Abstract
Human α and β-defensins are cationic antimicrobial peptides characterized by three disulfide bonds with a triple stranded β-sheet motif. It is presumed that interaction with the bacterial cell surface and membrane permeabilization by defensins is an important step in the killing process. In this study, we have compared interactions of three human α-defensins HNP3, HNP4, HD5 and human β-defensins HBD1-4 that are active against Escherichia coli, with its cell surface and inner membrane as well as negatively charged model membranes. We have also included the inactive α-defensin HD6 in the study. Among the α-defensins, HNP4, HD5 and HD6 were more effective in increasing the zeta potential as compared to HNP3. Among the β-defensins, HBD1 was the least effective in increasing the zeta potential. The zeta potential modulation data indicate variations in the surface charge neutralizing ability of α- and β-defensins. Comparison of E. coli inner membrane and model membrane permeabilizing abilities indicated that HD5, HD6 and HBD1 do not permeabilize membranes. Although HBD4 does not permeabilize model membranes, considerable damage to the inner membrane of E. coli is observed. Our data indicate that mammalian defensins do not kill E. coli by a simple mechanism involving membrane permeabilization though their antibacterial potencies are very similar.
Introduction
Human defensins are small cysteine rich cationic antimicrobial peptides with three disulfide bridges. Based on the disulfide connectivity, they have been classified into two major groups, α- and β-defensins [1]. The human genome contains five α-defensin genes, which codes for six α-defensins and approximately thirty β-defensin genes [2–6]. Human defensins show considerable variations in their amino acid sequences, except α-defensins HNP1-3 [7, 8]. The primary structures of HNP1-3 differ only by a single residue at the N-terminus [9]. The three dimensional structures of human α- and β-defensins are similar, consisting of a characteristic triple stranded antiparallel β-sheet structure connected by three disulfide bonds [10–15]. In the case of β-defensins, apart from the core β-sheet structure, a helix is also present at the N-terminal region [12–15]. A motif called “γ” core motif has been reported by Yount and Yeaman [16], which may have a role in modulating the activity of defensins [17]. Despite having similar structures, mammalian α- and β-defensins show considerable variations in their antibacterial potencies and spectrum of activity [18–23]. HNP1-3 are more active against certain strains of Staphylococcus aureus as compared to Escherichia coli, whereas HNP4 and HD5 show comparable activity against E. coli and S. aureus [18]. HD6 does not show antibacterial activity in vitro [18]. Human β-defensins also show variations in their activity. HBD1 and 2 are active predominantly against gram-negative bacteria, whereas HBD3 and 4 are active against gram-negative and gram-positive bacteria [19–24] In linear host defense peptides such as magainins [25, 26], cecropins [27, 28] and cathelicidins [29–31], their model membrane activity has been correlated to bacterial membrane permeabilization resulting in cell death. The interactions by α- and β-defensins with model membranes are highly variable [32–35] and their relevance to bacterial killing is not yet established unequivocally. Early investigations carried out with HNP1-3 showed sequential permeabilization of the outer and inner membranes of E. coli during killing of the bacteria [36]. Further, a membrane pore formation mechanism was proposed based on the crystal structure of HNP3 [11]. However, recent studies indicate that membrane activity of α-defensins may not necessarily correlate with bacterial killing [37]. The bactericidal mechanism of HNP1 against Staphylococcus aureus has been found to involve interaction with lipid II and inhibition of cell wall synthesis [37]. Human enteric α-defensins HD5 and HD6 do not permeabilize model membranes [38]. HD5 kills E. coli by localizing to the cytoplasm [38] and possibly interacting with DNA [39]. Unlike α-defensins, tertiary structures of human β-defensins (HBDs) do not favor mechanisms involving pore formation [12–14]. It appears that the electrostatic interaction between HBDs and bacterial membranes leads to destabilization of the membrane [40]. The exact mechanism by which human β-defensins permeabilize bacterial membranes is yet to be established unequivocally. Also, all human β-defensins do not form higher order oligomers in solution [12–14]. The activities of α- and β-defensins on model membranes or membranes of bacteria have not been compared in the same series of experiments. This would help direct comparison of their activities and also get better insights into the differences in their interaction with membranes. In this study, we compare membrane activities of four human α-defensins and four β-defensins against bacterial and model membranes. We also investigated whether human defensins form well defined aggregates by electron microscopy (EM). Primary structures of defensins used in this study are shown in Table 1. Our results indicate that contributions from membrane activity of human defensins to bacterial killing vary considerably.
Table 1. Primary structures of human defensins.
| Defensin | Sequencea | Net Charge |
|---|---|---|
| HNP3 | DC1YC2RIPAC3IAGERRYGTC2IYQGRLWAFC3C1 | +2 |
| HNP4 | VC1SC2RLVFC3RRTELRVGNC2LIGGVSFTYC3C1TRV | +4 |
| HD5 | ATC1YC2RTGRC3ATRESLSGVC2EISGRLYRLC3C1R | +4 |
| HD6 | AFTC1HC2RRSC3YSTEYSYGTC2TVMGINHRFC3C1L | +2 |
| HBD1 | DHYNC1VSSGGQC2LYSAC3PIFTKIQGTC2YRGKAKC1C3K | +4 |
| HBD2 | GIGDPVTC1LKSGAIC2HPVFC3PRRYKQIGTC2GLPGTKC1C3KKP | +6 |
| HBD3 | GIINTLQKYYC1RVRGGRC2AVLSC3LPKEEQIGKC2STRGRKC1C3RRKK | +11 |
| HBD4 | ELDRIC1GYGTARC2RKKC3RSQEYRIGRC2PNTYAC1C3LRKWDESLLNRTKP | +7 |
a Numbers in superscripts adjacent to cysteines denote disulfide connectivities.
Materials and methods
Materials
All human defensins used in this study were purchased from Peptides International, USA. N-(3-triethylammoniumpropyl)-4-(6-(4(diethylamino) phenyl-hexatrienyl)pyridinium dibromide) (FM4-64) and SYTOX green were obtained from Molecular Probes (Eugene, OR, US). Phospholipids POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine) and POPG (1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1-rac-glycerol)) (sodium salt) were from Avanti Polar Lipids (Alabaster, AL). Cholesterol and calcein were from Sigma-Aldrich. All the other chemicals used for this study were of the highest grade available.
Antibacterial activity
Minimum bactericidal concentration (MBC) of human defensins against E. coli MG1655 was determined as described elsewhere [39, 41]. In brief, cells collected from the mid-log-phase were washed and resuspended in 10 mM sodium phosphate buffer (pH 7.4) containing 1% tryptic soy broth. The final cell density was adjusted to 106 colony forming units (CFU)/mL. 100 μL of these cells were then incubated with varying concentration of defensins for 2 hours at 37°C. The cells were then spread onto nutrient-rich defensin free Luria-Bertani (LB) agar plates and incubated for 12–15 hrs at 37°C, colonies formed were counted and percentage of killing was calculated. The lowest concentration at which complete killing observed was taken as MBC.
Zeta potential measurements
Zeta potential of the E. coli in the presence of human defensins was measured as described previously [39]. The concentration of peptide used was 5μM, except for HBD3 and HD5. In the cases of HBD3 and HD5, 0.5 μM and 2.5 μM were found to neutralize the E. coli surface charge almost completely. Therefore, in the cases of HBD3 and HD5, 0.5 μM and 2.5 μM of peptides were used, respectively.
Time-lapse fluorescence confocal microscopy
Effect of defensins on the E. coli inner membrane was examined using time-lapse confocal fluorescence microscopy. The assay was carried out as follows. Bacteria from the mid-log-phase was collected and resuspended (final density was adjusted to 106 CFU/mL) in 10 mM sodium phosphate buffer (pH 7.4). Cells were then treated with 3 μM FM4-64 for 20 minutes at room temperature to stain the inner membrane. FM4-64 is a lipophilic dye which stains the bacterial inner membrane [42]. Excess dye was removed by centrifuging. The FM4-64 stained cells were then treated with sub-lethal concentrations of defensins in presence of 5 nM of SYTOX green. The concentrations of the defensins used were; HBD1: 3 μM, HBD2: 3 μM, HBD3: 1 μM, HBD4: 3 μM, HNP3: 8 μM, HNP4: 4 μM, HD5: 2.5 μM, HD6: 5 μM. Cells were immediately transferred to chambered slides and images were recorded using 100X oil immersion objective on a Leica Ultraspectral microscope SP8 (Leica Microsystems). Argon 488 and HeNe 561 lasers were used to excite SYTOX green and FM4-64, respectively. Signals ranging from 510 to 560 nm and 600 to 740 nm were collected for SYTOX green and FM4-64, respectively. Images were processed using LAS-AFver3.1.3 (Leica Microsystems).
SYTOX green uptake assay using fluorescent spectroscopy
Effect of defensins on the E. coli inner membrane was assessed by measuring the extent of intracellular accumulation of SYTOX green. Cells from mid-log-phase were collected, washed and resuspended in 10 mM phosphate buffer. The final density was adjusted to 5 × 106 CFU/mL. Cells were then treated with 2 × MBC of defensins in presence of 200 nM SYTOX green. Enhancement in SYTOX green fluorescence, a direct measure of the extent of membrane permeabilization was monitored in a Flurolog 3–22 fluorescence spectrophotometer (Jobin Yvon, USA). Excitation and emission wavelengths used were 503 nm and 523 nm, respectively.
Calcein release assay
Large unilamellar vesicles (LUVs) composed of POPC:POPG (1:1) containing 50 mM calcein were prepared by lipid extrusion method [43]. Desired amounts of lipids from their respective chloroform stocks were taken into a glass tube and dried under a nitrogen stream to form a thin uniform lipid film. The film was further dried under vacuum for 5 to 6 hours to remove trace amounts of organic solvents. After this step, the lipid film was hydrated with 10 mM sodium phosphate buffer (pH 7.4) containing 50 mM calcein at 4°C for 12 to 15 hours. Hydrated lipid films were then vortexed and passed through polycarbonate membrane having a pore diameter of 100 nm using a mini-extruder (Avanti Polar Lipid Inc.). Free calcein was removed by passing through a Sephadex G50 column. Calcein entrapped vesicles (25 μM) were then treated with increasing concentrations of defensins and extent of calcein released was measured in a Fluorolog 3–22 fluorescence spectrophotometer (Jobin Yvon, USA). Excitation and emission wavelengths used were 485 and 515 nm, respectively. 1% Triton X-100 was used for complete release of calcein.
Transmission electron microscopy
Defensins were diluted to a concentration of 20 μM in 10 mM sodium phosphate buffer (pH 7.4) from their respective stocks solutions (dissolved in Milli-Q water). From these solutions, 5 μL was deposited on a carbon-coated Formvar 200-mesh copper grid. The grid was left undisturbed for about 5 minutes; excess buffer was then removed using Whatman filter paper. Grids were stained with uranyl acetate (2% w/v) for 45 seconds. Excess stain was removed and grids were used for recording the images. Images were recorded using JAM-2100 LaB6 transmission electron microscope (JEOL, Tokyo, Japan) at 100 kV.
Results and discussion
Comparison of antibacterial activity of human defensins against E. coli
In order to compare the antibacterial activity and bacterial membrane permeabilizing abilities of human defensins, we determined the MBCs against E. coli MG1655. The MBC values of human defensins against E. coli MG1655 are summarized in Table 2. The antibacterial potencies of human defensins against E. coli are of following order: HBD3>HD5>HBD4>HBD2 = HBD1>HNP4>HNP3>HD6.
Table 2. Minimum Bactericidal Concentration (MBC) of human defensins against E. coli MG1655.
Electrostatic interaction of human defensins with the E. coli surface
Cationic antimicrobial peptides (CAMPs) initially interact with bacterial cell wall components, followed by subsequent interactions with bacterial inner membrane and other intracellular components [46]. It has been proposed that initial electrostatic interaction with bacterial cell wall components is critical in determining the efficacy of CAMPs, as the abrogation of these interactions leads to attenuation of antibacterial activity [47–49]. In fact, Alves et al., have reported a correlation between extent of surface charge neutralizing ability and minimum inhibitory concentrations for amphipathic CAMPs [50]. We have previously shown that effective interaction of human defensin analogs with bacterial surfaces enhances antibacterial potency [39, 51, 52]. With a view to understand, how human defensins vary in their ability to interact with bacterial surfaces, we compared the extent of electrostatic interactions between the E. coli cell envelope and human defensins by measuring changes in the zeta potential of the E. coli.
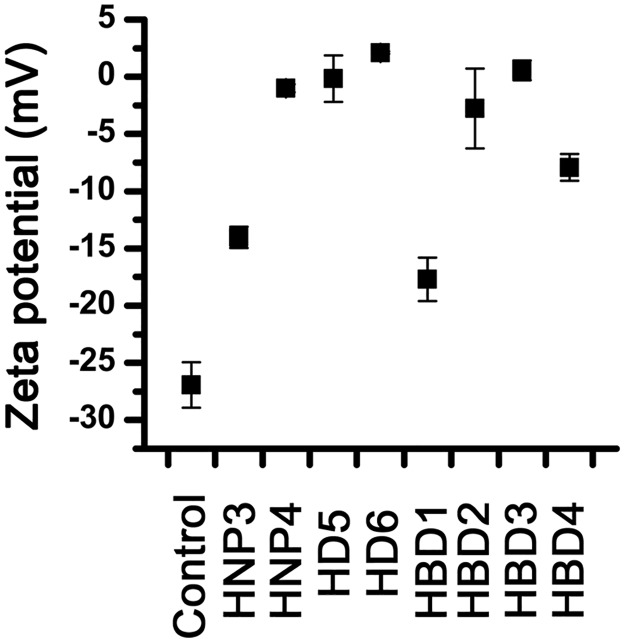
Changes in the zeta potential of the E. coli in the presence of human defensins are shown in Fig 1. Among the defensins tested, HBD3 exhibits the most efficient bacterial surface charge neutralizing ability, which is presumably due to its very high net positive charge (+11). HNP3 and HBD1 show poor surface charge neutralizing ability as compared to other human defensins. Intriguingly, HD6, which has the lowest net positive charge (+2) among the human defensins tested, neutralizes the E. coli surface charge more efficiently than HBD4 (+7), HBD1 (+ 4) and HNP3 (+2). Also, HD5 which has same net positive charge (+4) as HNP4, neutralizes the E. coli surface charge at much lower concentration than HNP4. Clearly, the ability of human defensins to interact with bacterial surfaces is not merely governed by net positive charge alone. The inactive defensin HD6, neutralizes bacterial surface charge more effectively as compared to defensins that possess potent bactericidal activity. This could arise due to rapid oligomerization of HD6 on bacterial surfaces [53], resulting in effective charge neutralization. Analysis of tertiary structures of human defensins indicate that there are differences in the arrangement of cationic residues in three dimensional structures [10]. The observed variations in the ability of human defensins to associate with bacterial surfaces via electrostatic interactions could arise due to differences in the distributions of cationic side chains in the three dimensional structures of defensins.
Fig 1. Effect of human defensins on the zeta potential of E. coli.
Cells were treated with 5 μM defensin and zeta potential was measured, except in the cases of HD5 and HBD3. In the cases of HD5 and HBD3, zeta potential was measured after treating with 2.5 μM and 0.5 μM of peptides, respectively. Control represent zeta potential of E. coli in the absence of any peptide. Error bars represents standard deviations of three independent experiments.
Effect of human defensins on the E. coli inner membrane
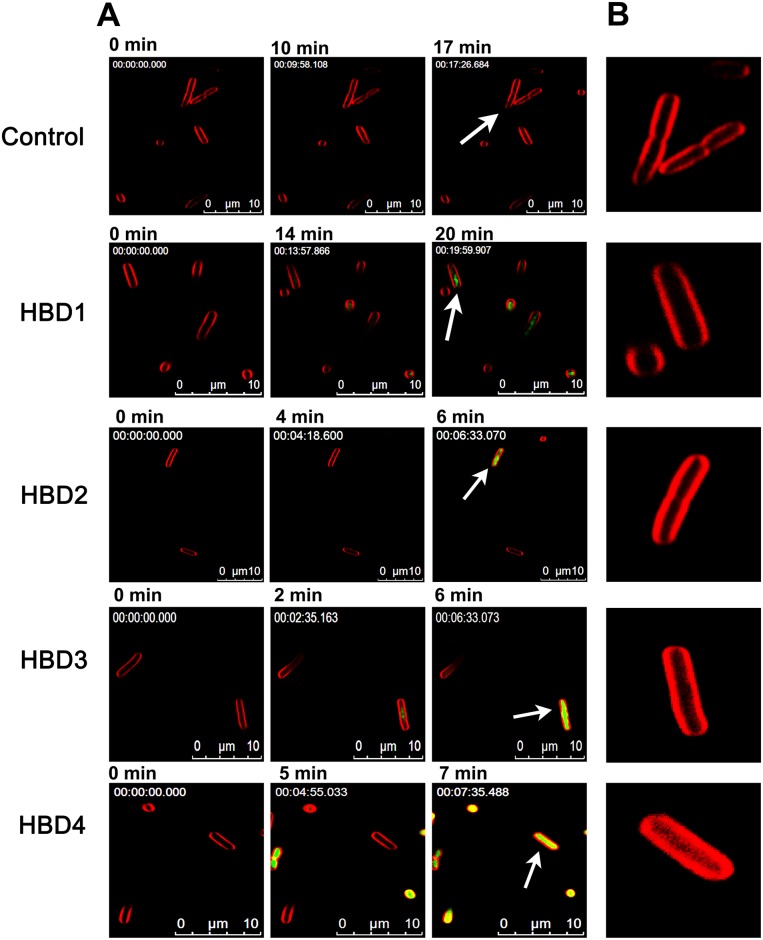
HNP1-3 kill E. coli by permeabilizing the outer and inner membranes in a sequential manner [36]. We next examined the effect human defensins on the E. coli inner membrane by monitoring intracellular accumulation of SYTOX green in defensin-treated cells using time-lapse fluorescence confocal microscopy. Confocal micrographs shown in Fig 2A indicate that not all human β-defensins permeabilize the E. coli inner membrane to the same extent. Intense accumulation of SYTOX green is observed only in the cases of HBD2-4. In the case of HBD1, faint accumulation of SYTOX green is evident at ~20min. However, the intensity is considerably low as compared to other membrane active defensins, suggestive of less extensive damage caused by HBD1 on the E. coli inner membrane. Intriguingly, both HBD1 and HBD2 show similar antibacterial efficacy against E. coli (Table 2). Therefore, the mechanism by which HBD1 kills E. coli is markedly different from HBD2. It has been proposed that bacterial killing mechanism of linear HBD1 is likely to involve interactions with cytoplasmic components [54]. Presumably, bacterial killing mechanism of HBD1 also follows similar pathways.
Fig 2. Effect of human β-defensins on the E. coli inner membrane.
(A), Intracellular accumulation of SYTOX green in defensin treated cells as function of time. The minute at which images were recorded is mentioned above the respective image. Numbers given in the upper left corner of the images represent the elapsed time (h:min:s:ms). (B), Morphological features of FM4-64 stained E. coli inner membrane of selected bacteria, as indicated by arrows.
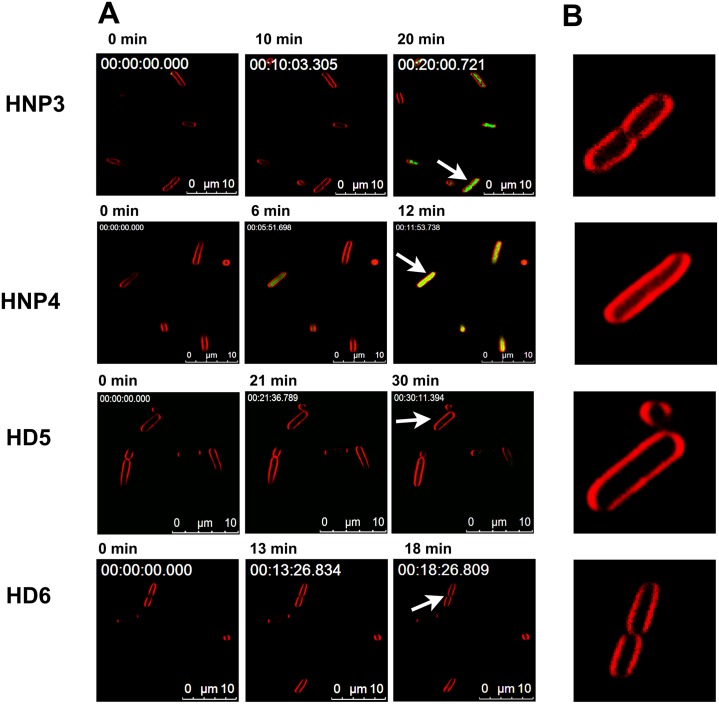
In the case of α-defensins, cytoplasmic accumulation of SYTOX green is evident only in cell treated with myeloid defensins HNP3 and HNP4 (Fig 3A). Though enteric defensin HD5 kills bacteria more efficiently than HNP3 and HNP4, HD5 does not permeabilize the E. coli inner membrane. Recent findings indicate HD5 exerts its activity against the E. coli by localizing to bacterial cytoplasm [38] and possibly interacting with DNA [39]. In a very recent study, Wang et al., have shown that HD5 permeabilizes the E. coli inner membrane [55]. However, prolonged incubation (~40 minutes) was required to observe membrane perturbation [55]. Interestingly, analysis of time-lapse microscopy data indicates that, although extensive damage does not occur, a faint localization of SYTOX green occurs in E. coli treated with HD5 at 30 min (S1 Fig). It has been reported that degradation or topological changes in the bacterial DNA can a ffect the SYTOX green fluorescence [56]. Considering the fact that HD5 possess strong affinity towards DNA [39], its interaction with DNA could influence the fluorescence of SYTOX green, which may possibly account for the variations in the observed and reported inner membrane permeabilizing ability. However, both the results unambiguously suggest that perturbation occurs only after prolonged incubation with HD5. Undoubtedly, HD5 kills E. coli without causing extensive damage to bacterial membrane.
Fig 3. Effect of human α-defensins on the E. coli inner membrane.
(A), Intracellular accumulation of SYTOX green in defensin treated cells as function of time. The minute at which images were recorded are mentioned above respective image. Numbers given in the upper left corner of the images represent the elapsed time (h:min:s:ms). (B), Morphological features of FM4-64 stained E. coli inner membrane of selected bacteria, as indicated by arrows. Numbers given in the upper left corner represent the elapsed time (h:min:s:ms).
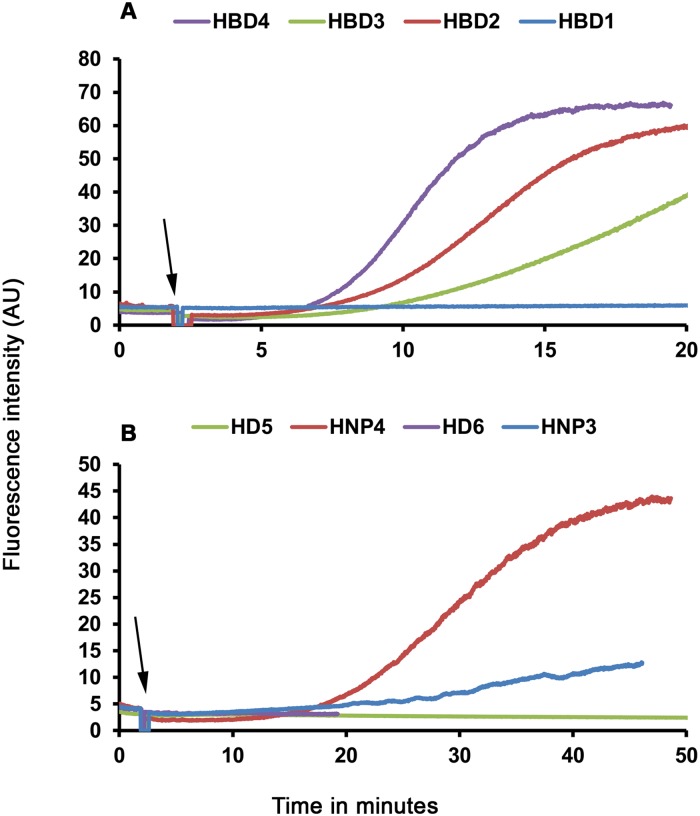
Analysis of kinetics of SYTOX green accumulation indicate that membrane active β-defensins cause more rapid influx of dye into the bacterial cytoplasm as compared to membrane active α-defensins. In the case of HBD2-4, intense accumulation is evident in less than 7 min while in the HNP4 and HNP3 treated cells, accumulation is evident only at 11 min and 20 min, respectively. Further, we also observe that morphological features of the FM4-64 stained E. coli membranes (Figs 2B and 3B) are largely intact, except for HBD4 treated cells. In the case of HBD4 treated cells, FM4-64 staining shows considerable diffusion into cytoplasm, suggestive of more extensive damage caused by HBD4 as compared to other defensins. To further validate the observed variations in the bacterial membrane permeabilizing abilities, we examined intracellular accumulation of SYTOX green in the E. coli treated with 2×MBC of defensins using fluorescent spectroscopy. Results shown in Fig 4 further confirm time-lapse microscopy results. Clearly, human α-defensins HD5, HD6, and human β-defensin HBD1 do not cause extensive damage to the E. coli inner membrane. Together, time-lapse microscopy and fluorescent spectroscopy experiments indicate that human defensins do not permeabilize the E. coli inner membrane to the same extent although their antibacterial potencies are similar. Thus, there appears to be no unifying mechanism of bacterial killing, particularly against E. coli.
Fig 4. Accumulation of SYTOX green in human defensins treated E. coli.
Cells were treated with 2 × MBC of (A), human β-defensins or (B), human α-defensins and intracellular accumulation of SYTOX green was monitored. The arrows indicate the point at which defensins were added.
The evolutionary and functional relevance of primary structures of human defensins have been a subject of extensive investigations [57, 58]. In an attempt to understand the evolutionary relationships between α-defensin genes, Das et al., classified primate α-defensins into three different phylogenitic classes, class I, II and III [58]. According to this classification, HD5 belongs to class I, HD6 belongs to class II and HNP1-4 belong to class III. Interestingly, based on the structural analysis, Das et al., even predicted that variations in the electrostatic surface distributions could lead to differences in their bacterial killing mechanisms [58]. Our observations on the bacterial membrane permeabilizing abilities of human defensins strongly support this hypothesis. Also, it appears that amino acid selection is favored by environmental niches, as the enteric defensins HD5 and HD6 are non-lytic in nature. Although such hypothesizes are absent for human β-defensins, it is presumable that there too evolutionary selection have played pivotal role in rendering heterogeneity to bacterial killing mechanisms. Clearly, the poorly conserved amino acids play a critical role in modulating the bacterial killing mechanism.
Interaction with lipid vesicles
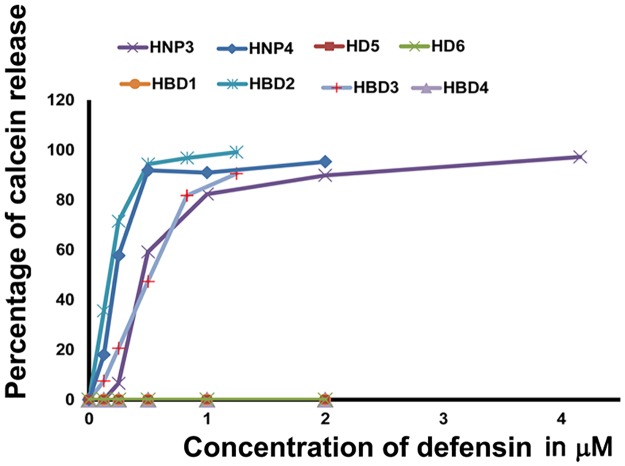
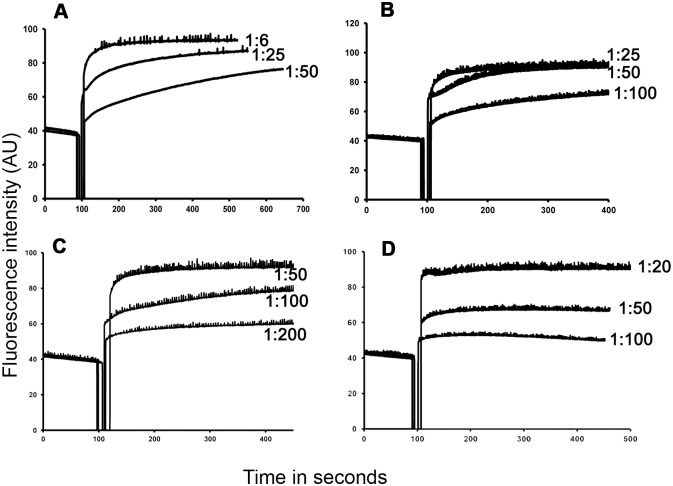
Model membranes have been used as a tool to study mechanisms by which defensins permeabilize microbial membranes. There have been reports, which suggest that defensins permeabilize model membranes and cause their destabilization [32–35,59, 60]. We have examined the effect of α- and β-defensins on calcein entrapped negatively charged vesicles composed of POPC:POPG (1:1). We observe that with the exception of HBD1, HBD4, HD5 and HD6, defensins permeabilize lipid vesicles (Fig 5), which correlates with their ability to permeabilize the E. coli inner membrane, except for HBD4. It is evident that even low concentrations of defensins can affect the release of calcein. Further, analysis of kinetics of calcein release indicate that defensins cause a rapid release of calcein, although there are variations in the peptide to lipid (P:L) ratio where the maximum release is observed (Fig 6).
Fig 5. Effect of human defensins on POPC:POPG (1:1) vesicles.
Calcein entrapped POPC: POPG vesicles (25 μM) were treated with increasing concentrations of human defensins and percentage of calcein released was calculated. Calcein released by 1% Triton X-100 was taken as 100%.
Fig 6. Kinetics of calcein release from POPC:POPG (1:1) vesicles treated with defensins.
(A), HNP3; (B), HNP4; (C), HBD2 and (D), HBD3. The peptide to lipid ratios (P:L) are mentioned along the respective spectra.
Based on the crystal structure and biophysical studies carried out with HNP1-3, it has been proposed that amphipathic dimer formed by human α-defensins form pores on bacterial membrane [11, 61]. Human enteric α-defensins, HD5 and HD6 also form amphipathic dimers similar to myeloid defensins [10]. However, they do not permeabilize model membranes [37]. Interestingly, detailed analysis of crystal structures indicated that amphipathic dimer formed by human enteric α-defensins are asymmetric in nature unlike HNP1-4 [10]. It is conceivable that the poorly conserved amino acids play a critical role in the observed variations in the dimer topology. Consequently, their ability to permeabilize lipid vesicles. Intriguingly, HBD4 which causes extensive damage to the E. coli inner membrane does not permeabilize PC:PG vesicles. The underlying physico-chemical reason behind this discrepancy is not clear at this point. It is possible that essential factors present on the microbial surface or membranes may be playing a critical role in determining the membrane activity of HBD4, as reported for other defensins [33, 62].
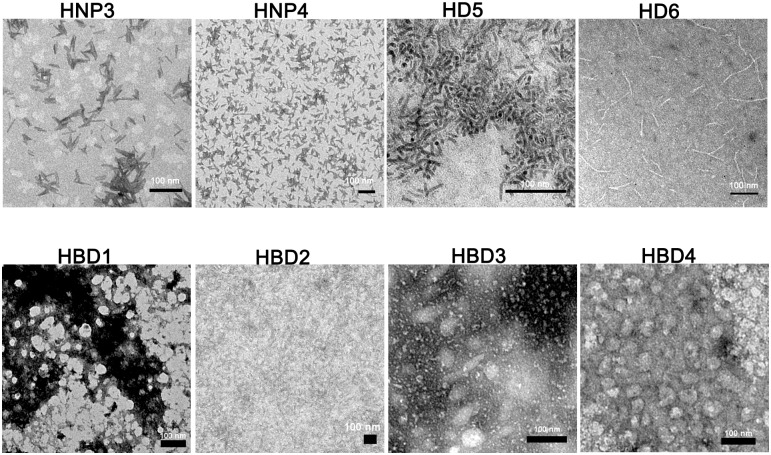
Human defensins form ordered aggregates
The ability of human α-defensin HD6 to inhibit bacterial infection of epithelial cells has been attributed to its ability to self-assemble and form fibrillar mesh-like structures [53, 63]. Since other human defensins have also been reported to form higher order oligomers under crystalline state and in solutions [10–12, 15, 64], we compared the self-assembling properties of α- and β-defensins. Electron micrographs of human defensins are shown in Fig 7. While distinctive fibrillar morphology is observed for HD6 similar structures are not observed for other defensins. HNP3 and HD5 show rod-like structures while others show amorphous structures. Considering the fact that HD6’s ability to form fibrils in vitro is critical to its ability to form “nanonets” on binding to bacterial surfaces [53, 63], it is presumable that rod-like aggregates formed by HD5 and HNP3 will also likely to play a deciding role in their antibacterial activity and bacterial killing mechanism. Even subtle changes in the dimer interface, which is likely to modulate self-assembling properties, has been reported to affect the antibacterial potency and selectivity of HNP1 and HD5 [65, 66]. The physico-chemical properties of self-assembled aggregates of defensins on bacterial surfaces might be playing critical roles in their ability to interact with bacterial surface and membranes.
Fig 7. Transmission electron micrographs of human defensin aggregates.
Conclusions
In this study, we have compared various aspects of bacterial and model membrane permeabilizing abilities of human defensins. There are considerable variations in their ability to interact with the E. coli cell surface and model membranes, suggesting differences in the mechanisms by which human defensins exert their antibacterial activity. Although the arguments presented in this paper are based on the observations on E. coli, similar variations in the mechanism of bacterial killing can be anticipated for other gram-negative species as well. Since all the defensins described in this study are not active against S. aureus, our investigations have been confined to E. coli. Defensins show differences in membrane destabilization which could result in variations in bacterial killing mechanisms despite having very similar three dimensional structures. It is evident that the topography of positively selected amino acids during the evolution play a critical role in rendering highly heterogeneous mechanisms of bacterial killing without affecting their overall three dimensional fold.
Supporting information
Arrows indicate the accumulation of SYTOX green. Numbers given in the upper left corner represent the elapsed time (h:min:s:ms).
(TIF)
Acknowledgments
We thank Ms. Nandini Rangaraj, CCMB for her assistance in the use of confocal microscope and A. Harikrishna for recording the electron microscopy images. RN is a recipient of the JC Bose fellowship from the Department of Science and Technology, India. RN is grateful to the National Academy of Science, India for NASI-Senior Scientist Platinum Jubilee Fellowship. BM was a recipient of Senior Research Fellowship from CSIR.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
RN is a recipient of the JC Bose fellowship from the Department of Science and Technology, India. RN is grateful to the National Academy of Science, India for NASI-Senior Scientist Platinum Jubilee Fellowship. BM was a recipient of Senior Research Fellowship from CSIR. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ganz T. Defensins: antimicrobial peptides of innate immunity. Nat Rev Immunol. 2003;3:710–720. 10.1038/nri1180 [DOI] [PubMed] [Google Scholar]
- 2.Radhakrishnan Y, Hamil KG, Yenugu S, Young SL, French FS, Hall SH. Identification, characterization, and evolution of a primate β-defensin gene cluster. Genes Immun. 2005;6:203–210. 10.1038/sj.gene.6364184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schutte BC, Mitros JP, Bartlett JA, Walters JD, Jia HP, Welsh MJ, et al. Discovery of five conserved β-defensin gene clusters using a computational search strategy. Proc Natl Acad Sci U S A. 2002;99:2129–2133. 10.1073/pnas.042692699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones DE, Bevins CL. Paneth cells of the human small intestine express an antimicrobial peptide gene. J Biol Chem. 1992;267:23216–23225. [PubMed] [Google Scholar]
- 5.Jones DE, Bevins CL. Defensin-6 mRNA in human Paneth cells: implications for antimicrobial peptides in host defense of the human bowel. FEBS Lett. 1993;315:187–192. [DOI] [PubMed] [Google Scholar]
- 6.Linzmeier R, Michaelson D, Liu L, Ganz T. The structure of neutrophil defensin genes. FEBS Lett. 1993;321:267–273. [DOI] [PubMed] [Google Scholar]
- 7.Pazgier M, Hoover DM, Yang D, Lu W, Lubkowski J. Human β-defensins. Cell Mol Life Sci. 2006;63:1294–1313. 10.1007/s00018-005-5540-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lehrer RI, Lu W. α-Defensins in human innate immunity. Immunol Rev. 2012;245:84–112. 10.1111/j.1600-065X.2011.01082.x [DOI] [PubMed] [Google Scholar]
- 9.Selsted ME, Harwig SS, Ganz T, Schilling JW, Lehrer RI. Primary structures of three human neutrophil defensins. J Clin Invest. 1985;76:1436–1439. 10.1172/JCI112121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szyk A, Wu Z, Tucker K, Yang D, Lu W, Lubkowski J. Crystal structures of human α-defensins HNP4, HD5, and HD6. Protein Sci. 2006;15:2749–2760. 10.1110/ps.062336606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hill C, Yee J, Selsted M, Eisenberg D. Crystal structure of defensin HNP-3, an amphiphilic dimer: mechanisms of membrane permeabilization. Science. 1991;251:1481–1485. [DOI] [PubMed] [Google Scholar]
- 12.Schibli DJ, Hunter HN, Aseyev V, Starner TD, Wiencek JM, McCray PB Jr, et al. The solution structures of the human β-defensins lead to a better understanding of the potent bactericidal activity of HBD3 against Staphylococcus aureus. J Biol Chem. 2002;277:8279–8289. 10.1074/jbc.M108830200 [DOI] [PubMed] [Google Scholar]
- 13.Sawai MV, Jia HP, Liu L, Aseyev V, Wiencek JM, McCray PB, et al. The NMR Structure of human β-defensin-2 reveals a novel α-helical Segment. Biochemistry. 2001;40:3810–3186. [DOI] [PubMed] [Google Scholar]
- 14.Hoover DM, Chertov O, Lubkowski J. The structure of human β-defensin-1: new insights into structural properties of β-defensins. J Biol Chem. 2001;276:39021–39026. 10.1074/jbc.M103830200 [DOI] [PubMed] [Google Scholar]
- 15.Hoover DM, Rajashankar KR, Blumenthal R, Puri A, Oppenheim JJ, Chertov O, et al. The structure of human β-defensin-2 shows evidence of higher order oligomerization. J Biol Chem. 2000;275:32911–3218. 10.1074/jbc.M006098200 [DOI] [PubMed] [Google Scholar]
- 16.Yount NY,Yeaman MR. Emerging themes and therapeutic prospects for anti-infective peptides. Annu Rev Pharmacol Toxicol. 2012;52: 337–360. 10.1146/annurev-pharmtox-010611-134535 [DOI] [PubMed] [Google Scholar]
- 17.Nigro E, Colavita I, Sarnataro D, Scudiero O, Zambrano G, Granata V, et al. An ancestral host defence peptide within human β-defensin 3 recapitulates the antibacterial and antiviral activity of the full-length molecule. Sci Rep. 20155:18450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ericksen B, Wu Z, Lu W, Lehrer RI. Antibacterial activity and specificity of the six human α-defensins. Antimicrob Agents Chemother. 2005;49:269–275. 10.1128/AAC.49.1.269-275.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harder J, Bartels J, Christophers E, Schroder JM. A peptide antibiotic from human skin. Nature. 1997;387:861 10.1038/43088 [DOI] [PubMed] [Google Scholar]
- 20.Harder J, Bartels J, Christophers E, Schroder JM. Isolation and characterization of human β-defensin-3, a novel human inducible peptide antibiotic. J Biol Chem. 2001;276:5707–5713. 10.1074/jbc.M008557200 [DOI] [PubMed] [Google Scholar]
- 21.Valore EV, Park CH, Quayle AJ, Wiles KR, McCray PB Jr, Ganz T. Human β-defensin-1: an antimicrobial peptide of urogenital tissues. J Clin Invest. 1998;101:1633–1642. 10.1172/JCI1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia JR, Krause A, Schulz S, Rodriguez-Jimenez FJ, Kluver E, Adermann K, et al. Human beta defensin 4: a novel inducible peptide with a specific salt-sensitive spectrum of antimicrobial activity. FASEB J. 2001; 15: 1819–1821. [PubMed] [Google Scholar]
- 23.Scudiero O, Nigro E, Cantisani M, Colavita I, Leone M, Mercurio FA, et al. Design and activity of a cyclic mini-β-defensin analog: a novel antimicrobial tool. Int J Nanomedicine. 2015;10:6523–39. 10.2147/IJN.S89610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ageitos JM, Sánchez-Pérez A, Calo-Mata P, Villa TG. Antimicrobial peptides (AMPs): Ancient compounds that represent novel weapons in the fight against bacteria. Biochem Pharmacol. 2016. September 20. pii: S0006-2952(16)30301-X. [DOI] [PubMed] [Google Scholar]
- 25.Matsuzaki K, Sugishita K-i, Harada M, Fujii N, Miyajima K. Interactions of an antimicrobial peptide, magainin 2, with outer and inner membranes of Gram-negative bacteria. Biochim Biophys Acta—Biomembranes. 1997;1327:119–130. [DOI] [PubMed] [Google Scholar]
- 26.Matsuzaki K, Harada M, Handa T, Funakoshi S, Fujii N, Yajima H, et al. Magainin 1-induced leakage of entrapped calcein out of negatively-charged lipid vesicles. Biochim Biophys Acta. 1989;981:130–134. [DOI] [PubMed] [Google Scholar]
- 27.Wang W, Smith DK, Moulding K, Chen HM. The Dependence of membrane permeability by the antibacterial peptide cecropin B and its analogs, CB-1 and CB-3, on liposomes of different composition. J Biol Chem. 1998;273:27438–27448. [DOI] [PubMed] [Google Scholar]
- 28.Wang W, Smith DK, Chen HM. The effect of pH on the structure, binding and model membrane lysis by cecropin B and analogs. Biochim Biophys Acta. 1999;1473:418–430. [DOI] [PubMed] [Google Scholar]
- 29.Sochacki KA, Barns KJ, Bucki R, Weisshaar JC. Real-time attack on single Escherichia coli cells by the human antimicrobial peptide LL-37. Proc Natl Acad Sci U S A. 2011;108:E77–E81. 10.1073/pnas.1101130108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee C-C, Sun Y, Qian S, Huang Huey W. Transmembrane pores formed by human antimicrobial peptide LL-37. Biophys J. 2011;100:1688–1696. 10.1016/j.bpj.2011.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henzler Wildman KA, Lee DK, Ramamoorthy A. Mechanism of lipid bilayer disruption by the human antimicrobial peptide, LL-37. Biochemistry. 2003;42:6545–6558. 10.1021/bi0273563 [DOI] [PubMed] [Google Scholar]
- 32.Lohner K, Latal A, Lehrer RI, Ganz T. Differential scanning microcalorimetry indicates that human defensin, HNP-2, interacts specifically with biomembrane mimetic systems. Biochemistry. 1997;36:1525–1531. 10.1021/bi961300p [DOI] [PubMed] [Google Scholar]
- 33.Böhling A, Hagge SO, Roes S, Podschun R, Sahly H, et al. Lipid-specific membrane activity of human β-defensin-3. Biochemistry. 2006;45:5663–5670. 10.1021/bi052026e [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y, Lu W, Hong M. The membrane-bound structure and topology of a human α-defensin indicate a dimer pore mechanism for membrane disruption. Biochemistry. 2010;49:9770–9782. 10.1021/bi101512j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmidt NW, Mishra A, Lai GH, Davis M, Sanders LK, Tran D, et al. Criterion for amino acid composition of defensins and antimicrobial peptides based on geometry of membrane destabilization. J Am Chem Soc. 2011;133:6720–6727. 10.1021/ja200079a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lehrer RI, Barton A, Daher KA, Harwig SS, Ganz T, Selsted ME. Interaction of human defensins with Escherichia coli. Mechanism of bactericidal activity. J Clin Invest. 1989;84:553–561. 10.1172/JCI114198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Leeuw E, Li C, Zeng P, Diepeveen-de Buin M, Lu WY, Breukink E, et al. Functional interaction of human neutrophil peptide-1 with the cell wall precursor lipid II. FEBS Lett. 2010;584:1543–1548. 10.1016/j.febslet.2010.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chileveru HR, Lim SA, Chairatana P, Wommack AJ, Chiang IL, Nolan EM. Visualizing attack of Escherichia coli by the antimicrobial peptide human defensin 5. Biochemistry. 2015; 54:1767–1777. 10.1021/bi501483q [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mathew B, Nagaraj R. Antimicrobial activity of human α-defensin 5 and its linear analogs: N-terminal fatty acylation results in enhanced antimicrobial activity of the linear analogs. Peptides. 2015;71:128–140. 10.1016/j.peptides.2015.07.009 [DOI] [PubMed] [Google Scholar]
- 40.Morgera F, Antcheva N, Pacor S, Quaroni L, Berti F, Vaccari L, et al. Structuring and interactions of human β-defensins 2 and 3 with model membranes. J Pept Sci. 2008;14:518–523. 10.1002/psc.981 [DOI] [PubMed] [Google Scholar]
- 41.Ganz T, Selsted ME, Szklarek D, Harwig SS, Daher K, Bainton DF, et al. Defensins. Natural peptide antibiotics of human neutrophils. J Clin Invest. 1985;7:1427–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fishov I, Woldringh CL. Visualization of membrane domains in Escherichia coli. Mol Microbiol. 1999;32:1166–1172. [DOI] [PubMed] [Google Scholar]
- 43.MacDonald RC, MacDonald RI, Menco BP, Takeshita K, Subbarao NK, Hu LR. Small-volume extrusion apparatus for preparation of large, unilamellar vesicles. Biochim Biophys Acta. 1991;1061:297–303. [DOI] [PubMed] [Google Scholar]
- 44.Krishnakumari V, Packiyanathan KK, Nagaraj R. Human β-defensins-1-3 and analogs do not require proton motive force for antibacterial activity against Escherichia coli. FEMS Microbiol Lett. 2013;348:52–57. 10.1111/1574-6968.12242 [DOI] [PubMed] [Google Scholar]
- 45.Sharma H, Nagaraj R. Antimicrobial activity of human β-defensin 4 analogs: insights into the role of disulfide linkages in modulating activity. Peptides. 2012;38:255–265. 10.1016/j.peptides.2012.08.024 [DOI] [PubMed] [Google Scholar]
- 46.Brogden KA. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol. 2005;3:238–250. 10.1038/nrmicro1098 [DOI] [PubMed] [Google Scholar]
- 47.Selsted ME, Szklarek D, Lehrer RI. Purification and antibacterial activity of antimicrobial peptides of rabbit granulocytes. Infect Immun. 1984;45:150–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Viljanen P, Koski P, Vaara M. Effect of small cationic leukocyte peptides (defensins) on the permeability barrier of the outer membrane. Infect Immun. 1988;56:2324–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bahar AA, Ren D. Antimicrobial Peptides. Pharmaceuticals. 2013;6:1543–1575. 10.3390/ph6121543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alves CS, Melo MN, Franquelim HG, Ferre R, Planas M, Feliu L, et al. Escherichia coli cell surface perturbation and disruption induced by antimicrobial peptides BP100 and pepR. J Biol Chem. 2010;285:27536–27544. 10.1074/jbc.M110.130955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sharma H, Mathew B, Nagaraj R. Engineering of a linear inactive analog of human β-defensin 4 to generate peptides with potent antimicrobial activity. J Pept Sci. 2015;21:501–511. 10.1002/psc.2770 [DOI] [PubMed] [Google Scholar]
- 52.Mathew B, Nagaraj R. Antimicrobial activity of human α-defensin 6 analogs: insights into the physico-chemical reasons behind weak bactericidal activity of HD6 in vitro. J Pept Sci. 2015;21:811–818. 10.1002/psc.2821 [DOI] [PubMed] [Google Scholar]
- 53.Chu H, Pazgier M, Jung G, Nuccio SP, Castillo PA, de Jong MF, et al. Human α-defensin 6 promotes mucosal innate immunity through self-assembled peptide nanonets. Science. 2012;337:477–481. 10.1126/science.1218831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schroeder BO, Wu Z, Nuding S, Groscurth S, Marcinowski M, Beisner J, et al. Reduction of disulphide bonds unmasks potent antimicrobial activity of human β-defensin 1. Nature. 2011;469:419–423. 10.1038/nature09674 [DOI] [PubMed] [Google Scholar]
- 55.Wang C, Shen M, Gohain N, Tolbert WD, Chen F, Zhang N, et al. Design of a potent antibiotic peptide based on the active region of human defensin 5. J Med Chem. 2015; 58;3083–3093. 10.1021/jm501824a [DOI] [PubMed] [Google Scholar]
- 56.Lebaron P, Catala P, Parthuisot N. Effectiveness of SYTOX Green stain for bacterial viability assessment. Appl Environ Microbiol. 1998;64:2697–2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lynn DJ, Lloyd AT, Fares MA, O'Farrelly C. Evidence of positively selected sites in mammalian α-defensins. Mol Biol Evol. 2004;21:819–827. 10.1093/molbev/msh084 [DOI] [PubMed] [Google Scholar]
- 58.Das S, Nikolaidis N, Goto H, McCallister C, Li J, Hirano M, et al. Comparative genomics and evolution of the α-defensin multigene family in primates. Mol Biol Evol. 2010;27:2333–2343. 10.1093/molbev/msq118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kagan BL, Selsted ME, Ganz T, Lehrer RI. Antimicrobial defensin peptides form voltage-dependent ion-permeable channels in planar lipid bilayer membranes. Proc Natl Acad Sci U S A. 1990;87:210–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fujii G, Selsted ME, Eisenberg D. Defensins promote fusion and lysis of negatively charged membranes. Protein Sci. 1993;2:1301–1312. 10.1002/pro.5560020813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang Y, Lu W, Hong M. The membrane-bound structure and topology of a human α-defensin indicate a dimer pore mechanism for membrane disruption. Biochemistry. 2010;49:9770–9782. 10.1021/bi101512j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Poon I, Baxter AA, Lay FT, Mills GD, Adda CG, Payne JA, et al. Phosphoinositide-mediated oligomerization of a defensin induces cell lysis. eLife. 2014;3:e01808 10.7554/eLife.01808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chairatana P, Nolan EM. Molecular basis for self-assembly of a human host-defense peptide that entraps bacterial pathogens. J Am Chem Soc. 2014;136:13267–13276. 10.1021/ja5057906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wommack AJ, Robson SA, Wanniarachchi YA, Wan A, Turner CJ, Wagner G, et al. NMR solution structure and condition-dependent oligomerization of the antimicrobial peptide human defensin 5. Biochemistry. 2012;51:9624–9637. 10.1021/bi301255u [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rajabi M, Ericksen B, Wu X, de Leeuw E, Zhao L, Pazgier M, et al. Functional determinants of human enteric α-defensin HD5: crucial role for hydrophobicity at dimer interface. J Biol Chem. 2012;287:21615–21627. 10.1074/jbc.M112.367995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pazgier M, Wei G, Ericksen B, Jung G, Wu Z, de Leeuw E, et al. Sometimes it takes two to tango: contributions of dimerization to functions of human α-defensin HNP1 peptide. J Biol Chem. 2012;287:8944–8953. 10.1074/jbc.M111.332205 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Arrows indicate the accumulation of SYTOX green. Numbers given in the upper left corner represent the elapsed time (h:min:s:ms).
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.