Abstract
The metabolism of poly(ADP-ribose) (PAR) is critical for genomic stability in multicellular eukaryotes. Here, we show that the failure to degrade PAR by means of disruption of the murine poly(ADP-ribose) glycohydrolase (PARG) gene unexpectedly causes early embryonic lethality and enhanced sensitivity to genotoxic stress. This lethality results from the failure to hydrolyze PAR, because PARG null embryonic day (E) 3.5 blastocysts accumulate PAR and concurrently undergo apoptosis. Moreover, embryonic trophoblast stem cell lines established from early PARG null embryos are viable only when cultured in medium containing the poly(ADP-ribose) polymerase inhibitor benzamide. Cells lacking PARG also show reduced growth, accumulation of PAR, and increased sensitivity to cytotoxicity induced by N-methyl-N′-nitro-N-nitrosoguanidine and menadione after benzamide withdrawal. These results provide compelling evidence that the failure to degrade PAR has deleterious consequences. Further, they define a role for PARG in embryonic development and a protective role in the response to genotoxic stress.
Keywords: poly(ADP-ribose) glycohydrolase, poly(ADP-ribose) polymerase, apoptosis
The metabolism of polyadenosine 5′-diphosphoribose [poly(ADP-ribose)] is a unique and ubiquitous biochemical pathway in multicellular organisms. Poly(ADP-ribose) (PAR) is synthesized by the PAR polymerase (PARP) family of enzymes as an immediate response to DNA damage using nicotinamide adenine dinucleotide (NAD+) as substrate to covalently modify protein acceptors (1). The transient nature of PAR is due to PAR glycohydrolase (PARG), a unique enzyme that catalyzes the hydrolysis of PAR into free ADP-ribose. Because PARP activation results in the modification of nuclear proteins, including chromatin structural proteins and DNA processing enzymes, PAR plays a role in DNA repair and replication (2, 3). In addition, PAR facilitates proper chromosome migration during cell division (4), regulates transcriptional activation after DNA damage (5), activates the proteasome (6), regulates telomere maintenance (7), and mediates cell death (8–10). Therefore, the emerging biological role for this biopolymer is to maintain genomic integrity (11).
Poly(ADP-ribosyl)ation plays an important role in cell death, primarily through the actions of PARP-1, the best understood PARP. Cellular injury activates PARP-1 by means of DNA strand breaks, and it modifies many nuclear proteins by using NAD+ as substrate, which leads to the consumption of cellular energy stores (10, 12). Animals treated with PARP inhibitors or mice lacking the PARP-1 gene are dramatically resistant to damage induced by myocardial infarction, cerebral ischemia, genotoxic stress, diabetic models, inflammation, and the 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) model of Parkinson's disease (8–10). Recent observations link PARP-1 activation with the translocation of apoptosis-inducing factor (AIF) to the nucleus (13) and suggest that AIF is an essential downstream effector of PARP-1-mediated cell death.
Poly(ADP-ribosyl)ation is dynamically regulated (2). There are at least 18 proposed PARP family members (14), but only one PARG has been identified to date (2). The PARG cDNA encodes a 110-kDa protein largely localized to the cytosol (15), and only a single PARG mRNA species has been identified (16). The single PARG gene encodes probably three cDNAs, which generate three PARG isoforms: the nuclear form, a 110-kDa form, and two cytoplasmic forms of 103 and 99 kDa, which are the most abundant (17, 18). Although PARG has a low cellular abundance, it has a high specific activity. Therefore, it is thought that PARG is always active, because the nuclear PAR concentration is approximately one to two times the KM of PARG (19). Photoderivatization studies using specific and selective inhibitors of bovine PARG suggest that Leu-771-Arg-801 contains the active site (20, 21), with Tyr-796 the likely residue for initial PAR binding (22). A detailed analysis of the biological role of PARG has been hindered by the lack of specific and selective cell-permeable inhibitors. Recently, the depletion of the 110-kDa full-length PARG isoform resulted in increased sensitivity to genotoxic stress in mice (23). PARG activity remained in these mice, however, due to the presence of alternatively spliced isoforms.
We have developed a complete PARG knockout to investigate the biological importance of PARG in PAR metabolism. Mice lacking PARG arrest during development at embryonic day (E) 3.5, indicating an essential role for PARG during mammalian embryogenesis. PARG null embryos and embryonic trophoblast stem (TS) cells accumulate PAR and undergo cell death. Moreover, PARG deficiency leads to enhanced cell death after genotoxic stress. Taken together, the results clearly demonstrate the deleterious consequences that occur due to the failure to degrade PAR and suggest that PARG is the major enzyme required to catalyze the hydrolysis of PAR.
Methods
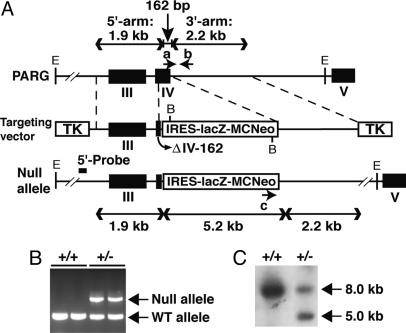
Gene Targeting and Generation of Knockout Mice. Two independent genomic λ phage clones were isolated from a 129/SvEv genomic library, and the murine PARG gene was sequenced and used as template for the targeting vector. A 1.9 kb of 5′-flanking sequence from exon 3 to the first 10 bp of exon 4 and a 2.2 kb of 3′-flanking region consisting of partial intron 5 sequence were amplified and, after sequence confirmation, ligated with an IRES-lacZ-MCneomycin cassette and a λ phage backbone harboring two thymidine kinase markers flanking the 5′ and 3′ arms of homology (Fig. 1). The final construct was verified by restriction digestion, PCR, and partial sequence determination. The linearized targeting vector was electroporated into mouse 129 SVJ embryonic stem (ES) cells. ES clones resistant to G418 and gancyclovir were selected and screened for the targeted PARG allele with an external 5′ probe and internal 3′ probe by Southern hybridization. One ES clone was injected into C57BLG/J blastocysts, mice carrying the targeted allele in the germ line were established, and PARG+/– mice were mated to generate PARG–/– embryos.
Fig. 1.
Targeted disruption of the PARG gene. (A) Restriction maps of the PARG target locus, targeting construct, and targeting allele. The targeting vector contains a total of 4.1 kb of PARG genomic sequence with the lacZ-Neo selection cassette replacing a 162-bp region in exon IV. a, oligonucleotide primer specific for WT allele; b, common primer for WT and targeted alleles; c, primer specific for targeted allele; 5′-probe, 258-bp external DNA probe for Southern blot (C); black boxes, exons; TK, thymidine kinase marker; E, EcoRI; B, BamHI restriction enzyme sites; Δ IV-162, exon IV missing the 3′ 162-bp terminus. (B) Identification of PARG+/+ and PARG+/– mice by competitive PCR assay. WT and null indicate PCR products amplified from the wild-type and targeted PARG alleles, respectively. (C) Identification of PARG+/+ and PARG+/– mice by Southern blot. WT (8.0-kb) and/or mutant (5.0-kb)-specific bands from adult mouse brain genomic DNA were obtained with the 5′-probe (EcoRI cut). These results (B and C) have been replicated at least three times with identical results.
Analyses of Marker Gene Expression. Targeted mice were screened by Southern blot as above to detect the WT and targeted alleles. For PCR of mice, embryos, and TS cells, one primer (5′-TAATGGAGAATGCTCTGAAGG) common to the targeted and WT allele and two specific to the WT (5′-AGTCACACAGTGACTGTTCGG) or targeted allele (5′-GTGGCGGACCGCTATCA) were combined in a reaction, and products were analyzed by agarose gel. For RT-PCR, total RNA was isolated by using TRIzol (Invitrogen), and cDNA was synthesized with SuperScript III Reverse Transcriptase (Invitrogen). Five microliters of cDNA was template in a PCR using the following primer pairs: PARG forward (5′-ACGCCACCTCGTTTGTTTTC), reverse (5′-GCAGTCTGCTGTTGGTTCAAAT) for Exon IV; PARG forward (5′-TTTCCAAGTCAGAGGACAGAAGAAA), reverse (5′-TCATCTTCTGTTTCAGGAGTGGTAT) for Exons III-IX; B2m forward (5′-ATACATACGCC-TGCAG), reverse (5′-TCAAATGAATCTTCAG).
Mouse Embryo Analysis. Embryos from PARG+/– mouse intercrosses were isolated and photographed by using a dissection microscope. Gestational age (±0.5 days) was estimated from vaginal plug dates. For the E3.5 stage, embryos were flushed from the uterus with M16 media, cultured in TS cell growth media [50% MEF conditioned media/15% FCS/DMEM/1 mM sodium pyruvate/2 mM l-glutamine/50 units/ml penicillin/streptomycin/0.1 mM monothioglycerol/25 ng/ml human recombinant FGF-4 (Sigma)/1 μg/ml heparin] and photographed under ×20 objective. Whole (E3.5) or pieces (>E3.5) of embryos were used for genotyping by PCR. For detection of apoptosis, E3.5 embryos were fixed in 4% paraformaldehyde for 20 min, permeabilized in 0.4% Triton X-100 for 15 min, washed three times in PBS plus 1 mg/ml polyvinylpyrrolidine for 2 min, and TUNEL assayed by using the In Situ Cell Death Detection Kit, Fluorescein (Roche). Embryos were counterstained with 1 mg/ml Hoechst 33342 (Molecular Probes) for 15 min to reveal nuclear morphology.
Immunohistochemistry and Fluorescence Microscopy. E3.5 embryos from PARG+/– intercrosses were washed in PBS, and fixed in 4% paraformaldehyde for 10 min, and, after blocking with 10% BSA in PBS, cells were probed with antibody against PAR (clone 96-10, 1:1,000 dilution) overnight at 4°C. Cells were washed with blocking solution three times and incubated with Cy3-conjugated secondary antibody (Jackson ImmunoResearch; 1:200 dilution) for 1 h. Microscopy was performed on a Nikon inverted fluorescence microscope by using the ×20 objective.
Embryonic TS Cell Cultures. PARG TS cells were derived from E3.5 embryos as described (24), except that embryos were cultured in TS cell growth media containing 0.5 mM benzamide (BZ, Sigma). After 6 days, embryonic outgrowths were repassaged with a portion of cells used for genotyping. PARG TS cell growth was determined by staining cultures with Hoechst and propidium iodide (PI) as described (25) and by counting dye-impermeable cells.
Induction and Assessment of Cytotoxicity. For detection of apoptosis, TS cells were grown with or without 0.5 mM BZ in TS cell growth medium for 3 days, harvested by trypsinization, washed with ice-cold PBS, stained with annexin V-FITC/PI (Vybrant Apoptosis Assay Kit No. 3, Molecular Probes), and analyzed by using a FACSCalibur flow cytometer (Becton Dickinson). For the induction of cell death, cultured TS cells were grown without BZ for 3 days, treated with N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) and menadione for 5 min, washed with PBS and growth medium, then incubated at 37°C. Cytotoxicity was assessed 24 h later by Hoechst/PI staining. From among a total of 200–1,200 cells per culture, dye-permeable and impermeable cells were counted.
Protein Extract and Immunoblots. Cells were lysed in buffer containing 10 mM Tris·HCl (pH 8.0), 0.3 M NaCl, 6 M urea, 1 mM EDTA and EGTA, and protease inhibitor tablets (Roche). The suspension was cleared by centrifugation (9,000 × g, 10 min, 4°C), and subjected to 7.5% SDS/PAGE, and the separated proteins were probed by polyclonal antibodies raised against PAR (clone 96-10) (26), monoclonal antibodies against TIM23 (1:2,500 dilution; Becton Dickinson), and polyclonal antibodies against amino acids 706–978 of murine PARG (M.F.P., V.L.D., and T.M.D., unpublished results) by using conditions described (26).
Results
Targeted Disruption of the Murine PARG Gene. The murine gene encoding PARG was cloned from a genomic library derived from 129/SvEv mice. Exon 4 was targeted and replaced with an IRES-lacZ-MCneomycin selection cassette (27). In the targeted allele, the selection cassette replaced the 3′ portion of exon 4, thereby creating a functional null allele (Fig. 1 A). Two embryonic stem clones containing the targeted PARG allele were isolated, and one independent line of genetically modified mice was generated from these clones. Genotypes were determined by PCR (Fig. 1B) and confirmed by Southern blot by using an oligonucleotide probe of 258 bases (Fig. 1C).
Loss of PARG Causes Peri-Implantation Lethality, Increased Amounts of PAR, and Apoptosis. Mice heterozygous for the target allele (PARG+/–) are viable and fertile, with no obvious abnormalities. However, no homozygous null mice (PARG–/–) are observed among offspring of PARG+/– intercrosses. To determine the gestational time of PARG lethality, we examined embryos from PARG+/– intercrosses at different developmental stages. No PARG–/– animals are observed as early as E6.5 (Table 1), which suggests that the PARG–/– embryos die before gastrulation. We then isolated E3.5 embryos resulting from PARG+/– intercrosses (24). In the first analysis of three litters (19 embryos), 79% of the blastocysts hatched whereas 21% failed to hatch (Table 2). None of the hatched and plated blastocysts are PARG–/–. The unhatched embryos could not be genotyped, probably due to cell death and genomic degradation after >72 h in culture. However, the unhatched embryo percentage (21%) corresponded to the expected Mendelian percentage of null embryos (25%), which suggests that the inability to hatch or develop past this stage could result from the disruption of PARG. An additional four litters were examined, genotyping was immediately performed, and all unhatched embryos were PARG–/– blastocysts (Table 2).
Table 1. The PARG mutation is embryonic lethal.
| Stage | Total | +/+ | +/- | -/- |
|---|---|---|---|---|
| Newborn | 279 | 96 | 183 | 0 |
| E13.5 | 9 | 1 | 8 | 0 |
| E11.5 | 12 | 4 | 8 | 0 |
| E10.5 | 17 | 5 | 12 | 0 |
| E8.5 | 7 | 1 | 6 | 0 |
| E6.5 | 30* | 7 | 22 | 0 |
Embryos were isolated at the indicated time of gestation and analyzed for viability. Genotypes of embryos were determined by PCR.
One embryo was not successfully assayed.
Table 2. Analysis of E3.5 blastocysts from intercrosses of PARG+/- mice.
| Hatched
|
Unhatched
|
||||||
|---|---|---|---|---|---|---|---|
| Litters | Total blastocysts | Unhatched/total blastocysts, % | PARG+/+ | +/- | ND | -/- | ND |
| 1-3 | 19* | 21 | 3 | 9 | 2 | 0 | 4 |
| 4-7 | 28 | 21 | 6 | 16 | 0 | 6 | 0 |
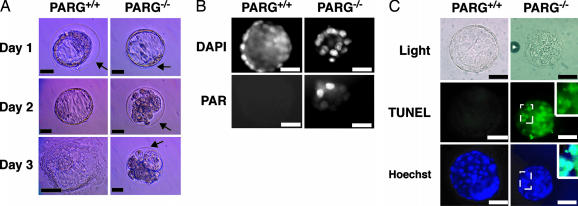
Microscopic inspection of each embryo reveals no gross morphologic defects or anomalies initially, but at the time of hatching of the WT blastocysts, the PARG–/– blastocysts are unhatched and the embryos begin to degenerate (Fig. 2A). At the time of plating of the WT blastocysts, PARG–/– blastocysts remain unhatched and continue to degenerate (Fig. 2 A). We then analyzed blastocysts that displayed these morphological differences for PAR staining (Fig. 2B) and cell death by TUNEL assay (Fig. 2C). Immunohistochemistry by using anti-PAR antibody showed that the degenerated, unhatched E3.5 blastocysts contained high levels of PAR as compared with WT, which contain no detectable PAR after 48 h (Fig. 2B). Further, the degenerated, unhatched E3.5 blastocysts positively stain for DNA fragmentation by means of the TUNEL assay, which indicates that the cells are apoptotic, whereas WT embryos have no detectable signal (Fig. 2C). TUNEL staining in the PARG–/– embryos was colocalized to the nucleus with Hoechst staining, which further verified the in situ cell death assay. Taken together, these results indicate that the disruption of PARG leads to peri-implantation lethality, which results in the failure of the embryo to hatch and the degeneration of the blastocyst. The apparent cause of this lethality is an accumulation of PAR, which leads to cell death by apoptosis.
Fig. 2.
Impaired ability to hatch, increased levels of PAR, and cell death by apoptosis in PARG–/– embryos. (A) Microscopic analysis of E3.5 PARG+/+ and PARG–/– embryos up to 72 h in culture. Arrows indicate an intact zona pellucida. (B) Fluorescence microscopic analysis of PAR levels in E3.5 embryos 24 h after culture. Embryos displaying the hatched/unhatched phenotype were analyzed for PAR levels. DAPI, 4′,6-diamidino-2-phenylindole. (C) Fluorescent immunostaining for TUNEL (Middle) and Hoechst 33342 (Bottom) in E3.5 embryos displaying the hatched/unhatched phenotype 24 h after culture. (Middle Right and Bottom Right Insets) Magnification of the area in the embryo indicated by the dotted box. The Bottom Right Inset represents the overlay of the TUNEL and Hoechst stained area. (Scale bars: 25 μm.) All photographs are representative of several analyzed blastocysts (Table 2).
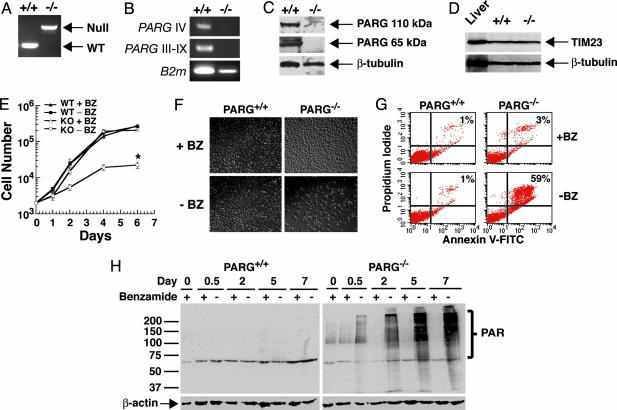
Decreased Proliferation, Accumulation of PAR, and Cell Death in PARG–/– Embryonic TS Cells. Because genetic inactivation of PARG resulted in embryonic lethality, we analyzed the role of PARG activity and PAR signaling in cells derived from WT and PARG–/– embryos. The ability of PARG–/– embryos to survive to at least E3.5 made it possible to culture embryonic TS cells from living embryos of both the PARG+/+ and PARG–/– genotypes (Fig. 3 and data not shown). Initial attempts at culturing PARG–/– cells were unsuccessful (data not shown) until the broad spectrum PARP inhibitor BZ was added to the cultures (Fig. 3). Under these conditions, we successfully established several lines of PARG–/– trophoblast cultures (Fig. 3). PCR analysis of the PARG–/– vs. PARG+/+ TS cells indicates that the PARG gene was successfully targeted in the PARG–/– TS cells (Fig. 3A). RT-PCR analysis of PARG–/– vs. PARG+/+ TS cell total RNA using primers that detect all reported splice variants of PARG reveals the complete absence of PARG mRNA in PARG–/– TS cells (Fig. 3B). There is also complete absence of PARG 110-kDa and 65-kDa protein in PARG–/– TS cells (Fig. 3C). Because the murine PARG gene shares a promoter with the mitochondrial inner translocase-23 (TIM23) gene (28), we examined TIM23 protein levels in both WT and PARG null TS cells and found that our knockout strategy had no effect on the expression levels of TIM23 (Fig. 3D). Growth analysis demonstrates the dependence of PARG–/– TS cells on BZ, because the PARG–/– TS cells remain viable and grow normally only when the PARP inhibitor is present. When BZ is withdrawn, the PARG–/– TS cells fail to exhibit normal logarithmic growth (Fig. 3 E and F). The withdrawal of BZ causes abnormal morphology with condensed nuclei in the PARG–/– TS cells after 6 days (Fig. 3F). The reduced proliferation in PARG–/– TS cells was further analyzed by FACS. After 3 days without BZ, ≈60% of the PARG–/– TS cells are apoptotic due to positive staining for PI and annexin-FITC (Fig. 3G), with no significant cell death in WT TS cells or PARG–/– TS cells treated with BZ. Analysis of PAR levels by Western blot reveals that PAR gradually accumulates in PARG–/– TS cells in the absence of BZ, and maximal PAR levels are observed 5–7 days after BZ withdrawal (Fig. 3H). These results suggest that the failure to degrade PAR leads to cell death by apoptosis. Furthermore, the time course of cell death in PARG–/– TS cells after BZ withdrawal is consistent with the timing of lethality in PARG–/– embryos, suggesting that functional PARG protein is required to maintain the rapid cellular proliferation of normal embryogenesis.
Fig. 3.
Decreased proliferation, increased cell death, and PAR accumulation in PARG–/– trophoblasts. (A) Identification of PARG+/+ and PARG–/– TS cells by competitive PCR assay. (B) RT-PCR assay of total RNA isolated from PARG+/+ and PARG–/– TS cells by using primers specific for Exons IV (PARG IV) and III-IX (PARG III-IX). (C) Immunoblot analysis of full-length (110-kDa) and catalytic fragment (65-kDa) PARG. (D) TIM23 protein levels in PARG+/+ and PARG–/– TS cells. Mouse liver lysate (Becton Dickinson) was used as positive control. (E and F) Decreased proliferation of PARG–/– TS cells in the absence of the PARP inhibitor BZ, and rescue of the growth defect with the addition of BZ. (E) *, P ≤ 0.01 between PARG–/– with or without BZ to WT with or without BZ (one-way ANOVA and unpaired Student's t test). (F) Photographs represent microscopic analysis of PARG+/+ and PARG–/– TS cells with or without BZ for 3 days. (G) Increased cell death in PARG–/– TS cells in the absence of BZ. TS cells were grown 3 days with or without BZ before determination of cell death by annexin-V-FITC/PI staining. (H) Increased PAR levels in PARG–/– TS cells. PAR levels were determined by immunoblot of PARG+/+ and PARG–/– TS cells grown with or without BZ for up to 7 days. These results (A–H) have been replicated at least twice with similar results. In E, error bars are shown as standard error of the mean (SEM).
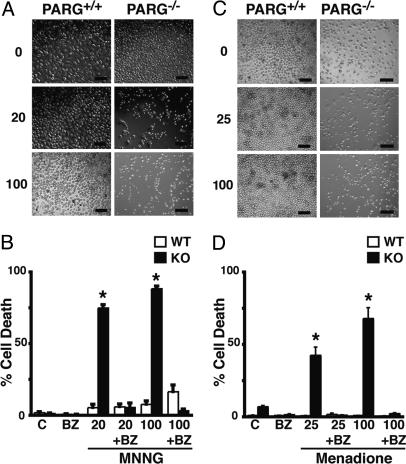
Increased Sensitivity of Cells Lacking PARG to PARP-1-Dependent Cytotoxicity. We next analyzed the susceptibility of PARG–/– TS cells to death stimuli. WT and PARG–/– TS cells were subjected to different types of treatment that activate the PARP-1-dependent cell death pathway in many cell types (Fig. 4) (29, 30). Cell death was quantified by Hoechst/PI staining (25). MNNG induces massive cell death in PARG–/– TS cells in the absence of BZ under conditions in which WT TS cells are relatively spared from the toxic effects of MNNG (Fig. 4 A and B). Consistent with the enhancement of PARP-1-dependent cell death, PARG–/– TS cells exhibit abnormal morphology, condensed nuclei, and a high level of cell death after MNNG treatment, which is reversed by the broad spectrum PARP inhibitor BZ (Fig. 4B). Oxidative stress-induced injury by menadione is also enhanced in PARG–/– TS cells compared with WT TS cells (Fig. 4 C and D). At these concentrations of menadione, which have minimal effects on WT TS cells, PARG–/– TS cells are dramatically sensitive to the toxic effects with abnormal cell morphology, nuclear condensation, and cell death. The PARP dependence of menadione-induced cell death is confirmed by the reversal of cell death by the broad spectrum PARP inhibitor BZ (Fig. 4D). Taken together, these results indicate that the PARG–/– TS cells have increased sensitivity to PARP-1-dependent cell death that is due to the failure to catabolize PAR.
Fig. 4.
Increased sensitivity to PARP-1-dependent cytotoxicity in PARG–/– TS cells. (A and C) Microscopic analysis of PARG+/+ and PARG–/– TS cells grown with or without BZ for 3 days were examined 24 h after 5 min treatment of 0, 20, or 100 μM MNNG (A), or 0, 25, or 100 μM menadione (C). (B and D) Cell death in PARG+/+ and PARG–/– TS cells. Cells were treated with MNNG (B) or menadione (D) as described above, and quantification of total cell death was performed by using Hoechst/PI staining. Numbers on the vertical axes (A and C) depict the doses of the cell death stimuli in micromolar. (Scale bars: 25 μm.) These results (A–D) have been replicated at least three times with similar results. (B and D) *, P ≤ 0.001 between PARG–/– with or without BZ to WT with or without BZ (one-way ANOVA and unpaired Student's t test). (B and D) Error bars are shown as SEM.
Discussion
The major findings of this report indicate that PARG plays a crucial role in mouse development and that the embryonic lethality in PARG null mutants results from the failure to degrade PAR polymer, which leads to cell death by apoptosis. PARG null TS cells are viable only in the presence of the broad spectrum PARP inhibitor BZ, indicating that the failure to degrade PAR results in embryonic lethality. Attempts were made to administer BZ to pregnant PARG heterozygous mice throughout the pregnancies, but no PARG null embryos were obtained (data not shown). This report also demonstrates that PARG null TS cells are remarkably sensitive to PARP-1-dependent cell death. This cell death seems to be predominantly apoptotic, but nonapoptotic mechanisms cannot be ruled out because necrotic and nonapoptotic mechanisms occur in PARP-1-dependent cell death. Our PARG gene disruption approach did not affect the expression of TIM23, the gene that shares a promoter with PARG (28). Thus, the specific disruption of PARG leads to embryonic lethality and increased susceptibility to PARP-1-dependent cell death. The ability of mice lacking full-length PARG to survive and reproduce reported elsewhere (23) is likely due to the presence of shorter PARG isoforms and the subsequent capacity to degrade PAR in these animals.
PARP activity plays an important role in modulating the cellular response to stress. The extent of poly(ADP-ribosyl)ation correlates with the severity of stress, and this phenomenon determines the cellular response. Poly(ADP-ribosyl)ation under severe stress leads to cell death, under moderate stress it plays important roles in DNA repair, and under mild stress it participates in the proinflammatory/cellular defense by means of transcriptional regulation (31). Inhibition of poly(ADP-ribosyl)ation and knockout of PARP-1 or PARP-2 alters the stress response at multiple levels (9, 10, 14, 32, 33), but normal cellular physiology is not affected to any appreciable degree, implying a very limited role for poly(ADP-ribosyl)ation in the absence of stress (31). The embryonic lethality of the PARG null mutant establishes a role for poly(ADP-ribosyl)ation in embryonic development, and it suggests that poly(ADP-ribosyl)ation is important in homeostatic cellular functions. Along these lines is the observation that the loss of PARG in Drosophila leads to lethality in larval stage at the normal developmental temperature (25°C). One-quarter of larva exposed to 29°C hatch, but develop progressive neurodegeneration, reduced locomotor activity, and a short lifespan. These animals accumulate PAR polymer (34), which may contribute to this phenotype.
The lethality of the PARG–/– mice indicates that the regulation of PAR levels is critical for cell survival and that there is little, if any, compensatory mechanism for the catabolism of PAR. The inactivation of cellular PARG activity leads to increases in PAR that are correlated with growth arrest and cell death. Consistent with this notion are observations in Saccharomyces, which are normally devoid of PARP and PARG activity. Overexpression of PARP-1 or PARP-2 in Saccharomyces leads to growth inhibition (35), but coexpression of PARP-1 or PARP-2 with PARG restores growth (36). Thus, accumulation of PAR is toxic to cells, and increased levels of PAR in embryonic tissue results in apoptosis. Moreover, the accumulation of PAR in PARG–/– TS cells in the absence of BZ demonstrates that PARG is responsible for most, if not all, of the hydrolysis of PAR. Therefore, our findings suggest that PARG is the primary enzyme that degrades PAR.
Knockdown of PARG was previously shown to increase sensitivity of mice to genotoxic stress (23). Our results also identify a protective role for PARG in the cellular response to DNA damage, but it also suggests that the complete absence of PARG creates a scenario where the cell is unable to withstand limited DNA damage because the doses of MNNG or menadione had little effect on WT TS cells but induced substantial cell death in PARG–/– TS cells. The enhanced cell death in the PARG–/– TS cells is PARP dependent because it can be reversed by the broad spectrum PARP inhibitor BZ. Thus, PARG activity is protective, and it is required for the cellular response to even sublethal levels of stress.
The exact mechanism of the toxicity resulting from the accumulation of PAR is not known. The failure to remove PAR from either covalently modified proteins or noncovalently bound PAR may lead to growth arrest and cell death, because many PAR-modified proteins are important for cell survival and DNA processing/repair (37–41). Recent evidence suggests that poly(ADP-ribosyl)ation regulates transcription (5) and that the failure to degrade PAR polymer may lead to transcriptional dysregulation. The absence of PARG activity leads to transcriptional dysregulation and altered circadian period length in Arabidopsis (42), consistent with the notion that PAR plays an important role in homeostatic cellular functions. Alternatively, PAR polymer may conceivably act as a death signal. Thus, the accumulation of PAR may directly and/or indirectly lead to cell death.
In conclusion, our study reveals that PARG plays a fundamental role in embryogenesis and that it plays a role as a cytoprotective protein against genotoxic and oxidative-induced cellular injury by regulating poly(ADP-ribosyl)ation. Moreover, these results suggest that PARG is the primary enzyme capable of hydrolyzing PAR and that this PAR catabolism is required for the maintenance of normal cell function. Although inhibition of PARG is unlikely to be of benefit in the therapy and prevention of toxicity after cellular stress, potent and selective inhibitors of PARG may ultimately be used to enhance the effectiveness of cytotoxic cancer chemotherapeutic agents.
Acknowledgments
This work was supported by National Institutes of Health Grant NS39148 and by the Christopher Reeve Paralysis Foundation (to D.W.K.). T.M.D. is the Leonard and Madlyn Abramson Professor in Neurodegenerative Diseases.
Author contributions: D.W.K., S.W., G.G.P., V.L.D., and T.M.D. designed research; D.W.K., A.M.L., M.F.P., M.S., S.W., and T.S. performed research; D.W.K., A.M.L., M.S., S.W., M.C.N., T.S., G.G.P., V.L.D., and T.M.D. analyzed data; and D.W.K., V.L.D., and T.M.D. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: PAR, poly(ADP-ribose); PARG, PAR glycohydrolase; PARP, PAR polymerase; TS, trophoblast stem; BZ, benzamide; MNNG, N-methyl-N′-nitro-N-nitrosoguanidine; En, embryonic day n; PI, propidium iodide.
References
- 1.Juarez-Salinas, H., Sims, J. L. & Jacobson, M. K. (1979) Nature 282, 740–741. [DOI] [PubMed] [Google Scholar]
- 2.Amé, J. C., Jacobson, E. L. & Jacobson, M. K. (2000) in From DNA Damage and Stress Signaling to Cell Death: Poly-ADP-Ribosylation Reactions, eds. de Murcia, G. & Shall, S. (Oxford Univ. Press, New York), pp. 1–34.
- 3.Dantzer, F., de La Rubia, G., Menissier-De Murcia, J., Hostomsky, Z., de Murcia, G. & Schreiber, V. (2000) Biochemistry 39, 7559–7569. [DOI] [PubMed] [Google Scholar]
- 4.Shall, S. & de Murcia, G. (2000) Mutat. Res. 460, 1–15. [DOI] [PubMed] [Google Scholar]
- 5.Hassa, P. O. & Hottiger, M. O. (1999) Biol. Chem. 380, 953–959. [DOI] [PubMed] [Google Scholar]
- 6.Ullrich, O., Reinheckel, T., Sitte, N., Hass, R., Grune, T. & Davies, K. J. (1999) Proc. Natl. Acad. Sci. USA 96, 6223–6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith, S., Giriat, I., Schmitt, A. & de Lange, T. (1998) Science 282, 1484–1487. [DOI] [PubMed] [Google Scholar]
- 8.Yu, S. W., Wang, H., Dawson, T. M. & Dawson, V. L. (2003) Neurobiol. Dis. 14, 303–317. [DOI] [PubMed] [Google Scholar]
- 9.Southan, G. J. & Szabo, C. (2003) Curr. Med. Chem. 10, 321–340. [DOI] [PubMed] [Google Scholar]
- 10.Szabo, C. & Dawson, V. L. (1998) Trends Pharmacol. Sci. 19, 287–298. [DOI] [PubMed] [Google Scholar]
- 11.Jacobson, M. K. & Jacobson, E. L. (1999) Trends Biochem. Sci. 24, 415–417. [DOI] [PubMed] [Google Scholar]
- 12.Ha, H. C. & Snyder, S. H. (1999) Proc. Natl. Acad. Sci. USA 96, 13978–13982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang, H., Yu, S. W., Koh, D. W., Lew, J., Coombs, C., Bowers, W. J., Federoff, H. J., Poirier, G. G., Dawson, T. M. & Dawson, V. L. (2004) J. Neurosci. 24, 10963–10973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Menissier-de Murcia, J., Ricoul, M., Tartier, L., Niedergang, C., Huber, A., Dantzer, F., Schreiber, V., Ame, J. C., Dierich, A., LeMeur, M., et al. (2003) EMBO J. 22, 2255–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winstall, E., Affar, E. B., Shah, R., Bourassa, S., Scovassi, I. A. & Poirier, G. G. (1999) Exp. Cell Res. 251, 372–378. [DOI] [PubMed] [Google Scholar]
- 16.Lin, W., Ame, J. C., Aboul-Ela, N., Jacobson, E. L. & Jacobson, M. K. (1997) J. Biol. Chem. 272, 11895–11901. [DOI] [PubMed] [Google Scholar]
- 17.Meyer-Ficca, M. L., Meyer, R. G., Coyle, D. L., Jacobson, E. L. & Jacobson, M. K. (2004) Exp. Cell Res. 297, 521–532. [DOI] [PubMed] [Google Scholar]
- 18.Bonicalzi, M. E., Vodenicharov, M., Coulombe, M., Gagne, J. P. & Poirier, G. G. (2003) Biol. Cell 95, 635–644. [DOI] [PubMed] [Google Scholar]
- 19.Davidovic, L., Vodenicharov, M., Affar, E. B. & Poirier, G. G. (2001) Exp. Cell Res. 268, 7–13. [DOI] [PubMed] [Google Scholar]
- 20.Koh, D. W., Patel, C. N., Ramsinghani, S., Slama, J. T., Oliveira, M. A. & Jacobson, M. K. (2003) Biochemistry 42, 4855–4863. [DOI] [PubMed] [Google Scholar]
- 21.Koh, D. W., Coyle, D. L., Mehta, N., Ramsinghani, S., Kim, H., Slama, J. T. & Jacobson, M. K. (2003) J. Med. Chem. 46, 4322–4332. [DOI] [PubMed] [Google Scholar]
- 22.Ramsinghani, S., Koh, D. W., Amé, J. C., Strohm, M., Jacobson, M. K. & Slama, J. T. (1998) Biochemistry 37, 7801–7812. [DOI] [PubMed] [Google Scholar]
- 23.Cortes, U., Tong, W. M., Coyle, D. L., Meyer-Ficca, M. L., Meyer, R. G., Petrilli, V., Herceg, Z., Jacobson, E. L., Jacobson, M. K. & Wang, Z. Q. (2004) Mol. Cell. Biol. 24, 7163–7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanaka, S., Kunath, T., Hadjantonakis, A. K., Nagy, A. & Rossant, J. (1998) Science 282, 2072–2075. [DOI] [PubMed] [Google Scholar]
- 25.Eliasson, M. J., Sampei, K., Mandir, A. S., Hurn, P. D., Traystman, R. J., Bao, J., Pieper, A., Wang, Z. Q., Dawson, T. M., Snyder, S. H. & Dawson, V. L. (1997) Nat. Med. 3, 1089–1095. [DOI] [PubMed] [Google Scholar]
- 26.Yu, S. W., Wang, H., Poitras, M. F., Coombs, C., Bowers, W. J., Federoff, H. J., Poirier, G. G., Dawson, T. M. & Dawson, V. L. (2002) Science 297, 259–263. [DOI] [PubMed] [Google Scholar]
- 27.Russ, A. P., Wattler, S., Colledge, W. H., Aparicio, S. A., Carlton, M. B., Pearce, J. J., Barton, S. C., Surani, M. A., Ryan, K., Nehls, M. C., et al. (2000) Nature 404, 95–99. [DOI] [PubMed] [Google Scholar]
- 28.Meyer, R. G., Meyer-Ficca, M. L., Jacobson, E. L. & Jacobson, M. K. (2003) Gene 314, 181–190. [DOI] [PubMed] [Google Scholar]
- 29.Jacobson, E. L., Smith, J. Y., Wielckens, K., Hilz, H. & Jacobson, M. K. (1985) Carcinogenesis 6, 715–718. [DOI] [PubMed] [Google Scholar]
- 30.Karczewski, J. M., Peters, J. G. & Noordhoek, J. (1999) Biochem. Pharmacol. 57, 19–26. [DOI] [PubMed] [Google Scholar]
- 31.Burkle, A. (2001) BioEssays 23, 795–806. [DOI] [PubMed] [Google Scholar]
- 32.Wang, Z. Q., Auer, B., Stingl, L., Berghammer, H., Haidacher, D., Schweiger, M. & Wagner, E. F. (1995) Genes Dev. 9, 509–520. [DOI] [PubMed] [Google Scholar]
- 33.de Murcia, J. M., Niedergang, C., Trucco, C., Ricoul, M., Dutrillaux, B., Mark, M., Oliver, F. J., Masson, M., Dierich, A., LeMeur, M., et al. (1997) Proc. Natl. Acad. Sci. USA 94, 7303–7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanai, S., Kanai, M., Ohashi, S., Okamoto, K., Yamada, M., Takahashi, H. & Miwa, M. (2004) Proc. Natl. Acad. Sci. USA 101, 82–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaiser, P., Auer, B. & Schweiger, M. (1992) Mol. Gen. Genet 232, 231–239. [DOI] [PubMed] [Google Scholar]
- 36.Perkins, E., Sun, D., Nguyen, A., Tulac, S., Francesco, M., Tavana, H., Nguyen, H., Tugendreich, S., Barthmaier, P., Couto, J., et al. (2001) Cancer Res. 61, 4175–4183. [PubMed] [Google Scholar]
- 37.Kanai, M., Tong, W. M., Sugihara, E., Wang, Z. Q., Fukasawa, K. & Miwa, M. (2003) Mol. Cell. Biol. 23, 2451–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Althaus, F. R., Bachmann, S., Hofferer, L., Kleczkowska, H. E., Malanga, M., Panzeter, P. L., Realini, C. & Zweifel, B. (1995) Biochimie 77, 423–432. [DOI] [PubMed] [Google Scholar]
- 39.Ohashi, S., Kanai, M., Hanai, S., Uchiumi, F., Maruta, H., Tanuma, S. & Miwa, M. (2003) Biochem. Biophys. Res. Commun. 307, 915–921. [DOI] [PubMed] [Google Scholar]
- 40.Ruscetti, T., Lehnert, B. E., Halbrook, J., Le Trong, H., Hoekstra, M. F., Chen, D. J. & Peterson, S. R. (1998) J. Biol. Chem. 273, 14461–14467. [DOI] [PubMed] [Google Scholar]
- 41.Simbulan-Rosenthal, C. M., Rosenthal, D. S., Luo, R. & Smulson, M. E. (1999) Cancer Res. 59, 2190–2194. [PubMed] [Google Scholar]
- 42.Panda, S., Poirier, G. G. & Kay, S. A. (2002) Dev. Cell 3, 51–61. [DOI] [PubMed] [Google Scholar]