Plant Systematics and Evolution. 2014; 301: 555–562. [Google Scholar]
- 18.Simpson CE. Pathways for introgression of pest resistance into A. hypogaea. Peanut Science. 1991; 18: 22–26. [Google Scholar]
- 19.Simpson CE, Starr JL. Registration of ‘Coan’ peanut. Crop Science. 2001; 41: 918–918. [Google Scholar]
- 20.Fávero AP, Pádua JG, Costa TS, Gimenes MA, Godoy IJ, Moretzsohn MC, et al. New hybrids from peanut (Arachis hypogaea L.) and synthetic amphidiploid crosses show promise in increasing pest and disease tolerance. Genetics and molecular research. 2015; 14: 16694–16703. 10.4238/2015.December.11.17 [DOI] [PubMed] [Google Scholar]
- 21.Fávero, AP: Cruzabilidade entre espécies silvestres de Arachis visando à introgressão de genes de resistência a doenças do amendoim cultivado. Ph.D Thesis, Universidade de São Paulo. 2004. http://www.teses.usp.br/teses/disponiveis/11/11137/tde-21092004-160937/pt-br.php
- 22.Fávero AP, Santos RF, Simpson CE, Valls JFM, Vello NA. Successful crosses between fungal-resistant wild species of Arachis (section Arachis) and Arachis hypogaea. Genetics and Molecular Biology. 2015; 38: 353–365. 10.1590/S1415-475738320140376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lüdke, DCW: Estudo das relações sistemáticas da espécie endêmica brasileira Arachis valida Krapov. e W.C. Greg. baseado em cruzamentos intra e interespecíficos no gênero Arachis L. Ph.D Thesis, Universidade de Brasília. 2015. http://repositorio.unb.br/handle/10482/17538
- 24.Grattapaglia D, Sederoff R. Genetic linkage maps of Eucalyptus grandis and Eucalyptus urophylla using a pseudo-testcross: mapping strategy and RAPD markers. Genetics. 1994; 137: 1121–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moretzsohn MC, Hopkins MS, Mitchell SE, Kresovich S, Valls JFM, Ferreira ME. Genetic diversity of peanut (Arachis hypogaea L.) and its wild relatives based on the analysis of hypervariable regions of the genome. BMC Plant Biology. 2004; 4: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferguson ME, Burow MD, Schulze SR, Bramel PJ, Paterson AH, Kresovich S, et al. Microsatellite identification and characterization in peanut (A. hypogaea L.). Theoretical and Applied Genetics. 2004; 108: 1064–1070. 10.1007/s00122-003-1535-2 [DOI] [PubMed] [Google Scholar]
- 27.Cuc LM, Mace ES, Crouch JH, Quang VD, Long TD, Varshney RK. Isolation and characterization of novel microsatellite markers and their application for diversity assessment in cultivated groundnut (Arachis hypogaea). BMC Plant Biology. 2008; 8: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moretzsohn MC, Gouvea EG, Inglis PW, Leal-Bertioli SCM, Valls JFM, Bertioli DJ. A study of the relationships of cultivated peanut (Arachis hypogaea L.) and its most closely related wild species using intron sequences and microsatellite markers. Annals of Botany. 2013; 111: 113–126. 10.1093/aob/mcs237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Creste S, Tulmann Neto A, Figueira A. Detection of single sequence repeat polymorphisms in denaturing polyacrylamide sequencing gel by silver staining. Plant Molecular Biology Reporter. 2002; 19: 299–306. [Google Scholar]
- 30.Nigam SN, Rao MJV, Gibbons RW. Artificial hybridization in groundnut. Andhra Prasdesh: International Crops Research Institute for the SemiArid Tropics; 1990. [Google Scholar]
- 31.Tallury SP, Stalker HT, Patee HE. Early reproductive ontogeny in interspecific crosses of Arachis hypogaea and Section Arachis species. Annals of Botany. 1995; 76: 397–404. [Google Scholar]
- 32.Faleiro FG. Genética e biotecnologia aplicada ao melhoramento genético vegetal: relatório de pós-doutorado. Embrapa Cerrados; 2012: 1–74. Available: http://ainfo.cnptia.embrapa.br/digital/bitstream/item/84605/1/doc-309.pdf [Google Scholar]
- 33.Pereira RC, Davide LC, Techio VH, Timbó ALO. Duplicação cromossômica de gramíneas forrageiras: uma alternativa para programas de melhoramento genético. Ciência Rural, 2012; 42: 1278–1285. [Google Scholar]
- 34.He G, Meng R, Newman M, Gao G, Pittman R, Prakash C. Microsatellites as DNA markers in cultivated peanut (Arachis hypogaea L.). BMC Plant Biology. 2003; 3: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bravo JP, Hoshino AA, Angelici C, Lopes CR, Gimenes MA. Transferability and use of microsatellite markers for the genetic analysis of the germplasm of some Arachis section species of the genus Arachis. Genetics and Molecular Biology. 2006; 29: 516–524. [Google Scholar]
- 36.Proite K, Leal-Bertioli SC, Bertioli DJ, Moretzsohn MC, da Silva FR, Martins NF, et al. ESTs from a wild Arachis species for gene discovery and marker development. BMC Plant Biology. 2007; 7: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
* Not evaluated.
(XLSX)
1See Descriptors details in Table 1. * Not evaluated.
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.