Abstract
Background
18F-fluoro-2-deoxy-glucose (18F-FDG) positron emission tomography (PET) is a functional imaging modality based on glucose metabolism. The correlation between EGFR or KRAS mutation status and the standardized uptake value (SUV) of 18F-FDG PET scanning has not been fully elucidated.
Methods
Correlations between EGFR or KRAS mutation status and clinicopathological factors including SUVmax were statistically analyzed in 734 surgically resected lung adenocarcinoma patients. Molecular causal relationships between EGFR or KRAS mutation status and glucose metabolism were then elucidated in 62 lung adenocarcinomas using cap analysis of gene expression (CAGE), a method to determine and quantify the transcription initiation activities of mRNA across the genome.
Results
EGFR and KRAS mutations were detected in 334 (46%) and 83 (11%) of the 734 lung adenocarcinomas, respectively. The remaining 317 (43%) patients had wild-type tumors for both genes. EGFR mutations were more frequent in tumors with lower SUVmax. In contrast, no relationship was noted between KRAS mutation status and SUVmax. CAGE revealed that 4 genes associated with glucose metabolism (GPI, G6PD, PKM2, and GAPDH) and 5 associated with the cell cycle (ANLN, PTTG1, CIT, KPNA2, and CDC25A) were positively correlated with SUVmax, although expression levels were lower in EGFR-mutated than in wild-type tumors. No similar relationships were noted with KRAS mutations.
Conclusions
EGFR-mutated adenocarcinomas are biologically indolent with potentially lower levels of glucose metabolism than wild-type tumors. Several genes associated with glucose metabolism and the cell cycle were specifically down-regulated in EGFR-mutated adenocarcinomas.
Introduction
Recently, driver oncogene mutations are being discovered at a rapid pace. Therapeutic agents targeting some of these driver oncogenes have been successfully developed. The somatic mutations in epidermal growth factor receptor (EGFR) and v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (KRAS) are the most frequently found in lung adenocarcinomas. The presence of an EGFR mutation is the most important predictor of the efficacy of EGFR tyrosine kinase inhibitors (TKIs) [1, 2]. In contrast, KRAS mutations are a useful biomarker of EGFR-TKI resistance [3]. It is therefore important to understand the occurrence of EGFR and KRAS mutations when deciding the initial treatment for lung cancer. However, to obtain sufficient tumor tissue to perform the genetic analyses is frequently difficult in lung cancer patients, especially those with unresectable disease. Non-invasive methods to estimate the probability of the EGFR/KRAS mutation status are helpful in clinical practice.
18F-fluoro-2-deoxy-glucose (18F-FDG) positron emission tomography (PET), a functional imaging modality based on glucose metabolism, has become a standard tool for the diagnosis, initial staging, and evaluation of treatment efficacy in lung cancer [4]. High 18F-FDG uptake reflects both the increased glucose metabolism and proliferative activity of tumor cells [5, 6]. EGFR mutations activate the EGFR-signaling pathway, inhibit apoptosis, and increase cell proliferation, angiogenesis and metastatic potential [7]. KRAS plays a key role in the downstream signaling RAS/MAPK pathway of EGFR and other growth factor receptors [7]. Point mutations of KRAS also play a critical role in cancer cell growth. Therefore, we hypothesized that there is a causal relationship between increased glucose metabolism and EGFR or KRAS mutation.
The emergence of next-generation sequencing technologies has enabled a wide range of protocols for more comprehensive and accurate genome-wide analysis. Among these, cap analysis gene expression (CAGE) is a genome-wide approach forming a comprehensive profile of the transcriptome by sequencing only the 5’-ends of capped RNAs [8]. Profiles represent promoter activities based on the frequencies of transcription starting sites (TSSs). CAGE has been used in genome-wide studies such as the ENCODE project [9] and FANTOM5 project [10–12]. Given that the transcriptome represents the molecular basis underlying cellular characteristics, we recently applied CAGE to the study of biomarkers to discriminate distinct types of lung cancer [13].To date, however, CAGE has not been used to study glucose metabolism in tumor cells.
Using transcriptome data from lung adenocarcinomas that monitor expression levels of genes that play important and specific roles in glucose metabolism, we investigated possible correlations between the standardized uptake value (SUV) of 18F-FDG PET and EGFR or KRAS mutation status in lung adenocarcinoma. Furthermore, we also investigated the specific molecular background of glucose metabolism in EGFR- or KRAS-mutated lung adenocarcinoma.
Materials and methods
Patients
Between February 2009 and May 2014, 1414 patients with primary lung cancers, including 1062 with adenocarcinomas, underwent pulmonary resection at our institution. Among these, we retrospectively reviewed 734 adenocarcinoma patients who underwent 18F-FDG PET-CT scanning within 2 months before surgery and whose surgically resected specimens were examined for EGFR and KRAS mutations. Patients who underwent induction chemotherapy and/or radiotherapy were excluded from this study. Patients were classified into three groups according to the mutation status of the tumors, namely EGFR mutation-positive (EGFR m+), KRAS mutation-positive (KRAS m+), and wild-type (WT) for both genes. Clinical characteristics such as age, gender, smoking status, preoperative serum carcinoembryonic antigen (CEA) level and SUVmax and pathological findings such as tumor size, nodal status, lymphatic permeation and vascular invasion of EGFR m+ and KRAS m+ tumors were compared to those of WT tumors.
This study was performed using surgical specimens in the tissue bank at our department, which was established with the approval of the institutional review board (IRB) of Juntendo University School of Medicine. Written consent was obtained from all patients prior to surgery for the procurement of tissue for the research purposes. The IRB approved the use of specimens stored in the tissue bank without obtaining new informed consent and deemed that the contents of this study were ethically acceptable.
18F-FDG PET-CT scanning
As detailed previously [14], PET-CT scan was carried out with a Discovery ST PET/CT scanner (GE Medical Systems; Waukesha, WI, USA) at the Yotsuya Medical Cube (Tokyo Japan). Two experienced nuclear medicine radiologists (W. K. and M. A.) evaluated the PET-CT images, side by side, and reached a consensus on the findings.
Mutation analyses for EGFR and KRAS
Genomic DNA was extracted from frozen lung cancer tissues sampled from surgically resected specimens. EGFR mutations were analyzed using the peptide nucleic acid-locked nucleic acid polymerase chain reaction (PCR) clamp method [15], and KRAS mutations using the peptide nucleic acid-mediated PCR clamping method [16].
Statistical analysis of the correlations between EGFR or KRAS mutation status and clinicopathological factors
The Steel-Dwass test was used to compare SUVmax among multiple groups based on EGFR and KRAS mutation patterns. Receiver operating characteristic (ROC) curves were generated to obtain a cut-off for SUVmax of the primary tumor which maximizes the sum of sensitivity and specificity for predicting EGFR or KRAS mutation status. Correlations between EGFR or KRAS mutation status and clinicopathological factors were evaluated. Univariate analyses between SUVmax and each clinicopathological factor were performed by a logistic regression model. All of the variables identified to be significant in the univariate analyses were subsequently entered into the multivariate analyses using a bidirectional (i.e., forward and backward) step-wise logistic regression model. A P-value of < 0.05 was considered statistically significant. All statistical analyses were performed using the R statistical software package (version 3.0.2, http://www.r-project.org/).
CAGE data
CAGE data generated using the previously described protocol [17] were obtained from a previous study [13]. In brief, double-stranded RNA/cDNA produced by reverse transcription from total RNA extracts was purified, oxidized with sodium periodate, and biotinylated with biotin hydrazide. The single-stranded cDNA was recovered after digestion of the single-stranded RNA with RNase I, and ligated with 3’-end and 5’-end adaptors specific to the samples. Double-stranded cDNAs were synthesized and mixed for sequencing in one lane of an Illumina HiSeq2500 sequencer (Illumina; San Diego, CA, USA). The CAGE reads were aligned to the reference genome (hg19) with high mapping quality of ≥ 20.
Differential and correlation analysis using the CAGE data
The aligned CAGE reads were counted in each region of the FANTOM5 robust peaks [11], a reference set of TSS regions, as raw signals for the promoter activities. Expression (activity) levels of individual promoters were quantified as counts per million (CPM) after normalization by the relative log expression method [18], and subjected to differential analysis using edgeR (version 3.2.4) [19] in R/Bioconductor [20]. Associations between expression levels and SUVmax and their statistical significance were assessed by Spearman’s rank correlation. Only results with a false discovery rate (FDR) less than 1% were considered statistically significant, in both the differential and correlation analyses.
Results
Patient characteristics and EGFR and KRAS mutation status
Patient characteristics are summarized in Table 1. Of 734 patients, 367 (50%) were male and 367 (50%) were female. Median age at the time of the operation was 68 years (range, 27–89 years). A total of 363 of 734 (49%) patients were smokers (pack-years > 5) and 371 (51%) were non-smokers (pack-years ≤ 5).
Table 1. Clinical characteristics of patients.
| Characteristic n (%) | ||
|---|---|---|
| Age (years) | ||
| ≤ 65 | 309 (42) | |
| > 65 | 425 (58) | |
| Sex | ||
| Male | 367 (50) | |
| Female | 367 (50) | |
| Smoking | ||
| ≤ 5 PY | 371 (51) | |
| > 5 PY | 363 (49) | |
| Serum CEA level | ||
| Normal | 386 (53) | |
| Elevated | 348 (47) | |
| Tumor size | ||
| < 30 mm | 514 (70) | |
| ≥ 30 mm | 220 (30) | |
| Pathological stage | ||
| IA/IB | 410/123 | |
| IIA/IIB | 40/36 | |
| IIIA/IIIB | 99/8 | |
| IV | 18 | |
| Pathological nodal status | ||
| N0 | 578 (79) | |
| N1 / N2 | 156 (21) | |
| Lymphatic permeation | ||
| Negative | 539 (73) | |
| Positive | 195 (27) | |
| Vascular invasion | ||
| Negative | 514 (70) | |
| Positive | 220 (30) | |
| SUVmax | ||
| Median (range) | 2.7 (0–33.2) | |
| EGFR mutation | ||
| Negative | 400 (54) | |
| Positive | 334 (46) | |
| exon 21 L858R | 194 | |
| exon 19 deletions | 120 | |
| minor mutations | 20 | |
| KRAS mutation | ||
| Negative | 651 (89) | |
| Positive | 83 (11) | |
| G to T/G to C | 60 | |
| G to A | 23 | |
PY = pack years.
Of the 734 lung adenocarcinomas, EGFR and KRAS mutations were detected in 334 (46%) and 83 (11%), respectively. The EGFR mutation spectra were distributed as follows. The point mutation L858R in exon 21 and deletions in exon 19 were detected in 194 and 120 tumors, respectively, which together accounted for 94% of all EGFR alterations. The remaining 6% of the minor EGFR mutations were exon 18 G719A in 8 tumors, exon 18 G719S in 5, exon 18 G719C in 2 and exon 21 L861Q in 3. Double mutations were found in 2 tumors; 1 harbored exon 21 L861Q and exon 20 T790M and the other had exon 18 G719A and exon 20 T790M, simultaneously. With regard to KRAS, a point mutation in codon 12 was found in 81 (98%) tumors, and a point mutation in codon 13 in 2 (2%). G to T, or G to C transversions were found in 60 (72%) tumors, and G to A transition in 23 (28%). EGFR and KRAS mutations were mutually exclusive.
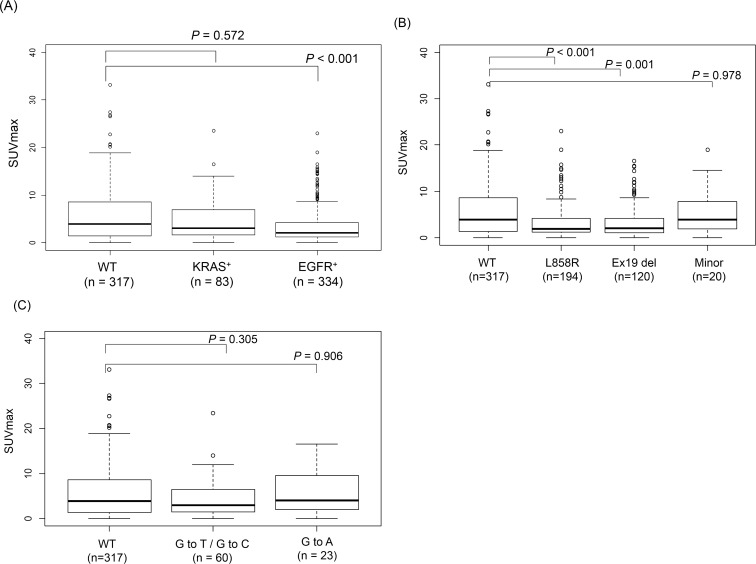
The median SUVmax of all primary tumors was 2.7 (range, 0–33.2). Median SUVmax in the EGFR m+ group, KRAS m+ group, and WT group were 2.1 (range, 0–23), 3.0 (range, 0–23.5), and 3.9 (range, 0–33.2), respectively. SUVmax of EGFR m+ tumors was significantly lower than that of WT and KRAS m+ tumors (Fig 1A). SUVmax of tumors with exon 21 L858R or exon 19 deletions was significantly lower than that of WT tumors. However, no significant differences were noted in SUVmax between tumors with minor mutations and WT tumors (Fig 1B). The SUVmax of KRAS m+ tumors did not significantly differ from that of WT tumors (Fig 1A). No significant differences were found in SUVmax between tumors with any KRAS mutation spectrum (G to T/G to C transversions or G to A transition) and WT tumors (Fig 1C).
Fig 1. Correlations between SUVmax of primary tumors and EGFR and KRAS mutation status.
(A) Box plot of SUVmax of primary tumors according to EGFR and KRAS mutation status, (B) Box plot of SUVmax of primary tumors according to EGFR mutation spectra, (C) Box plot of SUVmax of primary tumors according to KRAS mutation spectra.
ROC curve analyses of the cut-off values of SUVmax for the prediction of EGFR or KRAS mutations
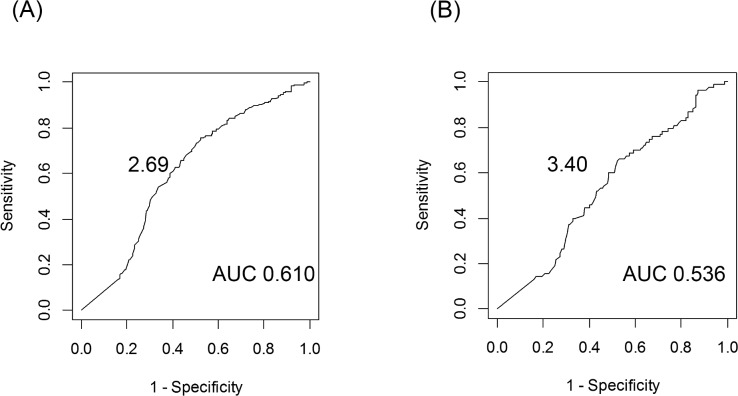
Next, we evaluated the prediction of EGFR or KRAS mutation using SUVmax. A cut-off value of SUVmax ≤ 2.69 provided the highest area under the curve (AUC; 0.610) for predicting EGFR mutation, while SUVmax ≤ 3.40 provided the highest AUC (0.536) for KRAS mutation (Fig 2). Using these cut-off values, parameters for the prediction of EGFR mutations were sensitivity, 60%; specificity, 61%; accuracy, 60%; positive predictive value (PPV), 62%; and negative predictive value (NPV), 59%; and parameters for the prediction of KRAS mutations were sensitivity, 54%; specificity, 54%; accuracy, 54%; PPV, 23%; and NPV, 82%.
Fig 2. Cut-off values of SUVmax in prediction of EGFR and KRAS mutation.
(A) EGFR mutation, (B) KRAS mutation.
Univariate and multivariate analysis of the predictors of EGFR or KRAS mutations
On univariate analysis, EGFR mutations were more frequent in females, non-smokers, patients with normal CEA levels, tumors without lymph node involvement or blood vessel invasion, and tumors with lower SUVmax. On multivariate analysis, significant predictors of EGFR mutation were smoking status and SUVmax (Table 2). The probability of EGFR mutation was inversely correlated with SUVmax. Univariate analyses showed that KRAS mutations were more frequent in males and smokers. On multivariate analysis, the only significant predictor of KRAS mutation was smoking history (Table 3). No relationship was found between the KRAS mutation status and SUVmax. The predictability of EGFR mutation status was compared between combinations of well-established clinical factors with or without SUVmax (Table 4). PPV of EGFR mutation status was increased by adding SUVmax to gender and smoking status.
Table 2. Univariate and multivariate analysis of predictors of EGFR mutation.
| Characteristic | WT | EGFR m+ | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|---|---|
| (n = 317) | Odds ratio (95% CI) | p-value | Odds ratio (95% CI) | p-value | |||
| Age (years) | |||||||
| ≤ 65 | 143 | 137 | 1 | ||||
| > 65 | 174 | 197 | 1.182 (0.866–1.613) | 0.292 | |||
| Sex | |||||||
| Female | 136 | 210 | 1 | ||||
| Male | 181 | 124 | 0.444 (0.323–0.607) | < 0.001 | |||
| Smoking | |||||||
| ≤ 5 PY | 131 | 229 | 1 | 1 | |||
| > 5 PY | 186 | 105 | 0.323 (0.234–0.444) | < 0.001 | 0.357 (0.256–0.494) | < 0.001 | |
| Serum CEA level | |||||||
| Normal | 157 | 197 | 1 | ||||
| Elevated | 160 | 137 | 0.682 (0.500–0.930) | 0.016 | |||
| Tumor size | |||||||
| < 30 mm | 218 | 243 | 1 | ||||
| ≥ 30 mm | 99 | 91 | 0.825 (0.587–1.156) | 0.264 | |||
| Pathological nodal status | |||||||
| N0 | 232 | 277 | 1 | ||||
| N1 / N2 | 85 | 57 | 0.562 (0.383–0.818) | 0.003 | |||
| Lymphatic permeation | |||||||
| Negative | 221 | 253 | 1 | ||||
| Positive | 96 | 81 | 0.737 (0.521–1.041) | 0.084 | |||
| Vascular invasion | |||||||
| Negative | 204 | 251 | 1 | ||||
| Positive | 113 | 83 | 0.597 (0.425–0.836) | 0.003 | |||
| SUVmax | |||||||
| ≤ 2.69 | 124 | 200 | 1 | 1 | |||
| > 2.69 | 193 | 134 | |||||
WT = wild-type; m+ = mutation-positive; PY = pack years.
Table 3. Univariate and multivariate analysis of predictors of KRAS mutation.
| Characteristic | WT | KRAS m+ | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|---|---|
| (n = 317) | (n = 83) | Odds ratio (95% CI) | p-value | Odds ratio (95% CI) | p-value | ||
| Age (years) | |||||||
| ≤ 65 | 143 | 29 | 1 | ||||
| > 65 | 174 | 54 | 1.530 (0.932–2.554) | 0.097 | |||
| Sex | |||||||
| Female | 136 | 21 | 1 | ||||
| Male | 181 | 62 | 2.218 (1.308–3.890) | 0.004 | |||
| Smoking | |||||||
| ≤ 5 PY | 131 | 12 | 1 | 1 | |||
| > 5 PY | 186 | 71 | 4.167 (2.248–8.359) | < 0.001 | 4.167 (2.248–8.359) | < 0.001 | |
| Serum CEA level | |||||||
| Normal | 157 | 32 | 1 | ||||
| Elevated | 160 | 51 | 1.564 (0.959–2.581) | 0.076 | |||
| Tumor size | |||||||
| < 30 mm | 218 | 53 | 1 | ||||
| ≥ 30 mm | 99 | 30 | 1.246 (0.745–2.059) | 0.394 | |||
| Pathological nodal status | |||||||
| N0 | 232 | 69 | 1 | ||||
| N1 / N2 | 85 | 14 | 0.554 (0.286–1.009) | 0.064 | |||
| Lymphatic permeation | |||||||
| Negative | 221 | 65 | 1 | ||||
| Positive | 96 | 18 | 0.637 (0.351–1.112) | 0.124 | |||
| Vascular invasion | |||||||
| Negative | 204 | 59 | 1 | ||||
| Positive | 113 | 24 | 0.734 (0.427–1.231) | 0.251 | |||
| SUV max | |||||||
| ≤ 3.4 | 147 | 45 | 1 | ||||
| > 3.4 | 170 | 38 | 0.730 (0.448–1.185) | 0.204 | |||
WT = wild-type; m+ = mutation-positive; PY = pack years.
Table 4. Predictability of the EGFR mutation status by the combinations of well-established clinical factors with or without SUVmax.
| EGFR mutation status | Sensitivity | Specificity | PPV | NPV | Accuracy | |||
|---|---|---|---|---|---|---|---|---|
| Clinical predictors | Positive | Negative | ||||||
| Female & Non-smoker * | Yes | 182 | 115 | 54% | 71% | 61% | 65% | 64% |
| No | 152 | 285 | ||||||
| Non-smoker & SUVmax ≤ 2.69 | Yes | 131 | 83 | 39% | 79% | 61% | 61% | 61% |
| No | 203 | 317 | ||||||
| Female & Non-smoker | Yes | 110 | 66 | 33% | 84% | 63% | 60% | 60% |
| & SUVmax ≤ 2.69 | No | 224 | 334 | |||||
* means pack-years ≤ 5.
PPV = positive predictive value; NPV = negative predictive value.
CAGE for the molecular background of glucose metabolism in EGFR or KRAS mutated lung adenocarcinoma
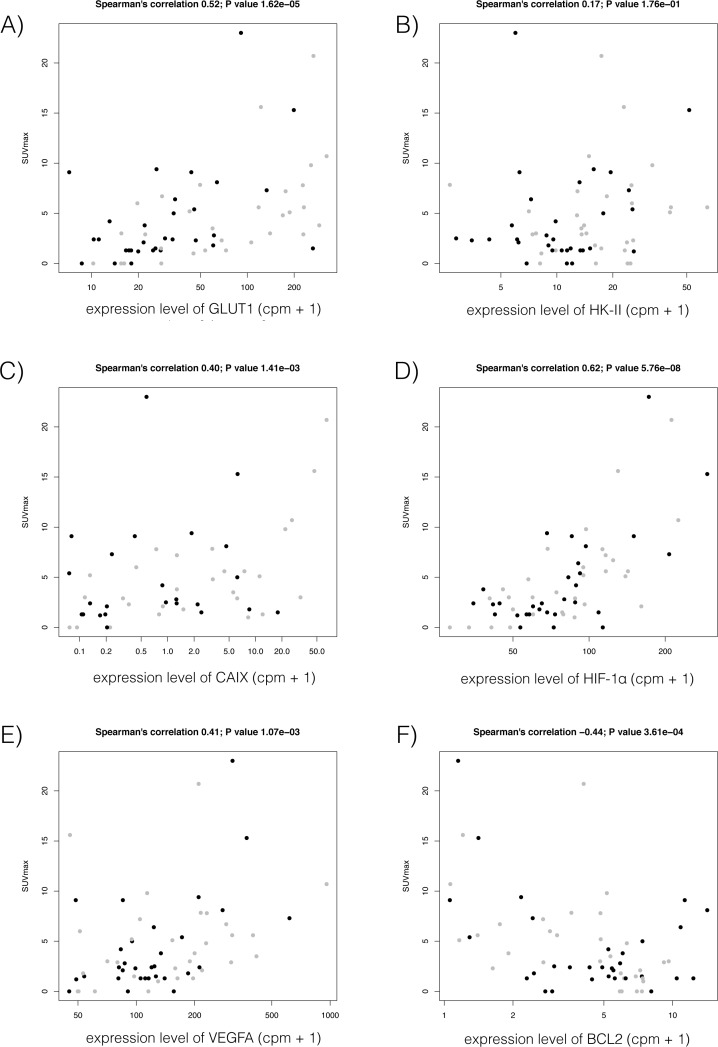
Further, we examined expression levels of genes based on the CAGE results (Takamochi et al., submitted), in particular those related to glucose metabolism and the cell cycle, in association with SUVmax. We manually selected 7 genes associated with glucose metabolism: class I glucose transporters (GLUT1, GLUT2, GLUT3, GLUT4), hexokinase-II (HK-II), hypoxia-inducible factor-1 alpha (HIF-1α), and carbonic anhydrase IX (CAIX). Of these, 4 genes (GLUT1, HK-II, HIF-1α, and CAIX) showed positive correlations between their expression levels monitored by CAGE with SUVmax across 62 lung adenocarcinomas (Fig 3). Next, we selected 5 genes associated with cell growth: TP53, CCND1, BCL2, vascular endothelial growth factor (VEGF), and MKI67. Of these, expression of VEGF showed a positive correlation with SUVmax, while BCL2 showed an inverse correlation with SUVmax (Fig 3).
Fig 3.
Scatter plots of association of SUVmax with expression levels of four genes associated with glucose metabolism (A-D) and two genes associated with cell proliferation (E and F): (A) GLUT-1, (B) HK-II, (C) CAIX, (D) HIF-1α, (E) VEGF, and (F) BCL2. Y-axis represents SUVmax and X-axis represents gene expression monitored by CAGE, in which the most correlated promoter activities are shown. Black and gray dots represent donors with EGFR mutation-positive (EGFR m+) and wild-type, respectively.
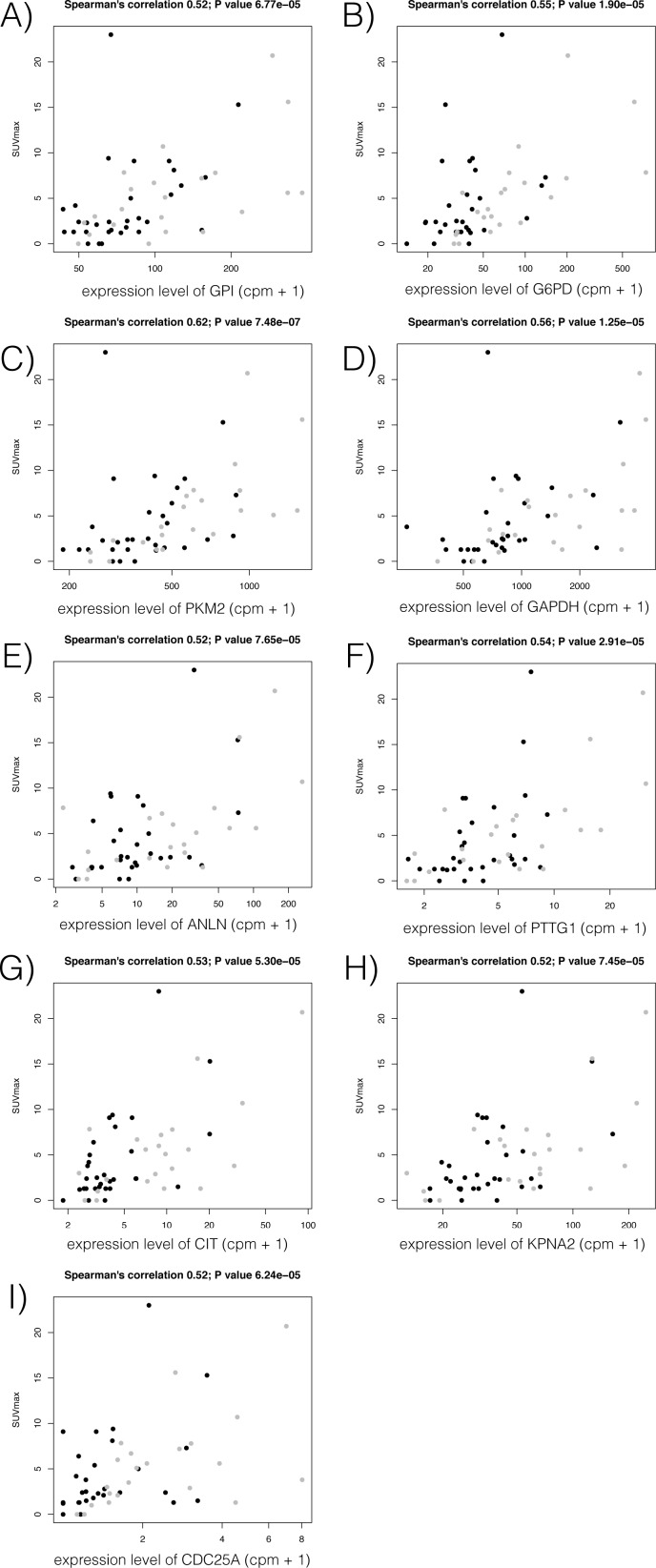
We expanded this expression analysis to examine genes involved in the 2 pathways. Among genes whose promoters were more significantly down-regulated in EGFR m+ tumors than WT tumors (FDR < 1%), we found that both glucose metabolism-related and cell cycle-related genes were enriched (P value < 5.2e-18 and 0.02, with GO term enrichment analysis with DAVID) [21, 22]. Of these, 4 genes associated with glucose metabolism (GPI, G6PD, PKM2, and GAPDH) and 5 genes associated with the cell cycle (ANLN, PTTG1, CIT, KPNA2, and CDC25A) showed a positive correlation between expression and SUVmax. (FDR < 1%; Fig 4). Notably, none of the genes down-regulated in KRAS m+ tumors showed significant correlation with SUVmax.
Fig 4.
Association of SUVmax with expression levels of genes associated with glucose metabolism (A-D) or the cell cycle (E-I), which were specifically down-regulated in EGFR-mutated tumors compared to wild-type tumors and correlated with SUVmax: (A) GPI, (B) G6PD, (C) PKM2, (D) GAPDH, (E) ANLN, (F) PTTG1, (G) CIT, (H) KPNA2, and (I) CDC25A. Y-axis represents SUVmax and X-axis represents gene expression monitored by CAGE, showing the most correlated promoter activities. Black and gray dots represent donors with EGFR mutation-positive (EGFR m+) and wild-type, respectively.
Discussion
In this study, we found that the probability of EGFR mutation in lung adenocarcinoma was inversely correlated with SUVmax. In contrast, the probability of KRAS mutation was not correlated with SUVmax. Further, several genes associated with glucose metabolism or the cell cycle were specifically down-regulated in EGFR m+ adenocarcinomas. These findings suggest that EGFR m+ adenocarcinomas are biologically indolent with potentially lower levels of glucose metabolism than wild-type tumors.
To our knowledge, this is the largest study to evaluate the correlations between 18F-FDG uptake and EGFR mutation status in lung cancer, and the first to investigate the correlation between the 18F-FDG uptake and KRAS mutation status. The 4 retrospective studies that previously investigated the correlation between the 18F-FDG uptake and EGFR mutation status in lung cancer [23–26] reported contradictory findings (Table 4). In their multivariate analysis, Huang et al.[23] and Ko et al.[26] showed that a higher SUVmax was a significant predictor of EGFR mutation, whereas Na et al.[25] and Mak et al.[24] reported that a lower SUVmax of the primary tumor was predictive of EGFR mutation. Our findings are compatible with those of the latter groups [24, 25]. These conflicting results may have resulted from differences in the ethnic background or the small size of the study populations (Table 5).
Table 5. Clinical studies of the role of 18F-FDG uptake on PET-CT scans in predicting EGFR mutation status.
| Author/year | Ethnicity | No. of patients | Histology | Stage | EGFR mutation | Results * |
|---|---|---|---|---|---|---|
| Huang et al./2010 | Asian (Taiwanese) | 77 | Ad | Clinical IIIB or IV | 49 (64%) | SUVmax ≥ 9.5, EGFR m+ 78% |
| Na et al./2010 | Asian (Korean) | 100 | 53 Ad, 47 non-Ad | Pathological I-IV | 21 (21%) | SUVmax < 9.2, EGFR m+ 40% |
| Mak et al./2011 | White (88% of all) | 100 | 90 Ad, 10 non-Ad | Clinical I-IV | 24 (24%) | SUVmax ≥ 5.0, WT 96% |
| Ko et al./2014 | Asian (Taiwanese) | 132 | Ad | Clinical I-IV | 69 (52%) | SUVmax ≥ 6.0, EGFR m+63% |
| Present study | Asian (Japanese) | 734 | Ad | Pathological I-IV | 334 (46%) | SUVmax ≤ 2.69, EGFR m+ 62% |
* shows threshold SUVmax and positive predictive value of EGFR mutation status.
Ad = adenocarcinoma; m+ = mutation-positive; WT = wild-type.
Consistent with numerous previous reports [27–29], EGFR mutations in the present study were more frequent in females and never-smokers. In addition, a higher probability of EGFR mutation was observed in tumors without lymph node involvement or blood vessel invasion and in those with a lower SUVmax. Higashi et al.[30] reported that the prevalence rates of lymphatic permeation and lymph node involvement were lower in primary tumors with low 18F-FDG uptake than those with a higher 18F-FDG uptake. These findings suggest that EGFR m+ adenocarcinomas are biologically indolent with potentially lower levels of glucose metabolism.
Although many factors have been reported to influence 18F-FDG uptake, the precise biological mechanism by which 18F-FDG accumulates in malignant cells remains to be clarified. In 1985, Mueckler et al.[31] initially reported that facilitative glucose transport across the plasma membrane was mediated by a family of structurally related proteins known as facilitated diffuse GLUTs. Among the 14 currently known GLUT isoforms [32], the overexpression of GLUT-1 has been shown to be most closely related to 18F-FDG uptake in lung cancer [33–35]. Sasaki et al.[36] reported that GLUT-1 overexpression evaluated by immunohistochemistry was significantly correlated with EFGR or KRAS mutation status, with overexpression in 18 (24%) of 76 EGFR m+ lung cancers and 20 (67%) of 30 KRAS m+ lung cancers. In our present patients, we found that the expression level of GLUT-1 was positively correlated with SUVmax, as were other genes related to glucose metabolism, namely HK-II, CAIX, and HIF-1α (Fig 3). This finding is consistent with previous reports [34, 37]. GO term analysis revealed that the glucose metabolism-related and the cell cycle-related genes were enriched among the down-regulated genes in EGFR m+ adenocarcinomas, which supports our results for 18F-FDG PET, with lower levels of SUVmax. Notably, 4 of the glucose metabolism-related genes, GPI, G6PD, PKM2, and GAPDH and 5 of the cell cycle-related genes, ANLN, PTTG1, CIT, KPNA2, and CDC25A, were significantly down-regulated in EGFR m+ adenocarcinomas, and showed a substantial correlation with SUVmax (Fig 4). These likely comprise a common subset of the pathway underlying EGFR mutation and glucose metabolism.
Several limitations of our study warrant mention. First, it was conducted under a retrospective design in patients who required surgical resection, most for early stage disease. Accordingly, the selected cases might not have reflected the overall features of lung adenocarcinoma. Second, the sample size of KRAS m+ tumors was too small to allow any firm conclusions. Although we found no significant relationship between 18F-FDG uptake and KRAS mutation status in lung adenocarcinoma and did not identify any genes specifically correlated with glucose metabolism in KRAS m+ tumors, a conclusive answer to this question would require a larger sample size.
In summary, the probability of EGFR mutation was inversely correlated with SUVmax. In contrast, the probability of KRAS mutation was not correlated with SUVmax. Several genes associated with glucose metabolism or the cell cycle were specifically down-regulated in EGFR m+ adenocarcinomas. These findings confirm that EGFR m+ adenocarcinomas are biologically indolent with potentially lower levels of glucose metabolism than wild-type tumors.
Data Availability
All relevant data are within the paper.
Funding Statement
This work is supported by a Grant-in-Aid for Scientific Research (C) to Kazuya Takamochi, a Smoking Research Foundation to Kazuya Takamochi, a Research Grant for the RIKEN Omics Science Center from MEXT to Yoshihide Hayashizaki, and a Research Grant to the RIKEN Preventive Medicine and Diagnosis Innovation Program from MEXT to Yoshihide Hayashizaki.
References
- 1.Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362(25):2380–8. Epub 2010/06/25. doi: 10.1056/NEJMoa0909530 [DOI] [PubMed] [Google Scholar]
- 2.Mitsudomi T, Morita S, Yatabe Y, Negoro S, Okamoto I, Tsurutani J, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol. 2010;11(2):121–8. Epub 2009/12/22. doi: 10.1016/S1470-2045(09)70364-X [DOI] [PubMed] [Google Scholar]
- 3.Eberhard DA, Johnson BE, Amler LC, Goddard AD, Heldens SL, Herbst RS, et al. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol. 2005;23(25):5900–9. doi: 10.1200/JCO.2005.02.857 [DOI] [PubMed] [Google Scholar]
- 4.Stroobants S, Verschakelen J, Vansteenkiste J. Value of FDG-PET in the management of non-small cell lung cancer. Eur J Radiol. 2003;45(1):49–59. [DOI] [PubMed] [Google Scholar]
- 5.Higashi K, Ueda Y, Yagishita M, Arisaka Y, Sakurai A, Oguchi M, et al. FDG PET measurement of the proliferative potential of non-small cell lung cancer. Journal of nuclear medicine: official publication, Society of Nuclear Medicine. 2000;41(1):85–92. Epub 2000/01/27. [PubMed] [Google Scholar]
- 6.Vesselle H, Schmidt RA, Pugsley JM, Li M, Kohlmyer SG, Vallires E, et al. Lung cancer proliferation correlates with [F-18]fluorodeoxyglucose uptake by positron emission tomography. Clinical cancer research: an official journal of the American Association for Cancer Research. 2000;6(10):3837–44. Epub 2000/10/29. [PubMed] [Google Scholar]
- 7.Baselga J. Why the epidermal growth factor receptor? The rationale for cancer therapy. The oncologist. 2002;7 Suppl 4:2–8. Epub 2002/08/31. [DOI] [PubMed] [Google Scholar]
- 8.Shiraki T, Kondo S, Katayama S, Waki K, Kasukawa T, Kawaji H, et al. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(26):15776–81. Epub 2003/12/10. PubMed Central PMCID: PMCPmc307644. doi: 10.1073/pnas.2136655100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nonaka D. A study of DeltaNp63 expression in lung non-small cell carcinomas. The American journal of surgical pathology. 2012;36(6):895–9. Epub 2012/03/01. doi: 10.1097/PAS.0b013e3182498f2b [DOI] [PubMed] [Google Scholar]
- 10.Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507(7493):455–61. Epub 2014/03/29. doi: 10.1038/nature12787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forrest AR, Kawaji H, Rehli M, Baillie JK, de Hoon MJ, Lassmann T, et al. A promoter-level mammalian expression atlas. Nature. 2014;507(7493):462–70. Epub 2014/03/29. doi: 10.1038/nature13182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arner E, Daub CO, Vitting-Seerup K, Andersson R, Lilje B, Drablos F, et al. Gene regulation. Transcribed enhancers lead waves of coordinated transcription in transitioning mammalian cells. Science (New York, NY). 2015;347(6225):1010–4. Epub 2015/02/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takamochi K, Ohmiya H, Itoh M, Mogushi K, Saito T, Hara K, et al. Novel biomarkers that assist in accurate discrimination of squamous cell carcinoma from adenocarcinoma of the lung. BMC Cancer. 2016;16(1):760 doi: 10.1186/s12885-016-2792-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hattori A, Suzuki K, Matsunaga T, Fukui M, Tsushima Y, Takamochi K, et al. Tumour standardized uptake value on positron emission tomography is a novel predictor of adenocarcinoma in situ for c-Stage IA lung cancer patients with a part-solid nodule on thin-section computed tomography scan. Interactive cardiovascular and thoracic surgery. 2014;18(3):329–34. Epub 2013/12/20. PubMed Central PMCID: PMCPMC3930213. doi: 10.1093/icvts/ivt500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagai Y, Miyazawa H, Huqun, Tanaka T, Udagawa K, Kato M, et al. Genetic heterogeneity of the epidermal growth factor receptor in non-small cell lung cancer cell lines revealed by a rapid and sensitive detection system, the peptide nucleic acid-locked nucleic acid PCR clamp. Cancer Res. 2005;65(16):7276–82. doi: 10.1158/0008-5472.CAN-05-0331 [DOI] [PubMed] [Google Scholar]
- 16.Thiede C, Bayerdorffer E, Blasczyk R, Wittig B, Neubauer A. Simple and sensitive detection of mutations in the ras proto-oncogenes using PNA-mediated PCR clamping. Nucleic Acids Res. 1996;24(5):983–4. Epub 1996/03/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murata M, Nishiyori-Sueki H, Kojima-Ishiyama M, Carninci P, Hayashizaki Y, Itoh M. Detecting expressed genes using CAGE. Methods Mol Biol. 2014;1164:67–85. Epub 2014/06/15. doi: 10.1007/978-1-4939-0805-9_7 [DOI] [PubMed] [Google Scholar]
- 18.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106 Epub 2010/10/29. PubMed Central PMCID: PMCPMC3218662. doi: 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics (Oxford, England). 2010;26(1):139–40. Epub 2009/11/17. PubMed Central PMCID: PMCPmc2796818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80 Epub 2004/10/06. PubMed Central PMCID: PMCPmc545600. doi: 10.1186/gb-2004-5-10-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4(1):44–57. Epub 2009/01/10. doi: 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 22.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic acids research. 2009;37(1):1–13. Epub 2008/11/27. PubMed Central PMCID: PMCPmc2615629. doi: 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang CT, Yen RF, Cheng MF, Hsu YC, Wei PF, Tsai YJ, et al. Correlation of F-18 fluorodeoxyglucose-positron emission tomography maximal standardized uptake value and EGFR mutations in advanced lung adenocarcinoma. Medical oncology (Northwood, London, England). 2010;27(1):9–15. Epub 2009/01/09. [DOI] [PubMed] [Google Scholar]
- 24.Mak RH, Digumarthy SR, Muzikansky A, Engelman JA, Shepard JA, Choi NC, et al. Role of 18F-fluorodeoxyglucose positron emission tomography in predicting epidermal growth factor receptor mutations in non-small cell lung cancer. The oncologist. 2011;16(3):319–26. Epub 2011/02/23. doi: 10.1634/theoncologist.2010-0300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Na II, Byun BH, Kim KM, Cheon GJ, Choe du H, Koh JS, et al. 18F-FDG uptake and EGFR mutations in patients with non-small cell lung cancer: a single-institution retrospective analysis. Lung cancer (Amsterdam, Netherlands). 2010;67(1):76–80. Epub 2009/04/18. [DOI] [PubMed] [Google Scholar]
- 26.Ko KH, Hsu HH, Huang TW, Gao HW, Shen DH, Chang WC, et al. Value of (1)(8)F-FDG uptake on PET/CT and CEA level to predict epidermal growth factor receptor mutations in pulmonary adenocarcinoma. European journal of nuclear medicine and molecular imaging. 2014;41(10):1889–97. Epub 2014/05/24. doi: 10.1007/s00259-014-2802-y [DOI] [PubMed] [Google Scholar]
- 27.Mitsudomi T, Kosaka T, Yatabe Y. Biological and clinical implications of EGFR mutations in lung cancer. Int J Clin Oncol. 2006;11(3):190–8. Epub 2006/07/20. doi: 10.1007/s10147-006-0583-4 [DOI] [PubMed] [Google Scholar]
- 28.Shigematsu H, Lin L, Takahashi T, Nomura M, Suzuki M, Wistuba II, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005;97(5):339–46. Epub 2005/03/03. doi: 10.1093/jnci/dji055 [DOI] [PubMed] [Google Scholar]
- 29.Tsao AS, Tang XM, Sabloff B, Xiao L, Shigematsu H, Roth J, et al. Clinicopathologic characteristics of the EGFR gene mutation in non-small cell lung cancer. J Thorac Oncol. 2006;1(3):231–9. Epub 2007/04/06. [DOI] [PubMed] [Google Scholar]
- 30.Higashi K, Ito K, Hiramatsu Y, Ishikawa T, Sakuma T, Matsunari I, et al. 18F-FDG uptake by primary tumor as a predictor of intratumoral lymphatic vessel invasion and lymph node involvement in non-small cell lung cancer: analysis of a multicenter study. Journal of nuclear medicine: official publication, Society of Nuclear Medicine. 2005;46(2):267–73. Epub 2005/02/08. [PubMed] [Google Scholar]
- 31.Mueckler M, Caruso C, Baldwin SA, Panico M, Blench I, Morris HR, et al. Sequence and structure of a human glucose transporter. Science (New York, NY). 1985;229(4717):941–5. Epub 1985/09/06. [DOI] [PubMed] [Google Scholar]
- 32.Suganuma N, Segade F, Matsuzu K, Bowden DW. Differential expression of facilitative glucose transporters in normal and tumour kidney tissues. BJU international. 2007;99(5):1143–9. Epub 2007/04/18. doi: 10.1111/j.1464-410X.2007.06765.x [DOI] [PubMed] [Google Scholar]
- 33.Higashi K, Ueda Y, Sakurai A, Wang XM, Xu L, Murakami M, et al. Correlation of Glut-1 glucose transporter expression with [18F]FDG uptake in non-small cell lung cancer. European journal of nuclear medicine. 2000;27(12):1778–85. Epub 2001/02/24. [DOI] [PubMed] [Google Scholar]
- 34.Mamede M, Higashi T, Kitaichi M, Ishizu K, Ishimori T, Nakamoto Y, et al. [18F]FDG uptake and PCNA, Glut-1, and Hexokinase-II expressions in cancers and inflammatory lesions of the lung. Neoplasia (New York, NY). 2005;7(4):369–79. Epub 2005/06/22. PubMed Central PMCID: PMC1501150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Baardwijk A, Dooms C, van Suylen RJ, Verbeken E, Hochstenbag M, Dehing-Oberije C, et al. The maximum uptake of (18)F-deoxyglucose on positron emission tomography scan correlates with survival, hypoxia inducible factor-1alpha and GLUT-1 in non-small cell lung cancer. European journal of cancer (Oxford, England: 1990). 2007;43(9):1392–8. Epub 2007/05/22. [DOI] [PubMed] [Google Scholar]
- 36.Sasaki H, Shitara M, Yokota K, Hikosaka Y, Moriyama S, Yano M, et al. Overexpression of GLUT1 correlates with Kras mutations in lung carcinomas. Molecular medicine reports. 2012;5(3):599–602. Epub 2011/12/28. doi: 10.3892/mmr.2011.736 [DOI] [PubMed] [Google Scholar]
- 37.Kaira K, Serizawa M, Koh Y, Takahashi T, Yamaguchi A, Hanaoka H, et al. Biological significance of 18F-FDG uptake on PET in patients with non-small-cell lung cancer. Lung cancer (Amsterdam, Netherlands). 2014;83(2):197–204. Epub 2013/12/25. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.