Abstract
Enterococcus faecalis is a Gram-positive, opportunistic, pathogenic bacterium that causes a significant number of antibiotic-resistant infections in hospitalized patients. The development of antibiotic resistance in hospital-associated pathogens is a formidable public health threat. In E. faecalis and other Gram-positive pathogens, correlations exist between lipid composition and antibiotic resistance. Resistance to the last-resort antibiotic daptomycin is accompanied by a decrease in phosphatidylglycerol (PG) levels, whereas multiple peptide resistance factor (MprF) converts anionic PG into cationic lysyl-PG via a trans-esterification reaction, providing resistance to cationic antimicrobial peptides. Unlike previous studies that relied on thin layer chromatography and spectrophotometry, we have performed liquid chromatography-tandem mass spectrometry (LC-MS/MS) directly on lipids extracted from E. faecalis, and quantified the phospholipids through multiple reaction monitoring (MRM). In the daptomycin-sensitive E. faecalis strain OG1RF, we have identified 17 PGs, 8 lysyl-PGs (LPGs), 23 cardiolipins (CL), 3 glycerophospho-diglucosyl-diacylglycerols (GPDGDAG), 5 diglucosyl-diacylglycerols (DGDAG), 3 diacylglycerols (DAGs), and 4 triacylglycerols (TAGs). We have quantified PG and shown that PG levels vary during growth of E. faecalis in vitro. We also show that two daptomycin-resistant (DapR) strains of E. faecalis have substantially lower levels of PG and LPG levels. Since LPG levels in these strains are lower, daptomycin resistance is likely due to the reduction in PG. This lipidome map is the first comprehensive analysis of membrane phospholipids and glycolipids in the important human pathogen E. faecalis, for which antimicrobial resistance and altered lipid homeostasis have been intimately linked.
Introduction
The normal habitat of enterococci is the gastrointestinal tract of animals and humans. As opportunistic pathogens, enterococci can cause bacteremia, meningitis, and endocarditis, and can also infect wound sites and the urinary tract. Commonly used medical devices such as intravascular and urinary catheters provide a surface on which enterococci can easily form biofilms, which are defined as “multilayer assemblages of bacteria that are held together by a sticky polysaccharide matrix” [1]. In the clinical setting, biofilms have been implicated in an estimated 65% of nosocomial infections, and it is when bacteria are in the biofilm form that they are more resistant to conventional antibiotics [2].
Enterococcus faecalis is a prominent causative agent of nosocomial and opportunistic infections, and has acquired resistance to many antibiotics. Daptomycin (DAP) is an antibiotic of last resort for multidrug resistant E. faecalis [3, 4]. It is a cationic lipopeptide that requires Ca2+ ions to target the cell membrane. Resistance to DAP is emerging in E. faecalis and has been linked to changes in phospholipid metabolism. Genes coding for glycerophosphoryl diester phosphodiesterase (gdpD) and cardiolipin synthase (cls) have been implicated in daptomycin resistance [3, 5, 6]. Besides playing a role in antibiotic resistance, lipid metabolism is involved in biomembrane synthesis and energy homeostasis during pathogen replication and persistence [7]. Homeostasis of membrane lipids is essential to bacterial viability [8–10]. The organization of lipids and proteins into membrane microdomains also helps to coordinate various bacterial processes [11–15], which include growth, genome replication, and cell division. These processes are dependent on the subcellular organization of biological molecules, including lipids [16, 17]. Both bacterial lipid composition and lipid organization into domains are important for normal physiology, secretion, signaling, virulence, and antibiotic resistance [10–12, 18–23].
Three major classes of phospholipids occur in bacterial membranes: phosphatidylglycerol (PG), cardiolipin (CL), and phosphatidylethanolamine (PE) [24]. All three classes are derived from the common precursor phosphatidic acid (PA) [8, 25]. The predominant phospholipid species in Gram-positive bacteria are phosphatidylglycerol (PG) and cardiolipin (CL) [26], whose relative composition varies from species to species [10]. The levels of individual phospholipids were shown to depend on growth conditions [26], indicating that phospholipid regulation is very dynamic and responsive to the environment. Bacteria are able to adjust the concentration of lipids [27] and thus alter their membrane composition as an adaptation to environmental stress [8]. Depending on the immediate physiological needs of the bacterium, fatty acids may be saturated or unsaturated [28] and in E. faecalis, fatty acids in the membrane are involved in adaptation to stressful conditions. Longer fatty acid chains decrease membrane fluidity [9], while the presence of one or more double bonds in these chains will increase fluidity [8]. A decrease in temperature leads to a higher level of unsaturated fatty acids in Bacillus subtilis [29], which is a specific example of how lipid composition is adjusted in response to temperature [8]. Unsaturated fatty acids in phospholipids increase resistance to bile salts and the antibiotic daptomycin. For example, the unsaturated fatty acid oleic acid protects E. faecalis against acute membrane stress caused by bile salts [30]. More recently, exogenous fatty acids were shown to confer protection against membrane stress caused by DAP despite the absence of LiaR, one of three components in the antibiotic-response regulator LiaFSR [31].
In recent years, several studies investigating enterococcal lipids in the context of resistance to antibiotics and antimicrobial peptides have been reported. The emergence of DAP resistance in enterococci has been correlated with lower fluidity and lower PG content and higher glycerophospho-diglucosyl-diacylglycerol (GPDGDAG) content. Total lysyl-PG (LPG) levels were unchanged. [3]. Lysyl-PG is a product of the bifunctional integral membrane enzyme known as multiple peptide resistance factor (MprF), which modifies PG with positively charged amino acids such as lysine in the inner CM leaflet, and then translocates or “flips” lysyl-PG to the outer leaflet. This modification mediates resistance to cationic antimicrobial peptides (CAMPs) such as colistin, nisin, human β-defensin 3 (hBD-3) and polymyxin B [22, 32–34]. DAP interacts with the septa of DAP-sensitive E. faecalis; however, changes in lipid composition and distribution in DAP-resistant strains are associated with changes in DAP interactions that move it away from the division septum to other cell membrane regions [6]. Thus, environmental factors and antimicrobial compounds exert considerable influences on lipid composition.
Bacterial cardiolipins (CLs) are compartmentalized into distinct membrane domains that play a role in cell division and membrane transport through lipid-protein interactions [35]. The first bacterial lipid domains reported were CL domains in B. subtilis [11, 36] and E. coli [11, 35, 37, 38]. CL domains are likely formed by the high negative curvature strain that is caused by CL in the lipid bilayer, owing to relative difference in size between its head group and its much larger hydrophobic domain [35].
To understand precisely how different lipid compositions contribute to antimicrobial resistance, we have performed a mass spectrometric (MS) analysis of the lipid composition of E. faecalis for the purpose of correlating individual lipid species with antibiotic resistance. Previous studies obtained the phospholipid content of daptomycin-resistant enterococci first by thin-layer chromatography (TLC) and then confirmation of TLC-separated products by tandem MS [3, 6] or solely by TLC [32]. These studies did not report the identities of individual lipid species. Here we have deployed MS-based methods directly on Enterococcal lipid extracts, and provide a detailed characterization of lipid species at the molecular level Our methods allow us to forgo TLC and analyze extracted lipids directly via MS, enabling the broadest possible analysis of lipids across various lipid classes. By taking advantage of high resolutions and sensitivities, we can obtain a snapshot of phospholipids and glycolipids that E. faecalis needs for growth and adaptation to antimicrobial challenge. Taking our lipidomic characterization one step further, we have employed the widely used approach of multiple reaction monitoring (MRM) for quantification of E. faecalis lipids. Using state-of-the-art MS platforms, our methods have better sensitivity, selectivity and accuracy, and can be used both for untargeted and targeted lipid analyses and rapid quantification. We have identified individual species of phosphatidylglycerols (PG), lysyl-PG (L-PG), cardiolipins (CL), glycerophospho-diglucosyl-diacylglycerols (GPDGDAG), diglucosyl-diacylglycerols (DGDAG), diacylglycerols (DAGs), and triacylglycerols (TAGs). Given the important role played by fatty acids in stress responses, we report fatty acid compositions for the lipids that we have identified. Furthermore, we have measured the concentrations of several PG species in E. faecalis OG1RF throughout the growth curve, and show that PG levels vary substantially as cells progress through the logarithmic phase to the late stationary phase. We also show that a subset of lysyl-PGs and PGs are substantially lower in two DAP-resistant (DapR) strains compared to DAP-sensitive wild type. The data and technical information that we provide in this study will lay the foundation for future lipidomics studies in E. faecalis and other bacteria, and provides a more detailed understanding of the lipidomic basis of DAP resistance.
Materials and methods
Bacterial strains and cultures
Enterococcus faecalis strains OG1RF [39], Dap21 and Dap22 were inoculated from single colonies grown on Brain Heart Infusion (BHI) agar at 37°C into BHI broth and was grown statically overnight for 15–18 h for all assays except the in vitro growth phase study. In addition, for the growth phase study, OG1RF was inoculated into BHI broth and grown statically until optical densities of 0.3 (early-logarithmic phase), 0.5 (mid-logarithmic phase), 0.8 (late-logarithmic phase) and 1.3 (late stationary phase) were achieved. Broth cultures were centrifuged, the cell pellets washed once with 1X PBS, and centrifuged again to harvest the cell pellets. Cell pellets were dried overnight on a lyophilizer and the dry cell pellets weighed.
In vitro evolution against DAP
The in vitro evolution protocol used here for E. faecalis OG1RF was adapted from that of E. faecalis V583 [40]. For each parent strain, multiple parallel lines of evolution were performed. The evolution experiment started 1:10 dilutions of overnight bacterial cultures into fresh BHI broth supplemented with calcium at 1.25 mM, and containing DAP at concentrations equivalent of the MIC, 2xMIC and 4xMIC. After 22 to 26-hour static incubation in 2 mL Eppendorf tubes at 37°C, cultures of each line were examined for visible bacterial growth. Bacterial cultures at the highest growth-permissive concentrations (HGPCs) were diluted 10 times into fresh DAP-containing medium at 0.5xHGPC, HGPC and 2xHGPC. We repeated the same incubation and dilution steps until the HGPC reached 512 μg/mL. Bacterial cultures were then passaged in plain BHI broth for 3 days, followed by a repeat test of the MIC to ensure the mutants were stably resistant to DAP. Passaged cultures were also streaked on selective media containing 25 μg/mL rifampicin (Sigma) plates and CHROMagar™ Orientation plates to ensure the absence of contamination by other microbes.
Whole-genome sequencing
Genomic DNA was extracted from overnight bacterial culture using PureLink Genomic DNA Mini Kit (Thermo Fisher Scientific). The samples were then sequenced on an Illumina MiSeq v3 platform. We analyzed the genomic sequence using CLC Genomics Workbench 8.0. The complete OG1RF reference genome (NC_017316) from NCBI database was used for mapping and annotation. The threshold variant frequency was set as 35%. Non-synonymous mutations within coding regions were filtered out. All structural variations were manually confirmed on the mapping track.
Daptomycin Minimal Inhibitory Concentration (MIC) with broth microdilution
The broth microdilution method was adapted from a published protocol [41]. Daptomycin (AG Scientific) was dissolved in ultrapure water and sterilized by filtration. The DAP stock solution was then further diluted in Brain Heart Infusion (BHI) broth (Acumedia, Neogen) supplemented with 50 mg/L Ca2+. Mid-log phase bacteria were diluted to 5 × 105 CFU/mL in a 96-well plate containing serial 2-fold dilutions of DAP or in the absence of DAP as a control. Technical triplicates were performed in each of 3 biological replicates. After overnight incubation at 37°C, wells were visually examined for presence of bacterial growth. The lowest DAP concentration with no visible bacterial growth was defined as the MIC. All incubations were static. The standard Mueller Hinton Broth (MHB) was not used because it significantly slows growth of E. faecalis OG1RF (data not shown).
Lipid extraction
Lipids were extracted from OG1RF cell pellets using a modified Bligh & Dyer method in which the extraction solvent contained chloroform/methanol in a ratio of 1:2 (v/v). For method validation, samples were spiked with known amounts of internal standards. 900 μl of chilled extraction solvent containing internal standards (purchased from Avanti polar lipids, Alabaster, AL, USA)) was added to the cell pellets (Table 1):
Table 1. Internal standards that were spiked into E. faecalis lipid extracts.
| Internal standard | Catalogue # | Stock conc. [mg/ml] | Final conc. [μg/ml] | Precursor ion (m1) m/z | Fragment ion (m3) m/z |
|---|---|---|---|---|---|
| PG 14:0 | 840445P | 1 | 5 | 665.5 [M-H]- | 153 |
| Lysyl PG 16:0 | 840520P | 0.1 | 4 | 849.6 [M-H]- | 145 |
| CL 14:0 | 750332P | 1 | 100 | 1239.9 [M-H]-, 619.5 [M-2H]2- | 153, 227 |
The suspension was vortexed for 15 seconds and incubated with agitation on a thermomixer at 1000 rpm at 4°C in the dark for 1 hour. Following a quick spin, 300 μl chilled chloroform and 250 μl chilled deionized water were added to the suspension, which was vortexed for 15 seconds. The suspension was centrifuged at 9,000 rpm for 2 min to achieve separation into aqueous and organic phases. The organic phase (containing lipid) was transferred into a clean 1.7 ml centrifuge tube. 500 μl of chilled chloroform was added to the remaining aqueous phase and residual lipids extracted by repeating the above steps. The first and second organic extracts were pooled together and were subsequently centrifuged in a vacuum concentrator (SpeedVac) until they were dry. The dried lipid extract was resuspended in a mixture of chloroform and methanol (1:1 v/v), to achieve a final lipid concentration of 10 mg/ml. This solution was stored at -80°C until the mass spectrometry analysis was performed.
Mass spectrometry
As a first approach, the lipid profile of E. Faecalis OG1RF was obtained through the use of high-resolution mass spectrometry. The OG1RF lipid extract was diluted 2:1 (v/v) with solvent containing isopropanol:methanol:chloroform (4:2:1, v/v) and 2 mM ammonium acetate. 10 μl of each sample was injected into an LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific, USA) using the Advion Nanomate interface. The nanospray voltage applied was −1.35kV in negative ionization mode. Mass spectra were acquired at a resolution of 30,000 and 100,000. Product ion scans (PIS) involving collision energies between 35 and 45 eV were used to identify α-alkyl chains.
Diacylglycerols (DAGs) and triacylglycerols (TAGs) analysis was undertaken by liquid chromatography coupled to mass spectrometry (LC-MS) with electrospray ionization (ESI) on an LTQ-Orbitrap XL (Thermo scientific, USA). A Zorbax Eclipse XDB-C18 column (3.0x150 mm, 1.8μm particle size, Agilent Technologies, USA) and a mobile phase consisting of a mixture of isopropanol:methanol:chloroform (4:2:1, v/v) and 2 mM ammonium acetate were used to separate lipids TAG from more polar lipids (DAG and glycerophospholipids). The separation was done isocratically at a flow rate of 130 μl/min. The total runtime was 30min. The LTQ-Orbitrap XL was operated with positive ionization at an electrospray voltage of 3.5 kV and mass-to-charge (m/z) range of 300–1400. Under these conditions, the TAG ionize as ammonium adducts and elute between 8 and 19min depending on chain length and unsaturation.
Phosphatidylglycerol (PG) and lysyl-phosphatidylglycerol (L-PG) in E. faecalis were quantified by LC-MSMS using multiple reaction monitoring (MRM). Lipid internal standards were added to the sample prior to lipid extraction. An Agilent 6460 QqQ mass spectrometer connected to a 1290 series chromatographic system was used. ESI was used to ionize lipids. Each lipid molecular species was analyzed using a targeted multiple reaction monitoring (MRM) approach containing transitions for known precursor/product mass-to-charge ratio (m1/m3) [42]. The experimental conditions and the MRM transitions required for lipid quantification are reported in the supplement.
Cardiolipins (CL) analysis was undertaken using the Agilent 6550 Q-ToF mass spectrometer connected to a 1200 series HPLC-Chip HILIC chip system. A customized HILIC-chip containing Amide-80 stationary phase (5 μm particle size, 80 Å pore size, Tosoh Bioscience, LLC. Montgomeryville, PA) was also prepared, along with a trapping column (160 nl) and an analytical column (75 μm × 150 mm) (Agilent Technologies Corp., Santa Clara CA). The solvents used for HPLC with the HILIC column were: 50% acetonitrile and 50% water containing 25 mM ammonium formate pH 4.6 (solvent A) and 95% acetonitrile and 50% water containing 25 mM ammonium formate pH 4.6. The pH value was adjusted with formic acid. Analytes spiked with internal standard were eluted with the following gradient: 100% solvent B from 0 to 1.5 minutes, 40% solvent B from 1.5 to 8.5 minutes, 30% solvent B from 8.5 to 10.5 minutes, 0% solvent B from 11.5 to 13.0 minutes, 100% solvent B from 13.1 to 19 minutes. The chip cube was operated with back flush mode. Each sample was injected through the enrichment column at a flow rate of 4 μl/min. 1.5 min after injection, the valve was switched to align the analytical column with the enrichment column at a flow rate of 400 nl/min. The QToF instrument was set to the negative ion mode, with an electrospray voltage of -1580 V (Vcap), a temperature of 185°C, a drying gas rate of 4 (or 12) l/min, and a fragmentor voltage of 150 V. Spectra were acquired in the targeted tandem MS mode. The MS acquisition rate was 2 spectra/sec and the tandem MS acquisition rate was 2 spectra/sec. Data are reported for 3–5 biological replicates.
Data processing
High-resolution MS spectra obtained on the LTQ-Orbitrap XL were processed using Xcalibur software (Thermo Fisher Scientific, USA). Lipid species were identified using an in-house database as well as online LipidMaps and Metlin lipid databases with an MS tolerance of 10 ppm. The identification was based on accurate mass and/or retention times (RT). Tandem MS data obtained on the Agilent 6550 Q-ToF were analyzed to obtain headgroup and/or fatty acid compositions and the RT for each lipid molecular species. For MRM data obtained on the Agilent 6460 QqQ, signal intensities were compared with the intensities from the spiked internal standards (PG 14:0 for PG and Lysyl-PG 16:0 for Lysyl-PG) and the retention times for the various classes were matched. Growth-phase MRM data were normalized to the dry cell pellet weights to ensure that comparisons were made per unit weight of bacterial sample. For the growth curve study, MRM peak areas for the early-logarithmic, mid-logarithmic and late-logarithmic phases were normalized to the peak areas for the late stationary phase to obtain a fold change. These fold changes were used to generate a heat map in R. The Student’s t test was performed to determine whether differences between OG1RF, Dap21 and Dap22 strains were statistically significant. p <0.05 was considered statistically significant.
Results
Identification of phospholipids: Phosphatidylglycerol (PG), Lysyl-PG (LPG) and Cardiolipin (CL)
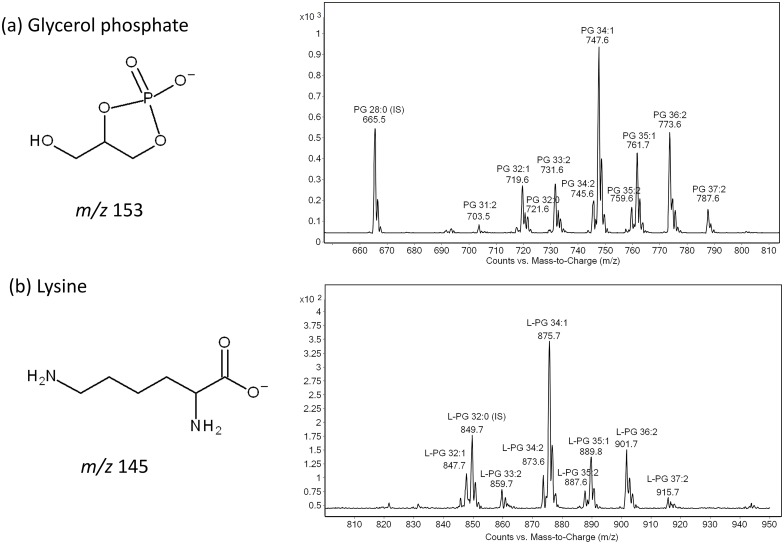
Anionic phosphatidylglycerol (PG) and cardiolipins (CL) are the major phospholipids in Gram-positive bacteria. In addition to PG and CL, Gram-negative bacteria contain zwitterionic phosphatidylethanolamine (PE) in abundance [13]. Based on this information, we performed a precursor ion scan (pre-IS) for the glycerol phosphate headgroup (m/z = 153) of PG on the Gram-positive bacterium, E. faecalis, and observed a number of peaks corresponding to PG (Fig 1a). The 665.5 peak corresponds to DMPG, the PG internal standard, whereas the endogenous E. faecalis peaks are seen at 703.5, 719.6, 721.6, 745.6, 747.6, 759.6, 761.6, 773.6, and 787.6.
Fig 1. Precursor ion scans for Enterococcus faecalis phospholipids.
Precursor ion scans for (A) 153 fragment ion (phosphatidylglycerol (PG))) and (B) 145 fragment ion (lysyl-phosphatidylglycerol (LPG)) reveal several species of PG (A) and of LPG (B). Each peak is labelled with the class (PG or LPG), fatty acid composition (e.g. 34:1), and the mass-to-charge (m/z) ratio (e.g. 747.6 for PG 34:1).
Since E. faecalis is known to produce the aminoacylated PG, lysyl-PG (LPG), we also performed a pre-IS for the lysine moiety (m/z = 145). In addition to the peak (849.7) corresponding to the LPG internal standard, lysyl-DPPG, we observed peaks at 847.7, 859.7, 875.7, 873.7, 887.6, 889.8, 901.7, and 915.7 (Fig 1b). Based on these data, we conclude that only a subset of PGs, i.e. PG 32:1, 33:2, 34:1, 34:2, 35:1, 35:2, 36:2 and 37:2, serve as substrates for the MprF enzyme that trans-esterifies amino acids to PG [43–45].
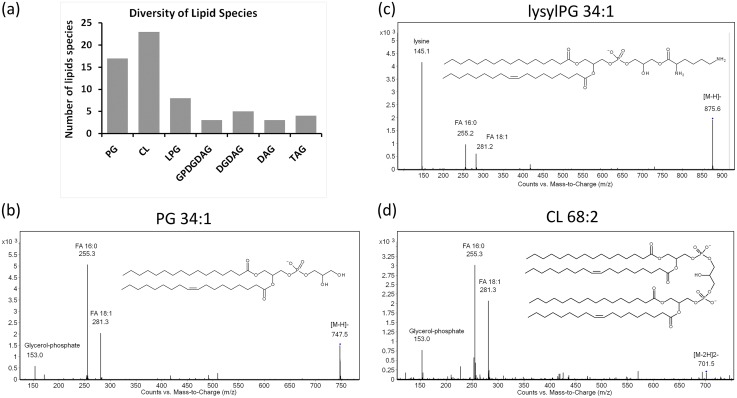
For a more detailed characterization of phosphatidylglycerol (PG) species in E. faecalis, we performed LC-MS on membrane lipids to achieve separation of lipid species according to class followed by species identification. and observed eight additional PGs, twenty three cardiolipins (CLs), three glycerophospho-diglucosyl-diacylglycerols (GPDGDAGs), five diglucosyl-diacylglycerols (DGDAGs), four diacylglycerols (DAGs), and four triacylglycerols (TAGs) (Fig 2a and S1–S3 Tables). Lipids of a given class (e.g. PG or LPG) differ from each other in the number of carbon atoms per fatty acid chain and the number of carbon-to-carbon double bonds in unsaturated chains. For example, PG 34:1 contains 34 carbon atoms in its two fatty acyl chains and one of these chains contains a single double bond (Fig 2b). Fig 2c shows the structure of LPG 34:1 which contains the same number of fatty acyl carbon atoms and double bonds as PG 34:1, but is conjugated to a lysine amino acid, which is characteristic of the LPG class. The structure of CL 68:2, shown in Fig 2d, is representative of the cardiolipin or di-PG class of phospholipids and contains twice as many carbon atoms and double bonds as PG 34:1.
Fig 2. The diversity of lipid species in E. faecalis strain OG1RF.
(a) The total number of lipid species identified is 63 across 7 classes. Representative MSMS spectra for the phospholipids (PG, lysyl-PG, and cardiolipin (CL)) are shown for (b) PG 34:1, (c) lysyl-PG 34:1, and (d) CL 68:2. Each species is identified based upon its characteristic fragmentation pattern: MSMS of (b) PG 34:1 gives rise the glycerol-phosphate headgroup (m/z = 153) and fatty acids 16:0 (m/z = 255.3) and 18:1 (m/z = 281.3); (c) lysyl-PG 34:1 gives rise to lysine (m/z = 145) and fatty acids 16:0 and 18:1; and CL (68:2) gives rise to the glycerol-phosphate headgroup and fatty acids 16:0 and 18:1.
The CL species appeared predominantly as [M-2H]2- ions (Fig 2d and S2 Table). The ionization of CL is more complex than that of PG because of CL’s larger structure [46]. The fact that the [M-2H]2- ions produce a characteristic signature within the MS spectrum in which the isotopic peaks are separated by a mass-to-charge (m/z) ratio of 0.5 [47] allowed us to identify CLs here (S1b Table). The number of carbon atoms in CL fatty acyl chains ranges from 64 to 70, and the degree of unsaturation ranges from 1 to 4 double bonds. Thus we see that the carbon atom number and degree of unsaturation of CLs is roughly twice that of PGs, which is consistent with PG being the substrate of cardiolipin synthase (Cls) 1 or 2.
Finally, using the same methodology, we detected three species of the phospholipid glycerophospho-diglucosyl-diacylglycerol (GPDGDAG) (Fig 2a and S1 Table). Tandem MS data from a previous study on GPDGDAG in Streptococcus mutans [48] guided us in the identification of three species of GPDGDAG in E. faecalis OG1RF. Although it is not a major bilayer-forming lipid, higher levels of GPDGDAG were nonetheless associated with daptomycin resistance in E. faecalis and E. faecium [3].
Identification of glycerolipids: Diacylglycerol (DAG) and Triacylglycerol (TAG) species
The synthesis of lipoteichoic acids (LTA) and membrane-derived oligosaccharides in bacteria leads to the production of diacylglycerols (DAGs) as secondary products [49]. DAG is a neutral lipid whose accumulation in the membrane leads to bilayer disruption [50]. For this reason, it is metabolized to form phosphatidic acid and other phospholipids [9, 49]. Little has been reported about bacterial triacylglycerol (TAG) [51], but its main function is as a reservoir of fatty acids [49]. TAG can also regulate membrane fluidity and serve as an electron sink [9]. We have identified three species of diacylglycerol (DAG) and four species of TAG in E. faecalis (S3 Table). As DAG and TAG do not ionize well on their own, a charged group or adduct needs to be attached to the parent DAG or TAG molecule. Therefore, for all species, we detected the positive ammonium adduct—[M+NH4]+ (S1 Fig and S3 Table). In addition, we have identified five species of diglucosyl-diacylglycerol (DGDAG) (Fig 2 and S1 Table). The DGDAG species were detected as negative formate ([M+HCOO]-) adducts.
Tracking PG and lysyl-PG levels during in vitro growth
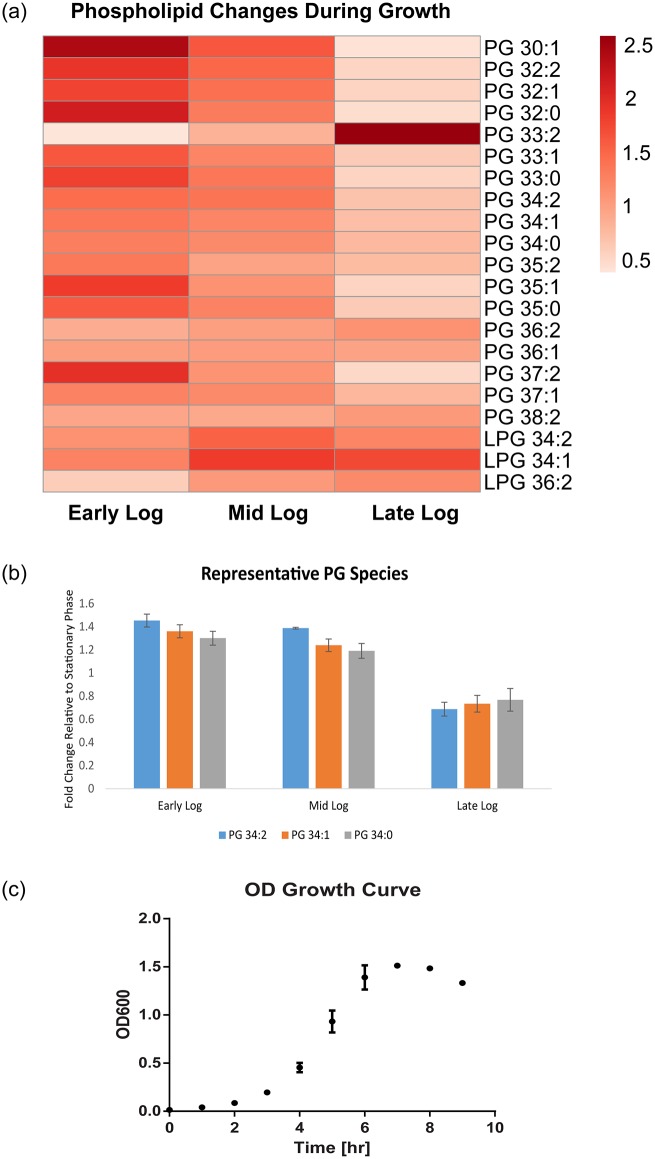
The quantification of E. faecalis phospholipids to obtain lipid concentrations via the mass spectrometry (MS) scanning mode known as multiple reaction monitoring (MRM) has not been previously reported. Earlier reports of enterococcal phospholipid quantification relied on spectrophotometric methods applied to excised TLC plate spots [3, 6]. Here, we have used MRM for direct quantification and tracked the levels of seventeen PG species during the in vitro growth of E. faecalis. We performed MRM on samples extracted during the exponential (early-logarithmic, mid-logarithmic, late-logarithmic) (Fig 3a) and late stationary (overnight) growth phases and quantified PG and lysyl-PG levels. In MRM, the m/z values of both the precursor and fragment ions are specified. Quadrupole Q1 isolates precursor ions, quadrupole Q2 fragments the precursor ions, and quadrupole Q3 allows selected fragment ions to pass through to the detector, which measures the abundance of each fragment [52]. The m/z of the precursor ion and the m/z of the fragment ion constitute one “MRM transition”, and each transition gives rise to a peak on the ion chromatogram. MRM transitions for E. faecalis PGs and LPGs are listed in S4 Table. Integrating a peak gives the peak area, which is proportional to the abundance of a single lipid species (e.g., PG 34:1). Dividing each peak area by the peak area of the internal standard gives a lipid concentration (μg/ml), and further dividing by the dry weight of the cell pellet gives a normalized lipid concentration. We further divided each normalized lipid concentration from early-log, mid-log or late-log phase by the corresponding late stationary normalized lipid concentration to obtain a fold change. We used these fold change values to generate the heat map in Fig 3a. Using this analysis protocol, we uncovered measurable differences in the levels of PGs and LPGs at different growth stages (Fig 3a). The short-chain PGs 30:1, 32:2, 32:1 and 32:0 follow similar trends in that they are abundant in early log phase and decrease through mid log phase to late log phase. PG 33:2 levels were unlike the other PGs in that it was low in early log phase and very high in late log phase. This stark difference between PG 33:2 and the more saturated PG 33:1 and 33:0 suggests that the two double bonds in PG 33:2 may be important for growth. Levels of PG 34:0, 34:1 and 34:2, which are also depicted in graphical form in Fig 3b, decreased throughout the exponential growth curve. There are slight differences in the late-log phase concentrations of PG 35:0, 35:1 and 35:2, but they otherwise also show a downward trend from early to late log phase. PG 36:1, PG 36:2 and PG 38:2 levels remain almost constant throughout the growth curve. The trend for PG 37:2 almost mirrors that of PG 33:2:PG 37:2 is highest in early log phase and lowest in late log phase. PG 37:1, which has one fewer double bond than PG 37:2, remains constant in early and mid log phase but decreases slightly in late log phase. The overall trend for PGs is that their amounts decrease during exponential growth to a certain minimum level in late-log phase, and then increase to a level above this minimum as they progress into stationary phase. The overall trend for the LPGs is opposite to that of the PGs. LPG levels tend to increase throughout the growth curve, suggesting that E. faecalis may fortify its membrane with positively-charged phospholipids as it propagates. As the bacteria divide exponentially, they may need to maintain a certain optimum ratio of anionic PG and cationic LPG in order to ensure that various membrane-dependent processes continue to function optimally under the prevailing growth conditions.
Fig 3. The levels of E. faecalis strain OG1RF PG and LPG can be tracked throughout the growth curve.
(A) Heat map depicting phosphatidylglycerol (PG) and lysyl-PG levels in the early-, middle- and late-logarithmic growth phases relative to the late stationary phase. The colour scale (top right) indicates fold change of phospholipids (PG and lysyl-PG) in early-, mid-, and late-logarithmic phase cells with respect to late stationary phase cells. (B) Bar chart showing representative fold change values for PG 34:0, 34:1 and 34:2. (C) The three logarithmic phases corresponded to optical density (OD) values of 0.3, 0.5 and 0.8 on an OD growth curve.
Phospholipid levels in daptomycin-resistant strains
In order to study the lipid profiles of antibiotic-resistant E. faecalis strains, we generated two daptomycin-resistant (DapR) strains via in vitro evolution from the DAP-sensitive OG1RF strain. We performed whole genome sequencing to identify the single nucleotide polymorphisms (SNPs) in these DapR strains (Table 2). Dap21 harbours mutations in three different genes: cardiolipin synthase 1 (cls1; RF10364), a gene encoding a putative chaperone protein (RF11464), and a gene encoding hypothetical membrane protein (RF11507). Dap22 harbours mutations in the same chaperone- and membrane protein-encoding genes that are mutated in Dap21, as well as mutations in cardiolipin synthase 2 (cls2; RF11324) and a putative metal-dependent HD-domain-containing hydrolase (RF11901).
Table 2. Single Nucleotide Polymorphisms (SNPs) in the DapR strains Dap21 and Dap22.
| Strain | Gene/locus | |||||
|---|---|---|---|---|---|---|
| cls1 (OG1RF_10364) | cls2 (OG1RF_11324) | OG1RF_11464 | OG1RF_11507 | OG1RF_11901 | ||
| Dap21 | Mutation type | Substitution | - | Frameshift | Frameshift | - |
| Mutation location | R267C | - | 272 | 19 | - | |
| Dap22 | Mutation type | - | Substitution | Frameshift | Nonsense | Inframe insertion |
| Mutation location | - | P181H | 272 | L4 | 84TARA85 | |
The minimum inhibitory concentrations (MICs) of DAP for both of these DapR strains are significantly higher than the MIC for OG1RF (Table 3).
Table 3. Minimum Inhibitory Concentrations (MICs) of daptomycin for three E. faecalis strains.
| Strain | MIC [μg/ml] |
|---|---|
| OG1RF | 1–2 |
| Dap21 | 128 |
| Dap22 | 128 |
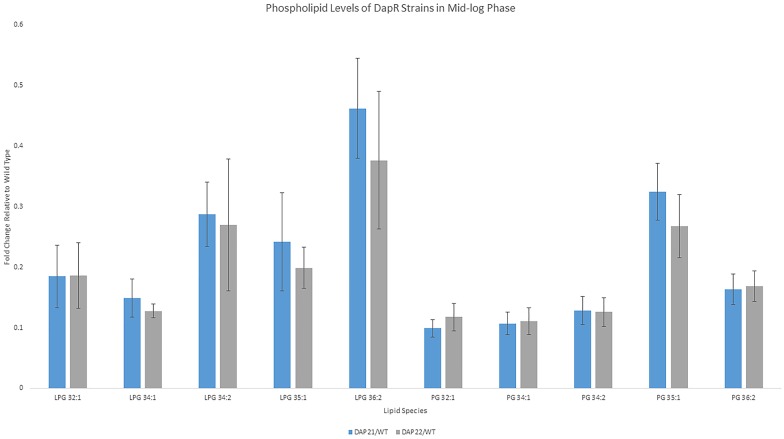
We quantified PG and LPG levels in Dap21 and Dap22, as compared to wild type, and observed that that at mid-log phase (OD = 0.5), levels of 5 LPGs and the corresponding PGs were lower in both DapR strains than in OG1RF (Fig 4). The differences in PG and LPG levels between OG1RF and Dap21, and between OG1RF and Dap22, are all statistically significant, with all p values being less than 0.05. The phospholipid profiles of Dap21 and Dap22 do not differ significantly from each other another, however, suggesting that the emergence of DAP resistance is associated with an overall decrease in LPG and PG levels.
Fig 4. The levels of PG and LPG in two daptomycin-resistant strains, Dap21 to Dap22, compared to non-resistant OG1RF.
The bar chart shows the fold change of 5 LPGs and their corresponding PGs from Dap21 and Dap22 grown to mid-logarithmic phase, with respect to the non-resistant OG1RF parental strain. Data are expressed as mean ± standard deviation (SD) (calculated from 5 replicates) with the SDs represented as error bars.
Discussion & conclusion
There is great diversity in lipid composition across bacterial species, and environmental conditions can affect the lipid composition of a single species [10]. Membrane lipid homeostasis and adaptation to changing environmental conditions are essential for bacterial survival. Variations may occur in response to temperature, pH, osmolarity, salinity and the presence of chemical compounds [8]. In response to these changes, bacteria modify membrane-forming phospholipids by altering specific properties of their fatty acyl chains, e.g. changing the degree of unsaturation (i.e. the number of double bonds), adopting either a cis or trans configuration for unsaturated chains, adjusting the ratio of iso to anteiso chains, or introducing a cyclopropane ring [8]. Lipid changes affect membrane proteins whose functions can be affected in two main ways: 1) by the chemical properties of lipids, e.g. length of fatty acid chains, number of carbon-carbon double bonds, nature of the head group, net charge and chelation; 2) by the ability of lipids to associate with themselves as a result of bulk properties, e.g. molecular shape, viscosity/fluidity, molecular packing and thickness of the lipid bilayer [53]. Hence, to fully appreciate lipid-protein interactions in the membrane, we need information about lipids as well. Lipidomics is thus essential to understanding bacterial physiology.
Besides these physiological roles for lipids, changes in bacterial lipid composition have been correlated with antibiotic resistance. In 1970, Dunnick and O’Leary showed that for Gram-negative bacterial strains resistant to polymyxin, tetracyclines and penicillin, the proportion of fatty acids containing cyclopropane was lower than that in sensitive strains [54]. More recently, a drop in PG levels was correlated with DAP resistance in E. faecalis and E. faecium [3]. In E. faecalis, DAP resistance was also related to mutations in genes involved in lipid metabolism, namely, glycerophosphoryldiester-phosphodiesterase (gdpD), cardiolipin synthetase (cls). Multiple peptide resistance factor (mprF) in Staphylococcus aureus and enterococci has also been associated with DAP-resistance [5, 40, 55–58]. The comprehensive lipidomic analysis that we report here reveals that DAP resistance in E. faecalis is accompanied by a decrease in both lysyl-PG and PG levels in strains that harbour a single point mutation in a cardiolipin synthase. Since DAP is known to bind to PG, we would expect lower PG levels to confer resistance to DAP, and our results support this notion. On the other hand, the reduction in lysyl-PG is counterintuitive since this cationic phosphospholipid would help repel cationic DAP from the cell membrane. However, E. faecalis strains harbouring deletions in genes encoding multiple peptide resistance factor (mprF), the enzyme that synthesizes LPG, show a minimal change in the DAP MIC and suggesting that lysyl-PG alone does not determine DAP susceptibility [32].
The compartmentalization of bacterial lipids into discrete membrane domains is also of growing interest and is likely to depend on both the chemical and bulk properties of lipids [59, 60]. These domains include PG domains, CL domains, and lipid rafts [12–15, 35, 61]. These domains coordinate a range of processes, including growth, cell division, and antibiotic resistance [11, 12, 17]. One example of a domain-specific process is the secretion and biogenesis of cell-surface proteins [18, 62]. In E. faecalis, these proteins include virulence factors including pili, which are secreted via the general secretion (Sec) machinery, assembled by sortase C, and attached by sortase A to the cell wall within focal domains [18, 22]. Extracellular secretion is essential to bacterial pathogenesis because it allows bacteria to release virulence factors and toxins into the host [63]. In E. faecalis, virulence factors that are assembled in distinct domains include pili and aggregation substance, both of which contribute to disease progression [64–67]. In B. subtilis, SecA becomes delocalized when anionic PG are depleted [19], while in S. pyogenes, the focal localization of a protease called HtrA is physically linked to domains containing anionic lipids [20]. In B. subtilis, and S. aureus, lipid rafts play roles in sporulation, cell division, secretion and biofilm formation, and can also serve as drug targets [68–71]. To better understand the role of lipids in the physiology, pathogenesis and antibiotic resistance of E. faecalis, we must begin with a detailed characterization of its lipids. The methods we report here could be used to study the lipid composition of E. faecalis membrane domains. Since DAP is known to bind focally to E. faecalis [6], MRM-based quantification of phospholipids within these target domains would reveal what quantities of phospholipids confer resistance to DAP.
Previous studies characterizing the lipids of E. faecalis were performed by thin layer chromatography and subsequent MS of lipid spots, and revealed several classes of lipid molecules including phosphatidylglycerol (PG), cardiolipins (CL), glycerophospho-diglucosyl-diacylglycerol (GPDGDAG), and aminoacyl-PG [6, 43, 72, 73]. Here, using liquid-chromatography mass spectrometry (LC-MS), we have detected previously described lipids, i.e. PG, LPG, GPDGDAG, and identified previously unidentified E. faecalis lipids including DGDAG, DAG and TAG. Previous reports on E. faecalis lipids are summarized in Table 4 and are compared against the findings achieved in the present study. The methodology we report here provides high selectivity, sensitivity and accuracy and enabled us to identify several PG and CL species in E. faecalis strain OG1RF by accurate mass determination and tandem MS fragmentation analysis. We provide the most detailed analysis of E. faecalis lipids performed to date.
Table 4. Previous biochemical analyses of Enterococcus faecalis compared to the present study.
| Bao et al. (2012) | Mishra et al. (2012) | Tran et al. (2013) | Fozo et al. (2014) | This Study | |
|---|---|---|---|---|---|
| Growth phase | Late stationary | Late stationary | Late stationary | Late stationary | Exponential, Late stationary |
| GPL | |||||
| PG | + | + | + | - | + |
| CL | + | + | + | - | + |
| Lys-PG | - | + | + | - | + |
| Glycerolipids | |||||
| DAG | - | - | - | - | + |
| TAG | - | - | - | - | + |
| DGDAG | + | - | - | - | + |
| GPDGDAG | - | + | + | - | + |
| Fatty acids | - | + | - | + | + (acyl chains by MSMS) |
| Methodology | TLC & molybdenum, ninhydrin staining | TLC, GC-FAME & ESI-MSMS | TLC, ESI-MSMS | GC-FAME | TLC, ESI-MSMS |
| Resolution of molecular species | - | - | - | - | + |
| Quantification | - | - | - | - | + |
| Number of molecular species quantified by MRM | 0 | 0 | 0 | 0 | 20 |
In bacteria, membrane lipid biosynthesis begins with fatty acid synthesis via the type II fatty acid biosynthetic pathway (FASII), and is followed by the transfer of fatty acid chains to glycerol-3-phosphate in the membrane to form phosphatidic acid (PA). PA is the common precursor to PG, LPG and CL [8]. GPDGDAG is one of the glycolipid precursors of lipoteichoic acid (LTA), a component of the Gram-positive cell envelope [3]. Another component of the Gram-positive cell envelope is DGDAG, which is formed when a uridine diphosphate sugar reacts with DAG [74]. Both DGDAG and MGDAG serve as membrane anchors for LTA in the cell envelope of many Gram-positive bacteria [75]. DAG is formed during phospholipid turnover, while TAG is formed from DAG by the enzyme diacylglycerol acyltransferase [8, 51]. We quantified the PG and LPG species by multiple reaction monitoring (MRM). The lipid GPDGDAG is important because daptomycin-resistant strains of E. faecium and E. faecalis were reported to contain a higher GPDGDAG concentration [3]. In a daptomycin-resistant (DapR) clinical isolate of E. faecalis, CL was re-distributed away from the septum but total CL levels remain unchanged [6]. The same DapR isolate had lower levels of PG [3]. LPG confers resistance to E. faecalis against the antimicrobial peptide human β-defensin 2 (hBD2) [22, 32]. In contrast to previous studies on DapR E. faecalis strains that showed LPG levels were unchanged, highly sensitive MRM analysis used here reveals that LPG levels are lower in the two DapR strains that we generated from a susceptible parental strain via in vitro evolution. One result of decreased LPG synthesis might be that more of the PG substrate can be consumed in an alternative pathway, e.g CL and GPDGDAG biosynthesis.
Mass spectrometric identification of cardiolipin has been challenging due to its complex structure and complex ionization [46]. The cardiolipin structure is complex because it contains a polyglycerophosphate backbone connected to four acyl chains [76], which is twice as large as a PG molecule with only two acyl chains connected to one glycerophosphate backbone. Historically, CLs as a class were studied using radiochemical labelling [77] or another method that coupled high-performance liquid chromatography (HPLC) to fluorescence detection [78]. Fatty acid analysis of the total CL pool was then carried out [79, 80]. Since individual CL molecular species were not directly analyzed, these methods could not determine the number of different CL species present. Despite the technical challenges, we were able to identify the [M-2H]2- ions of CL species by LC-MS. CLs have been reported to predominantly form [M-2H]2- ions [81] and we thus searched for these doubly charged ions in our own spectra, where we observed a [M-2H]2- ion “signature”. CL mass spectrometry studies are ongoing to develop optimum assays for CL quantification by multiple reaction monitoring (MRM).
Using MRM quantification of PG and LPG levels, we tracked changes in the levels of these species during in vitro growth of E. faecalis. Previously, the levels of selected PG and PE species were monitored by MS in the Gram-positive B. subtilis and shown to remain constant during exponential growth. During stationary growth, the PG level decreased while the PE level increased. Neither the LPGs nor the glycolipids changed significantly during growth [82]. We show that the levels of different PGs change during logarithmic growth and entry into stationary phase. These PGs differ from each other both in terms of carbon chain length and the number of carbon-to-carbon double bonds, and thus our analysis reveals that E. faecalis favors PGs with specific chemical properties at different growth phases. Since higher LPG levels have been correlated with increased resistance to cationic antimicrobial peptides, the higher LPG levels observed in mid and late log phase E. faecalis may suggest that these growth phases may be most resistant to cationic antimicrobial peptides. Based on our DapR data, it is plausible that resistance to cationic antimicrobials may be further enhanced by a decrease in PG levels. E. faecalis might thus use a concomitant increase in LPG and decrease in PG as a strategy to resist CAMPs and daptomycin.
In this study, we have used a combination of mass spectrometry techniques to identify phospholipids and glycerolipids and we now have a detailed lipid profile for this organism. With the quantification of PG and LPG in hand, future studies are now possible to study lipid level dynamics under a variety of environmental conditions as well as exposure to antimicrobials. Thus, lipidomics has promising applications in the areas of microbiology, infectious diseases, and biofilms. We expect that lipidomics will play a pivotal role in the development of new and effective therapies against E. faecalis and other human pathogens.
Supporting information
Each peak is labelled with the class (DAG or TAG), fatty acid composition (e.g. 34:1), and the mass-to-charge (m/z) ratio (e.g. 612.5554 for DAG 34:1). The spectrum was acquired in positive ionization mode.
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
R.R., I.H.G., Z.J.N., and K.A.K. were supported by the National Research Foundation and Ministry of Education Singapore under its Research Centre of Excellence Programme and by the National Research Foundation under its Singapore NRF Fellowship programme awarded to K.A.K. (NRF-NRFF2011-11). A.C.G., J.K.K., L.G. and M.R.W. were supported by grants from the National University of Singapore via the Life Sciences Institute (LSI), the National Research Foundation (NRFI2015-05) and a BMRC-SERC joint grant (BMRC-SERC 112 148 0006) from the Agency for Science, Technology and Research (A*Star).
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Authors RR, IHG, ZJN, and KAK were supported by the National Research Foundation and Ministry of Education Singapore under its Research Centre of Excellence Programme and by the National Research Foundation under its Singapore NRF Fellowship programme awarded to KAK (NRF-NRFF2011-11). ACG, JKK, LG, and MRW were supported by grants from the National University of Singapore via the Life Sciences Institute (LSI), the National Research Foundation (NRFI2015-05), and a BMRC-SERC joint grant (BMRC-SERC 112 148 0006) from the Agency for Science, Technology and Research (A*Star). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Wilson B, Salyers A, Whitt D, Winkler M. Bacterial pathogenesis: A molecular approach. Washington, DC, USA: ASM Press; 2011. [Google Scholar]
- 2.Römling U, Balsalobre C. Biofilm infections, their resilience to therapy and innovative treatment strategies. J Intern Med. 2012;272(6):541–61. 10.1111/joim.12004 [DOI] [PubMed] [Google Scholar]
- 3.Mishra N, Bayer A, Tran T, Shamoo Y, Mileykovskaya E, Dowhan W, et al. Daptomycin resistance in enterococci is associated with distinct alterations of cell membrane phospholipid content. PLoS One. 2012;7(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diaz L, Tran T, Munita J, Miller W, Rincon S, Carvajal L, et al. Whole-genome analyses of Enterococcus faecium isolates with diverse daptomycin MICs. Antimicrob Agents Chemother. 2014;58(8):4527–34. 10.1128/AAC.02686-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arias C, Panesso D, McGrath D, Qin X, Mojica M, Miller C, et al. Genetic basis for in vivo daptomycin resistance in enterococci. N Engl J Med. 2011;365(10):892–900. 10.1056/NEJMoa1011138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tran T, Panesso D, Mishra N, Mileykovskaya E, Guan Z, Munita J, et al. Daptomycin-resistant Enterococcus faecalis diverts the antibiotic molecule from the division septum and remodels cell membrane phospholipids. MBio. 2013;4(4):e00281–13. 10.1128/mBio.00281-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wenk M. Lipidomics of host-pathogen interactions. FEBS Letters. 2006;580(5541–5551):5541–51. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y, Rock C. Membrane lipid homeostasis in bacteria. Nature Reviews Microbiology. 2008;6:222–33. 10.1038/nrmicro1839 [DOI] [PubMed] [Google Scholar]
- 9.Parsons J, Rock C. Bacterial lipids: metabolism and membrane homeostasis. Prog Lipid Res. 2013;52(3):249–76. 10.1016/j.plipres.2013.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sohlenkamp C, Geiger O. Bacterial membrane lipids: diversity in structures and pathways. FEMS Microbiology Reviews. 2015. Epub 2015 Apr 9. [DOI] [PubMed] [Google Scholar]
- 11.Barák I, Muchová K. The Role of Lipid Domains in Bacterial Cell Processes. Int J Mol Sci. 2013;14(2):4050–65. 10.3390/ijms14024050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bramkamp M, Lopez D. Exploring the existence of lipid rafts in bacteria. Microbiol Mol Biol Rev. 2015;79(1):81–100. 10.1128/MMBR.00036-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Epand R, Epand R. Domains in bacterial membranes and the action of antimicrobial agents. Mol Biosyst. 2009;5(6):580–7. 10.1039/b900278m [DOI] [PubMed] [Google Scholar]
- 14.Epand R, Epand R. Lipid domains in bacterial membranes and the action of antimicrobial agents. Biochim Biophys Acta. 2009;1788(1):289–94. 10.1016/j.bbamem.2008.08.023 [DOI] [PubMed] [Google Scholar]
- 15.Farnoud A, Toledo A, Konopka J, Del Poeta M, London E. Raft-like membrane domains in pathogenic microorganisms. Curr Top Membr. 75 2015. p. 233–68. 10.1016/bs.ctm.2015.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.White D, Drummond J, Fuqua C. The Physiology and Biochemistry of Prokaryotes. Fourth ed: Oxford; 2012. [Google Scholar]
- 17.Govindarajan S, Nevo-Dinur K, Amster-Choder O. Compartmentalization and spatiotemporal organization of macromolecules in bacteria. FEMS Microbiology Reviews. 2012;36(5):1005–22. 10.1111/j.1574-6976.2012.00348.x [DOI] [PubMed] [Google Scholar]
- 18.Kline K, Kau A, Chen S, Lim A, Pinkner J, Rosch J, et al. Mechanism for sortase localization and the role of sortase localization in efficient pilus assembly in Enterococcus faecalis. Journal of Bacteriology. 2009;191(10):3237–47. 10.1128/JB.01837-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Campo N, Tjalsma H, Buist G, Stepniak D, Meijer M, Veenhuis M, et al. Subcellular sites for bacterial protein export. Mol Microbiol. 2004;53(6):1583–99. 10.1111/j.1365-2958.2004.04278.x [DOI] [PubMed] [Google Scholar]
- 20.Rosch J, Hsu F, Caparon M. Anionic lipids enriched at the ExPortal of Streptococcus pyogenes. J Bacteriol. 2007;189(3):801–06. 10.1128/JB.01549-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.López D, Kolter R. Functional microdomains in bacterial membranes. Genes Dev. 2010;24(17):1893–902. 10.1101/gad.1945010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kandaswamy K, Liew T, Wang C, Huston-Warren E, Meyer-Hoffert U, Hultenby K, et al. Focal targeting by human β-defensin 2 disrupts localized virulence factor assembly sites in Enterococcus faecalis. Proc Natl Acad Sci U S A. 2013;110:20230–35. 10.1073/pnas.1319066110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsui H, Keen S, Sham L, Wayne K, Winkler M. Dynamic distribution of the SecA and SecY translocase subunits and septal localization of the HtrA surface chaperone/protease during Streptococcus pneumoniae D39 cell division. MBio. 2011;2(5):e00202–11. 10.1128/mBio.00202-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Foss M, Eun Y, Weibel D. Chemical−Biological Studies of Subcellular Organization in Bacteria. Biochemistry. 2011;50:7719–34. 10.1021/bi200940d [DOI] [PubMed] [Google Scholar]
- 25.Mileykovskaya E, Dowhan W. Role of membrane lipids in bacterial division-site selection. Curr Opin Microbiol. 2005;8:135–42. 10.1016/j.mib.2005.02.012 [DOI] [PubMed] [Google Scholar]
- 26.Epand R, Epand R. Domains in bacterial membranes and the action of antimicrobial agents. Molecular BioSystems. 2009;5:580–7. 10.1039/b900278m [DOI] [PubMed] [Google Scholar]
- 27.Martinez M, Schujman G, Gramajo H, de Mendoza D. Regulation of Membrane Lipid Homeostasis in Bacteria In: Timmis K, editor. Handbook of Hydrocarbon and Lipid Microbiology. Berlin; Heidelberg: Springer-Verlag; 2010. p. 510–17. [Google Scholar]
- 28.Heath R, Jackowski S, Rock C. Fatty acid and phospholipid metabolism in prokaryotes In: Vance D, Vance J, editors. Biochemistry of Lipids, Lipoproteins and Membranes. 5th ed Amsterdam; Boston: Elsevier; 2002. p. 59–96. [Google Scholar]
- 29.Aguilar P, Hernandez-Arriaga A, Cybulski L, Erazo A, de Mendoza D. Molecular basis of thermosensing: a two-component signal transduction thermometer in Bacillus subtilis. EMBO J. 2001;20(7):1681–91. 10.1093/emboj/20.7.1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saito H, Harp J, Fozo E. Incorporation of exogenous fatty acids protects Enterococcus faecalis from membrane-damaging agents. Appl Environ Microbiol. 2014;80(20):6527–38. 10.1128/AEM.02044-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harp J, Saito H, Bourdon A, Reyes J, Arias C, Campagna S, et al. Exogenous Fatty Acids Protect Enterococcus faecalis from Daptomycin-Induced Membrane Stress Independently of the Response Regulator LiaR. Appl Environ Microbiol. 2016;82(14):4410–20. 10.1128/AEM.00933-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bao Y, Sakinc T, Laverde D, Wobser D, Benachour A, Theilacker C, et al. Role of mprF1 and mprF2 in the pathogenicity of Enterococcus faecalis. PLoS One. 2012;7(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ernst C, Staubitz P, Mishra N, Yang S, Hornig G, Kalbacher H, et al. The bacterial defensin resistance protein MprF consists of separable domains for lipid lysinylation and antimicrobial peptide repulsion. PLoS Pathog. 2009;5(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geiger O, González-Silva N, López-Lara I, Sohlenkamp C. Amino acid-containing membrane lipids in bacteria. Prog Lipid Res. 2010;49(1):46–60. 10.1016/j.plipres.2009.08.002 [DOI] [PubMed] [Google Scholar]
- 35.Mileykovskaya E, Dowhan W. Cardiolipin membrane domains in prokaryotes and eukaryotes. Biochim Biophys Acta. 2009;1788(10):2084–91. 10.1016/j.bbamem.2009.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawai F, Shoda M, Harashima R, Sadaie Y, Hara H, Matsumoto K. Cardiolipin domains in Bacillus subtilis marburg membranes. Journal of Bacteriology. 2004;186(5):1475–83. 10.1128/JB.186.5.1475-1483.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mileykovskaya E, Dowhan W. Visualization of phospholipid domains in Escherichia coli by using the cardiolipin-specific fluorescent dye 10-N-nonyl acridine orange. J Bacteriol. 2000;182(4):1172–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dowhan W, Mileykovskaya E, Bogdanov M. Diversity and versatility of lipid-protein interactions revealed by molecular genetic approaches. Biochim Biophys Acta. 2004;1666(1–2):19–39. 10.1016/j.bbamem.2004.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dunny G, Brown B, Clewell DB. Induced cell aggregation and mating in Streptococcus faecalis: Evidence for a bacterial sex pheromone. Proc Natl Acad Sci U S A. 1978;75(7):3479–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palmer K, Daniel A, Hardy C, Silverman J, Gilmore M. Genetic basis for daptomycin resistance in enterococci. Antimicrob Agents Chemother. 2011;55(7):3345–56. 10.1128/AAC.00207-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wiegand I, Hilpert K, Hancock R. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc. 2008;3(2):163–75. 10.1038/nprot.2007.521 [DOI] [PubMed] [Google Scholar]
- 42.Shui G, Bendt A, Pethe K, Dick T, Wenk M. Sensitive profiling of chemically diverse bioactive lipids. J Lipid Res. 2007;48(9):1976–84. 10.1194/jlr.M700060-JLR200 [DOI] [PubMed] [Google Scholar]
- 43.Bao Y, Sakinc T, Laverde D, Wobser D, Benachour A, Theilacker C, et al. Role of mprF1 and mprF2 in the pathogenicity of Enterococcus faecalis. PLoS One. 2012;7(6):e38458 10.1371/journal.pone.0038458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ernst C, Staubitz P, Mishra N, Yang S, Hornig G, Kalbacher H, et al. The bacterial defensin resistance protein MprF consists of separable domains for lipid lysinylation and antimicrobial peptide repulsion. PLoS One. 2009;5(11):e1000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roy H, Ibba M. Broad range amino acid specificity of RNA-dependent lipid remodeling by multiple peptide resistance factors. J Biol Chem. 2009;284(43):29677–83. 10.1074/jbc.M109.046367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garrett T, Kordestani R, Raetz C. Quantification of cardiolipin by liquid chromatography-electrospray ionization mass spectrometry. Methods Enzymol. 2007;433:213–30. 10.1016/S0076-6879(07)33012-7 [DOI] [PubMed] [Google Scholar]
- 47.Garrett T, Kordestani R, Raetz C. Quantification of cardiolipin by liquid chromatography-electrospray ionization mass spectrometry. Methods Enzymol. 2007;433:213–30. 10.1016/S0076-6879(07)33012-7 [DOI] [PubMed] [Google Scholar]
- 48.Sallans L, Giner J, Kiemle D, Custer J, Kaneshiro E. Structural identities of four glycosylated lipids in the oral bacterium Streptococcus mutans UA159. Biochim Biophys Acta. 2013;1831(7):1239–49. [PubMed] [Google Scholar]
- 49.Christie W. AOCS Lipid Library 2011 [cited 2015 October 26]. http://lipidlibrary.aocs.org/.
- 50.Takahashi H, Watanabe M, Quinn P, Kato S, Murayama S, Ohki K, et al. Effects of diacylglycerol on the structure and phase behaviour of non-bilayer forming phospholipid. Biophy Chem. 1999;77(1):37–48. [DOI] [PubMed] [Google Scholar]
- 51.Alvarez H, Steinbüchel A. Triacylglycerols in prokaryotic microorganisms. Appl Microbiol Biotechnol. 2002;60(4):367–76. 10.1007/s00253-002-1135-0 [DOI] [PubMed] [Google Scholar]
- 52.Jung H, Sylvänne T, Koistinen K, Tarasov K, Kauhanen D, Ekroos K. High throughput quantitative molecular lipidomics. Biochim Biophys Acta. 2011;1811(11):925–34. 10.1016/j.bbalip.2011.06.025 [DOI] [PubMed] [Google Scholar]
- 53.Geiger O, Sohlenkamp C, Lopez-Lara I. Chapter 27: Formation of Bacterial Membrane Lipids: Pathways, Enzymes, Reactions In: Timmis K, editor. Handbook of hydrocarbon and lipid microbiology. Verlag Berlin Heidelberg: Springer; 2010. [Google Scholar]
- 54.Dunnick J, O'Leary W. Correlation of bacterial lipid composition with antibiotic resistance. J Bacteriol. 1970;101(3):892–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hachmann A, Sevim E, Gaballa A, Popham D, Antelmann H, Helmann J. Reduction in membrane phosphatidylglycerol content leads to daptomycin resistance in Bacillus subtili. Antimicrob Agents Chemother. 2011;55(9):4326–37. 10.1128/AAC.01819-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mehta S, Cuirolo A, Plata K, Riosa S, Silverman J, Rubio A, et al. VraSR two-component regulatory system contributes to mprF-mediated decreased susceptibility to daptomycin in in vivo-selected clinical strains of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2012;56(1):92–102. 10.1128/AAC.00432-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peleg A, Miyakis S, Ward D, Earl A, Rubio A, Cameron D, et al. Whole genome characterization of the mechanisms of daptomycin resistance in clinical and laboratory derived isolates of Staphylococcus aureus. PLoS One. 2012;7(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang S, Xiong Y, Dunman P, Schrenzel J, François P, Peschel A, et al. Regulation of mprF in daptomycin-nonsusceptible Staphylococcus aureus strains. Antimicrob Agents Chemother. 2009;53(6):2636–7. 10.1128/AAC.01415-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rashid R, Veleba M, Kline K. Focal Targeting of the Bacterial Envelope by Antimicrobial Peptides. Front Cell Dev Biol. 2016;4:55 10.3389/fcell.2016.00055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mitra S, Afonina I, Kline K. Right Place, Right Time: Focalization of Membrane Proteins in Gram-Positive Bacteria. Trends Microbiol. 2016;24(8):611–21. 10.1016/j.tim.2016.03.009 [DOI] [PubMed] [Google Scholar]
- 61.Barák I, Muchová K, Wilkinson A, O'Toole P, Pavlendová N. Lipid spirals in Bacillus subtilis and their role in cell division. Mol Microbiol. 2008;68(5):1315–27. 10.1111/j.1365-2958.2008.06236.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rosch J, Caparon M. The ExPortal: an organelle dedicated to the biogenesis of secreted proteins in Streptococcus pyogenes. Mol Microbiol. 2005;58(4):959–68. 10.1111/j.1365-2958.2005.04887.x [DOI] [PubMed] [Google Scholar]
- 63.Lee V, Schneewind O. Protein secretion and the pathogenesis of bacterial infections. Genes Dev. 2001;15(14):1725–52. 10.1101/gad.896801 [DOI] [PubMed] [Google Scholar]
- 64.Nallapareddy S, Singh K, Sillanpää J, Garsin D, Höök M, Erlandsen S, et al. Endocarditis and biofilm-associated pili of Enterococcus faecalis. J Clin Invest. 2006;116(10):2799–807. 10.1172/JCI29021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nielsen H, Guiton P, Kline K, Port G, Pinkner J, Neiers F, et al. The metal ion-dependent adhesion site motif of the Enterococcus faecalis EbpA pilin mediates pilus function in catheter-associated urinary tract infection. mBio. 2012;3(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nallapareddy S, Singh K, Sillanpää J, Zhao M, Murray B. Relative contributions of Ebp Pili and the collagen adhesin ace to host extracellular matrix protein adherence and experimental urinary tract infection by Enterococcus faecalis OG1RF. Infect Immun. 2011;79(7):2901–10. 10.1128/IAI.00038-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dunny G, Leonard B, Hedberg P. Pheromone-inducible conjugation in Enterococcus faecalis: interbacterial and host-parasite chemical communication. J Bacteriol. 1995;177(4):871–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Donovan C, Bramkamp M. Characterization and subcellular localization of a bacterial flotillin homologue. Microbiology. 2009;155(Pt 6):1786–99. 10.1099/mic.0.025312-0 [DOI] [PubMed] [Google Scholar]
- 69.Yepes A, Schneider J, Mielich B, Koch G, García-Betancur J, Ramamurthi K, et al. The biofilm formation defect of a Bacillus subtilis flotillin-defective mutant involves the protease FtsH. Mol Microbiol. 2012;86(2):457–71. 10.1111/j.1365-2958.2012.08205.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bach J, Bramkamp M. Flotillins functionally organize the bacterial membrane. Mol Microbiol. 2013;88(6):1205–17. 10.1111/mmi.12252 [DOI] [PubMed] [Google Scholar]
- 71.Liu C, Liu G, Song Y, Yin F, Hensler M, Jeng W, et al. A cholesterol biosynthesis inhibitor blocks Staphylococcus aureus virulence. Science. 2008;319(5868):1391–4. 10.1126/science.1153018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dos Santos Mota J, Op Den Kamp J, Verheij H, Van Deenen L. Phospholipids of Streptococcus faecalis. Journal of Bacteriology. 1970;104(2):611–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mishra N, Bayer A, Tran T, Shamoo Y, Mileykovskaya E, Dowhan W, et al. Daptomycin resistance in enterococci is associated with distinct alterations of cell membrane phospholipid content. PLoS One. 2012;7(8):e43958 10.1371/journal.pone.0043958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jorasch P, Wolter F, Zähringer U, Heinz E. A UDP glucosyltransferase from Bacillus subtilis successively transfers up to four glucose residues to 1,2-diacylglycerol: expression of ypfP in Escherichia coli and structural analysis of its reaction products. Mol Microbiol. 1998;29(2):419–30. [DOI] [PubMed] [Google Scholar]
- 75.Neuhaus F, Baddiley J. A continuum of anionic charge: structures and functions of D-alanyl-teichoic acids in gram-positive bacteria. Microbiol Mol Biol Rev. 2003;67(4):686–723. 10.1128/MMBR.67.4.686-723.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Garrett T, O'Neill A, Hopson M. Quantification of cardiolipin molecular species in Escherichia coli lipid extracts using liquid chromatography/electrospray ionization mass spectrometry. Rapid Commun Mass Spectrom. 2012;26(19):2267–74. 10.1002/rcm.6350 [DOI] [PubMed] [Google Scholar]
- 77.Nishijima M, Raetz C. Membrane lipid biogenesis in Escherichia coli: identification of genetic loci for phosphatidylglycerophosphate synthetase and construction of mutants lacking phosphatidylglycerol. J Biol Chem. 1979;254(16):7837–44. [PubMed] [Google Scholar]
- 78.Houtkooper R, Turkenburg M, Poll-The BT, Karall D, Pérez-Cerdá C, Morrone A, et al. The enigmatic role of tafazzin in cardiolipin metabolism. Biochim Biophys Acta. 2009;1788(10):2003–14. 10.1016/j.bbamem.2009.07.009 [DOI] [PubMed] [Google Scholar]
- 79.Schlame M. Cardiolipin synthesis for the assembly of bacterial and mitochondrial membranes. J Lipid Res. 2008;49(8):1607–20. 10.1194/jlr.R700018-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schlame M, Rua D, Greenberg M. The biosynthesis and functional role of cardiolipin. Prog Lipid Res. 2000;39(3):257–88. [DOI] [PubMed] [Google Scholar]
- 81.Garrett T, Kordestani R, Raetz C. Quantification of Cardiolipin by Liquid Chromatography-Electrospray Ionization Mass Spectrometry In: Brown H, editor. Methods in Enzymology. 433: Elsevier; 2007. p. 213–30. 10.1016/S0076-6879(07)33012-7 [DOI] [PubMed] [Google Scholar]
- 82.Gidden J, Denson J, Liyanage R, Ivey D, Lay J. Lipid Compositions in Escherichia coli and Bacillus subtilis During Growth as Determined by MALDI-TOF and TOF/TOF Mass Spectrometry. Int J Mass Spectrom. 2009;283(1–3):178–84. 10.1016/j.ijms.2009.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Each peak is labelled with the class (DAG or TAG), fatty acid composition (e.g. 34:1), and the mass-to-charge (m/z) ratio (e.g. 612.5554 for DAG 34:1). The spectrum was acquired in positive ionization mode.
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.