Abstract
The rise of antimicrobial resistance limits therapeutic options for infections by methicillin-resistant staphylococci. The staphylococcal cassette chromosome mec (SCCmec) is a mobile genetic element as the only carrier of the methicillin-resistance determinants, the mecA or mecC gene. The use of antibiotics increases the spread of antibiotic resistance, but the mechanism by which antibiotics promote horizontal dissemination of SCCmec is largely unknown. In this study, we demonstrate that many antibiotics, including β-lactams, can induce the expression of ccrC1 and SCCmec excision from the bacterial chromosome. In particular, three widely used antibiotics targeting DNA replication and repair (sulfamethoxazole, ciprofloxacin and trimethoprim) induced higher levels of ccrC1 expression and higher rates of SCCmec excision even at low concentrations (1/8 × minimum inhibitory concentration). LexA was identified as a repressor of ccrC1 and ccrAB by binding to the promoter regions of ccrC1 and ccrAB. The activation of RecA after antibiotic induction alleviated the repression by LexA and increased the expression of ccrC1 or ccrAB, consequently increasing the excision frequency of the SCCmec for SCCmec transfer. These findings lead us to propose a mechanism by which antimicrobial agents can promote horizontal gene transfer of the mecA gene and facilitate the spread of methicillin resistance.
INTRODUCTION
Methicillin-resistant staphylococci (MRS) are major causes of bacterial infections in humans and animals. The mecA or mecC gene responsible for methicillin resistance is located on a mobile genomic island, the staphylococcal cassette chromosome mec, which is a major gene acquisition machine and is responsible for the horizontal dissemination of many resistance genes (1).
Coagulase-negative staphylococci (CoNS) may serve as gene reservoirs facilitating the conversion of methicillin-susceptible Staphylococcus aureus (MSSA) to methicillin-resistant S. aureus (MRSA) (2). The transmission of staphylococcal cassette chromosome mec (SCCmec) from MR-CoNS to MSSA has been proposed based on the following: (i) the original source of the mecA gene appears to be S. sciuri or S. fleurettii (3,4); (ii) the IS1272 element inserted in SCCmec is observed much more frequently in Staphylococcus haemolyticus than in S. aureus (5) and (iii) methicillin resistance is more frequent among CoNS than S. aureus (6,7). Although there is no evidence for SCCmec transfer from MRSA to CoNS, the restriction of the SCCmec element to the Staphylococcus genus and the homology of SCCmec DNA sequences between different staphylococcal species highly suggest that the transfer of SCCmec among different staphylococci is possible (8).
As a mobile genetic element, SCCmec integrates and excises from a unique location in the staphylococcal chromosome within the 3΄ end of the highly conserved ribosomal methyltransferase gene orfX, which is also known as rlmH (9,10). For the movement, SCCmec carries specific gene complexes containing the cassette chromosome recombinase genes AB or C (ccrAB or ccrC), which encode proteins belonging to the large serine recombinase family and recognizing the specific recombination sites of SCCmec (11,12). The ccr genes with nucleotide identities of <50% are distinguished as distinct types. Within ccrA and ccrB types, allotypes ccrA1–7 and ccrB1–6 have been characterized based on their nucleotide identities (>85% within each allotype). In the ccrC type, only two allotypes have been identified: ccrC1 and ccrC2 (8,13). Eleven alleles of ccrC1 have been assigned (14). The Ccr proteins mediate integration and excision of the SCCmec into and from the chromosome. The integration of SCCmec occurs via Ccr-mediated recombination between this unique sequence (designated attB) and a specific site in the circularized SCCmec element (designated attSCC). When SCCmec is inserted, a new pair of sites is generated, referred to as attL and attR flanking the element's left and right ends, respectively. During SCCmec excision the reverse happens-attR and attL sites are recombined to regenerate the original sites, attB and attSCC (9). β-lactam antibiotics have been recently shown to increase expression of ccrAB (15). However, the regulatory mechanisms by which antibiotics promote expression of ccrAB and ccrC1 and initiate SCCmec transfer have not been elucidated. In the studies by Cuirolo et al. (16) and Plata et al. (17), oxacillin (OXA) induced the SOS response in MRSA and mitomycin C (MMC) induced mecA expression, showing that SOS was important for the resistant phenotype.
The SOS pathway is employed by bacteria to respond to various bactericidal agents, including β-lactam antibiotics (18,19). Governed by the products of lexA and recA genes, the SOS pathway coordinates a comprehensive response against DNA lesions (20). Under basal conditions, the transcription of dozens of genes involved in the SOS response is repressed (21). When chromosomes are damaged, persisting regions of single-stranded DNA allow the assembly of activated RecA nucleoprotein filaments called RecA*. The presence of RecA* activates the transcriptional upregulation of SOS genes by facilitating the cleavage of the LexA repressor (22). The SOS response is involved not only in cell division inhibition, nucleotide excision repair and recombination repair (23), but also in horizontal gene transfer (HGT) to neighboring cells (24), such as the transfer of pathogenicity island-encoded virulence factors in staphylococci (25,26), the transfer of integrating conjugative elements in Vibrio cholerae (27) and the recombination of chromosomal integrons in many Gram-negative pathogens (28–30). In this study, we provide evidence that antibiotics trigger the initiation of SCCmec transfer by inducing SOS responses.
MATERIALS AND METHODS
Bacterial strains, media and culture conditions
The bacterial strains and plasmids used in this study are listed in Supplementary Table S1. Strain NW19 is an OXA-susceptible, mecA positive S. haemolyticus with ccrC1 allele 2 and 8 in type V(5C2&5) SCCmec (GenBank accession number, MRUY00000000). MRSA strain Mu50 has ccrA2B2 genes in its type II SCCmec. E. coli DH5α was cultured in the Luria–Bertani medium supplemented with 100 mg/L ampicillin at 37°C. S. aureus and S. haemolyticus strains were grown in the trypticase soy broth (TSB) at 37°C with the appropriate antibiotic agents (chloramphenicol, 10 mg/L; Zeocin, 25 mg/L). MMC was used at a final concentration of 0.5 mg/L. The minimum inhibitory concentratidfons (MICs) of antibiotics were determined by the agar dilution method according to Clinical and Laboratory Standards Institute recommendations (31).
Transcript determinations
One colony of each sample was inoculated in 2 ml of the TSB medium and incubated at 37°C overnight. The overnight culture was diluted to OD600 = 0.05 and grown with constant aeration at 37°C until being treated with antibiotics at the mid-log phase of growth (OD600 = 0.6). After incubating for an additional 15 min, the cells were harvested by centrifugation at 4°C. Total RNA extraction was performed using a TaKaRa MiniBEST Universal RNA Extraction kit (Takara, Dalian, China). The cDNA was subsequently synthesized by a TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix kit (Takara, Dalian, China). SYBR Premix Ex Taq II (Takara, Dalian, China) was used for real-time polymerase chain reaction (RT-PCR) analyzes. The primers used for RT-PCR are listed in Supplementary Table S2. Each sample was analyzed in triplicate.
Western blot analysis
Total protein was prepared using a ProteoPrep® Total Extraction Sample kit (Sigma-Aldrich, Zwijndrecht, The Netherlands). To detect the amount of green fluorescent protein (GFP), 60 μg of total protein was resolved by 8% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to 0.45 μm polyvinylidene fluoride membranes at 4°C for 1.5 h. A monoclonal anti-GFP primary antibody (GenScript Co., Nanjing, China) diluted by 1/5000 was used to probe the blots according to the manufacturer's protocol. The secondary antibody was horseradish peroxidase-conjugated goat anti-mouse IgG (GenScript Co., Nanjing, China). The immune complexes were detected using an ECL western blot detection system (Pierce, Thermo Fisher Scientific, Waltham, MA, USA).
Electrophoretic mobility shift assays
The lexA gene was amplified from the NW19 and Mu50 genomes using primers (LexAexpF/LexAexpR and Lex50expF/Lex50expR) and cloned into the pCold II vector. The corresponding LexA proteins were overexpressed and purified in E. coli (Supplementary Data Materials and Methods for detail). The wild-type ccrC1 promoter was amplified using the primers (ccrCOPF and ccrCOPR) and cloned into the pEAZY-T vector (GenScript Co., Nanjing, China). Site-directed mutants of the ccrC1 promoter were constructed using the primers (mut1F/R, mut2F/R and mut3F/R) by overlap extension PCR. Electrophoretic mobility shift assay (EMSA) probes were obtained by amplification using the oligonucleotides BIO-ccrCOPF and ccrCOPR. The ccrA promoter was amplified using the primers (BIO-ccrAOPF and ccrAOPR). The EMSA experiments were performed using the LightShift EMSA Optimization and Control Kit according to the manufacturer's instructions (Pierce, Thermo Fisher Scientific, Waltham, MA, USA). All samples were loaded in 6% non-denaturing Tris–glycine polyacrylamide gels. Biotin-labeled DNA–protein complexes were detected using the Chemiluminescent Nucleic Acid Detection Module Kit (Pierce, Thermo Fisher Scientific).
Construction of gene overexpressing and reducing reporter strains
Detailed protocols are described in SuppIementary Data Materials and Methods.
Excision frequency detection
Genomic DNA from staphylococci was extracted using a Column Bacterial DNAout kit according to the manufacturer's instructions (GenScript, Nanjing, China) and used as a template for the detection of excision, as previously described (32). Primer pairs from the two sides of SCCmec (Supplementary Table S2) were used to determine the ratio of the SCC-excised genome. One copy of the housekeeping gene tpi (triosephosphate isomerase gene) was used as a reference. Each sample was determined in triplicate. All PCR products were confirmed by nucleotide sequence determination.
Gene deletion and complementation in Mu50 strain
Deletions of the recA gene were obtained using the plasmid pKZΔrecA50, which was constructed using pKOR1 with some modifications; a standard protocol (33) allowed marker-less recA replacement in Mu50 and the construction of Mu50ΔrecA (for details, see Supplementary Data Materials and Methods).
UV treatment
Cells grown overnight were inoculated into the TSB medium and re-suspended in 0.9% NaCl when OD600 = 0.6. Cells were treated for 10 min with UV irradiation and kept in the dark before RNA and DNA preparation. Irradiation was conducted by placing the plate containing the suspension 30 cm below a UV germicidal lamp and the UV dose was ∼35 J/m2.
RESULTS
DNA damage caused by antibiotics promotes the expression of ccrC1
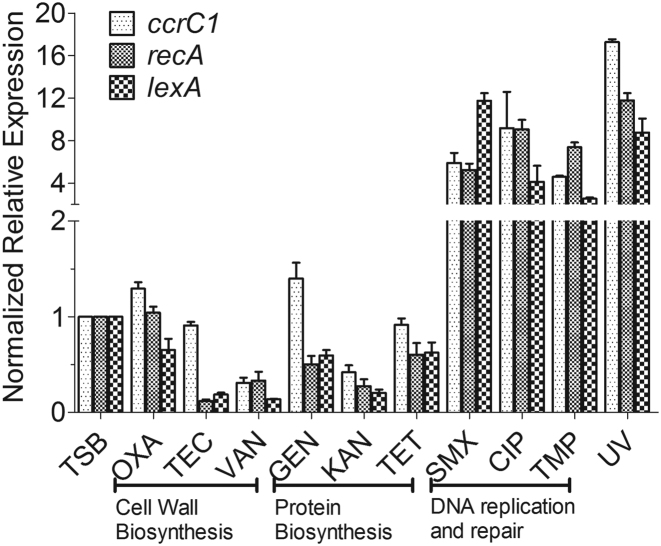
Stress induction by antibiotics is expected to occur under concentrations close to the MIC. Therefore, a ccrC1 promoter-gfp fusion was constructed to evaluate the activity of the ccrC1 promoter under 1/8 × MIC, 1/4 × MIC, 1/2 × MIC and 1 × MIC concentrations. The MICs of each antibiotic under our experimental conditions for the strain S. haemolyticus NW19 were first determined (Supplementary Table S3). The activity of the ccrC1 promoter tended to be higher upon exposure to DNA synthesis-targeting antibiotics exposure than those with other antibiotic treatments (data not shown). The effects of nine antibiotics on the expression of ccrC1 at 1/8 × MIC concentrations were also investigated by qPCR. The targets of these antibiotics are cell wall synthesis (OXA, teicoplanin (TEC) and vancomycin (VAN)), protein synthesis (gentamicin (GEN), kanamycin (KAN) and tetracycline (TET)) and DNA replication or repair (sulfamethoxazole (SMX), ciprofloxacin (CIP) and trimethoprim (TMP)). No significant differences in ccrC1 expression were observed for four antibiotics (TEC, VAN, KAN and TET) compared to the control (Figure 1). OXA and GEN showed a light induction of the promoter (Figure 1), although their effects were much lower than those observed with DNA-targeting antibiotics. Exposure to the three DNA synthesis-targeting antibiotics significantly increased the transcription of ccrC1.
Figure 1.
Antibiotics and Ultraviolet (UV) exposure promote ccrC1 expression in Staphylococcus haemolyticus NW19. Transcriptional levels of ccrC1, recA and lexA in NW19 with different antibiotic induction (1/8 MIC) and UV exposure. Gene expression levels were normalized and are presented relative to the trypticase soy broth (TSB) cultured control.
Agents or Stimuli that cause DNA damage promote the excision of SCCmec
To identify the CcrC1-mediated excision of SCCmec, genomic DNA of NW19 was whole-genome sequenced (BGI-Shenzhen, Shenzhen, China). The SCC composite island in strain NW19 consists of five recombination sites, identical to the genome island in another strain (NW19A, GenBank accession no. KM369884) (34). The elements SCCmec, ΨSCC and ΨSCCcad/ars/cop are flanked by different direct repeats (DR1 and DR2, DR2 and DR3-4, DR3-4 and DR5, respectively). There are 83 base pairs between DR3 and DR4. The SCCmec element consists of the ccrC1 allele 8 gene complex, the mec gene complex, the ccrC1 allele 2 gene complex and the ydhk gene cluster (Supplementary Figure S2A). The recombinations between different DRs in the SCC-excised genome were identified by PCR. Three strong positive occurrences were identified (DR1 + DR2, DR1 + DR3 and DR1 + DR5; Supplementary Figure S2A). Detailed information on the different DR site sequences in the variant SCC-excised genome is shown in Supplementary Figure S2B. To assess the effects of antibiotics on the excision of SCC, RT-PCR was performed to evaluate the frequency of excision under 1/8 × MIC antibiotic exposure. The three SCC-excised genomes shown in Supplementary Figure S2A were detected, and the SCCmec-excised genome (DR1 + DR2 shown in Supplementary Figure S2B) exhibited the highest frequency. Therefore, the detection of the SCCmec-excised genome was chosen to evaluate the influence of different antibiotic treatments. The frequency of SCCmec excision in NW19 was <10−11 without treatment (Figure 2). Treatment with SMX, CIP or TMP, three antibiotics targeting DNA replication and repair, led to significantly higher excision frequencies (3.4 × 10−5, 2.3 × 10−6 and 1.1 × 10−5, respectively) than the control. OXA and GEN showed an ∼100-fold increase in excision rates (8.8 × 10−9 and 1.0 × 10−9 respectively), which were much less than those observed with SMX, CIP or TMP, but still significant.
Figure 2.
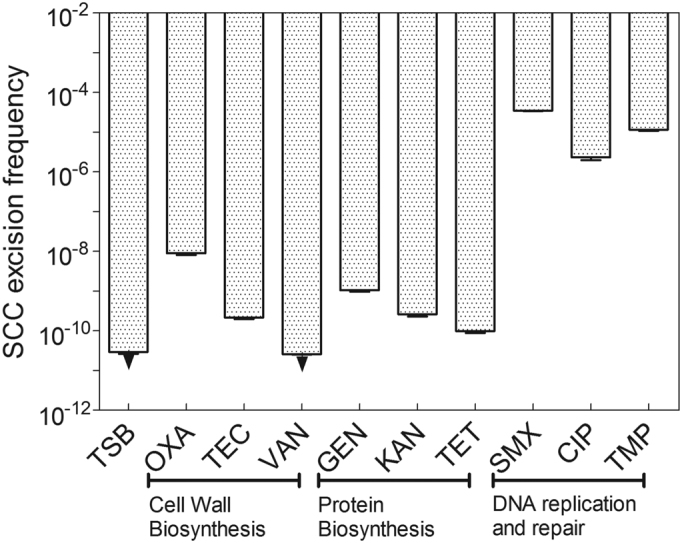
Antibiotics promote SCC excision via ccrC1. Primer pairs corresponding to the two sides of SCCmec (EGRT-F and EGRT-R) were used to determine the ratio of the SCC-excised genome. The arrowheads indicate values below the limit of detection (<2.5 × 10−11). One copy of the gene tpi (triosephosphate isomerase gene) was used as a reference. The bars represent the averages of three independent measurements in three different cultures, and the error bars represent the standard error of the mean. OXA, oxacillin; TEC, teicoplanin; VAN, vancomycin; GEN, gentamicin; KAN, kanamycin; TET, tetracycline; SMX, sulfamethoxazole; CIP, ciprofloxacin; TMP, trimethoprim.
After showing that certain antibiotics initiated the expression of ccrC1 and the excision of SCCmec, the effects of nine antibiotics on the expression of recA and lexA, two important and essential genes involved in DNA damage repair, were determined. Under antibiotic exposure at a concentration of 1/8 × MIC, only three antibiotics (SMX, CIP and TMP) induced a significant increase in the expression of recA-lexA compared to the control (Figure 1). In previous studies, it was shown that OXA could induce SOS response in MRSA (16,17,26). Thus, we further determined the expression of recA and ccrC1 in strain NW19 with different concentrations of OXA exposure by RT-PCR. The results are showed in Supplementary Figure S1. Increased expression of gene recA and ccrC1 was observed, when the concentrations of OXA at MIC and 2 MIC were used. The involvement of DNA damage in the effects of these antibiotics was confirmed by UV exposure. UV exposure promoted the expression of recA-lexA and SOS initiation. The transcription of ccrC1 increased by ∼17-fold compared to the control (Figure 1). These results suggest that DNA damage is responsible for the expression of ccrC1.
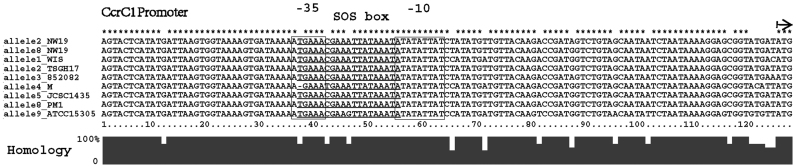
An SOS box is highly conserved in the ccrC1 promoter
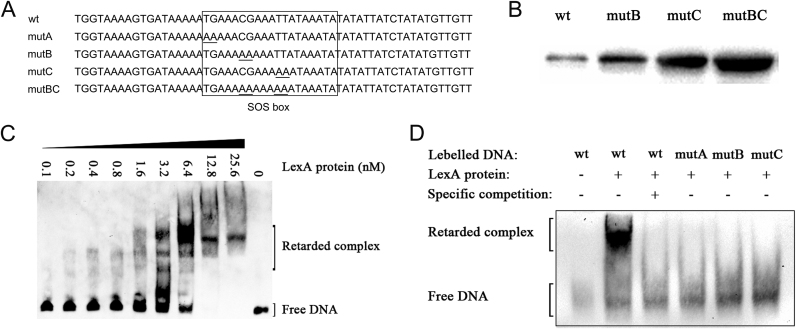
In many bacterial species, DNA damage caused by DNA-targeting antibiotics triggers the SOS response, which involves induction of expressing SOS response genes, including recA and lexA. LexA is the repressor of the SOS response for all the genes of the chromosomal SOS regulon, except for some horizontally transferred genes such as phage lambda (35). Analysis and alignment of the promoter sequences of ccrC1 and several other SOS genes in S. aureus (Table 1) revealed three putative overlapping LexA-binding motifs in the promoter of ccrC1 in S. haemolyticus NW19. A search of ccrC1 promoter sequences from seven different ccrC1 allele types available in the NCBI GenBank indicated that all these allele types harbor the SOS box, which covers the partial −35 region and partial −10 region (Figure 3). All LexA-binding sequences are located 79 bp upstream of the translation initiation site of ccrC1. To further confirm the LexA binding sites, plasmids pCL::ccr-gfp and pCL::mutccr-gfp (mutB, mutC and mutBC in Figure 4A), which contain mutations in the putative SOS box of the ccrC1 promoter, were constructed. MutA site is in the −35 region of ccrC1 promoter and mutation in this site affects binding of the RNA polymerase to the promoter and reduces the expression of GFP. Analysis of GFP levels indicated that the substitution of the putative SOS box greatly increased the expression of the GFP reporter (Figure 4B). The strain containing the original promoter also had a detectable level of GFP expression, which suggested a weak activity of the ccrC1 promoter in the absence of environmental stress. These results indicate that mutations in the putative SOS box affect the transcription activity of the ccrC1 promoter.
Table 1. Comparison of LexA binding sites in S. aureus and our isolate.
| Strain and Gene | SOS box | Reference | |||
|---|---|---|---|---|---|
| S. aureus RA1 uvrA | CG | AAA | GATTT AG | AT | (37) |
| S. aureus RA1 uvrC | CG | AAG | ATGTT GA | TT | (37) |
| S. aureus COL dmpI | CG | AAC | ACGTG TT | CT | (38) |
| S. aureus COL ssb2 | CG | AAC | ATATG TT | CT | (38) |
| S. aureus COL recA | CG | AAC | AAATA TT | CG | (38) |
| S. aureus COL lexA | CG | AAC | AAATG TT | TG | (38) |
| S. aureus COL 2162 | GA | AAC | ATATT TT | CG | (38) |
| S. heamolyticus NW19 ccrC1a | GA | AAC | GAAAT TA | TA | This study |
| S. heamolyticus NW19 ccrC1a | TG | AAA | CGAAA TT | AT | This study |
| S. heamolyticus NW19 ccrC1a | CG | AAA | TTATA AA | TA | This study |
| S. aureus Mu50 ccrA2B2b | TG | AAA | GGAGGAG | CC | This study |
aThree putative LexA-binding sequences on the promoter of ccrC1 are overlapped and cover 19 bp in total (5΄-TGAAACGAAATTATAAATA-3΄).
bA putative LexA-binding sequences on the promoter of ccrA2B2 in Mu50 (5΄-TGAAAGGAGGAGCC-3΄).
Figure 3.
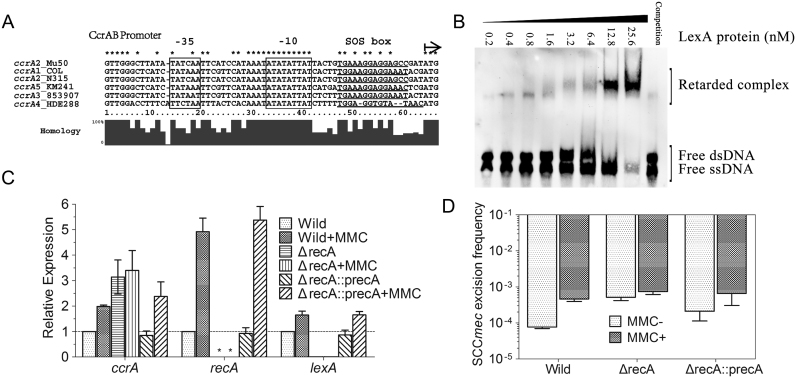
Alignment of the promoter regions of ccrC1 genes from Staphylococcus aureus WIS (accession number AB121219), S. aureus TSGH17 (accession number AB512767), S. aureus 85/2082 (accession number AB037671), S. aureus M (U10927), Staphylococcus haemolyticus JCSC1435 (AP006716), S. aureus PM1 (AB462393) and Staphylococcus saprophyticus ATCC15305 (AP008934). Type V(5C2&5) SCCmec in S. haemolyticus NW19 contains two ccrC-carrying gene complexes on either side of mec gene complex. The ccrC1 allele 8 is located in the region between orfX and mec gene complex, while ccrC1 allele 2 is located between mec gene complex and ydhK complex (as showed in Supplementary Figure S2A). Two allele genes were 92.5% identical. The putative LexA-binding sequences are underlined. Promoter elements (−35 and −10) are framed. The start codon of ccrC1 gene is indicated by a black arrow.
Figure 4.
(A) Partial sequences of the ccrC1 promoter region (wt) and SOS box mutants (mutA, mutB, mutC and mutBC) used in the electrophoretic mobility shift assays (EMSAs). (B) Western blotting detection of green fluorescent protein (GFP) under the wild-type and mutant promoters of ccrC1. The plasmids pCL::ccr-gfp and pCL::mutccr-gfp, which encode the wild-type and mutant promoters of ccrC1, respectively, followed by gfp, were transformed into NW19. Cells were grown until OD600 = 0.6 and total bacterial protein was prepared. (C) EMSA experiments performed with the LexA protein and wild-type ccrC1 promoter. (D) EMSA of the wild-type or mutant ccrC1 promoter and LexA protein.
To confirm binding ability and specificity, purified LexA protein was used to examine the binding ability to the ccrC1 promoter by an EMSA. The probes were titrated with increasing concentrations of purified LexA and subjected to gel-shift analysis. LexA specifically retarded the mobility of the ccrC1 promoter in a dose-dependent manner (Figure 4C). The formation of a specific LexA–DNA complex was preceded by a detectable shift of the free probe but indistinct complex formation. This observation reflects the binding of the initial LexA monomer followed by the stable dimerization of LexA on DNA. Furthermore, the presence of the 100-fold specific competitor (unlabeled DNA of the lexA promoter) and mutations in key sites of the putative SOS box prevented the interaction between LexA and the wild-type promoter and mutant promoters, respectively (Figure 4A and D). These results indicate that the putative SOS box on the ccrC1 promoter (5΄-TGAAACGAAATTATAAATA-3΄) is the LexA binding site.
Expression of ccrC1 and excision of SCCmec are influenced by recA and lexA in vivo
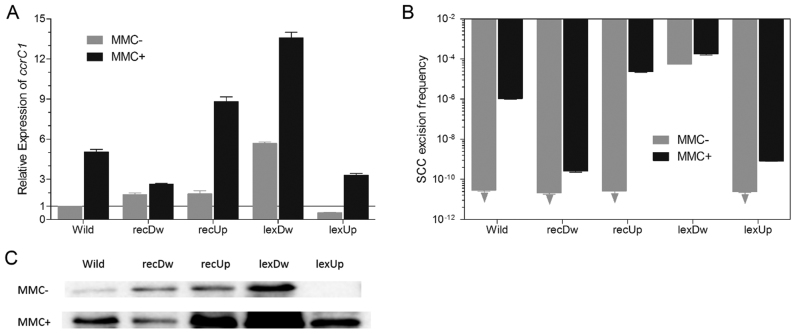
To confirm the roles of RecA and LexA in the regulation of ccrC1 expression in vivo, increased and reduced expression of recA and lexA were achieved by plasmid-mediated overexpression and antisense RNA as described previously (36). Cells were grown in TSB to OD600 = 0.6 and treated with MMC (an inducer of DNA damage) at a concentration of 0.5 mg/L for 30 min. In the absence of MMC, overexpression or reduction of recA had little or no effect on ccrC1 expression (Figure 5A). Overexpression of lexA in strain 19lexUp caused reduced transcription of ccrC1, and antisense RNA-mediated reduction of lexA transcripts in strain 19lexDw led to upregulation of ccrC1. We also analyzed the expression of the GFP reporter in wild and mutant strains (Figure 5C), and the results were in agreement with the transcript detection of ccrC1. Compared to wild-type NW19, SCC excision frequency was much higher in strain 19lexDw (Figure 5B). By contrast, in strains NW19, 19recDw, 19recUp and 19lexUp, detection of SCC excision by the RT-PCR method was limited and frequencies of <2.5 × 10−11 were not detectable. In the presence of MMC, the expression level of ccrC1 was upregulated to varying degrees in all five strains (Figure 5A). Compared to wild-type NW19, recA overexpression and lexA reduction caused a higher induction of ccrC1 expression. Reduction of recA and overexpression of lexA led to lower induction of ccrC1 expression. Compared to wild-type NW19, the SCC excision frequency was higher in strains 19recUp and 19lexDw and lower in strains 19recDw and 19lexUp (Figure 5B). These results suggest that LexA is the transcriptional repressor of ccrC1 and that SCC transfer is initiated by the activated SOS response.
Figure 5.
Effects of recA and lexA on the expression of ccrC1 and SCC excision frequency in NW19. (A) Relative expression of ccrC1 in different strains compared to the control (wild-type without mitomycin C (MMC)). (B) SCC excision frequency detection in different strains with and without MMC treatment. (C) Detection of GFP using ccr-gfp reporter strategy. Wild, wild-type NW19; recDw, NW19 with reduced expression of recA; recUp, NW19 with increased expression of recA; lexDw, NW19 with reduced lexA expression; lexUp, lexA transcript increased NW19; MMC−, without MMC exposure; MMC+, with MMC exposure. Arrowhead, below the limit of detection (<2.5 × 10−11). The relative expression of ccrC1 and the SCC excision frequency were detected with and without MMC exposure (0.5 mg/L). Each sample was determined in triplicate. The data represent the mean ± Standard Error Mean (SEM).
Expression of ccrAB and excision of SCCmec in Mu50 are dependent on the SOS pathway
The excision of the type II SCCmec in Mu50 is mediated by CcrA2B2 recombinases; CcrA2 allotype and CcrB2 allotype are encoded in the same operon. To further determine whether the expression of ccrAB is also controlled by the RecA-LexA system, different allotypes of the ccrAB promoter sequences were aligned. The putative LexA binding site (5΄-TGAAAGGAGGAGCC-3΄) overlapped with the transcription start sites (Figure 6A). In vitro EMSA experiments revealed specific retardation of the ccrA promoter (Figure 6B). The expression of ccrA was ∼3-fold higher in the recA deletion mutant than in the wild-type of Mu50 (Figure 6C). Expression of lexA is regulated by LexA itself. In the recA deletion mutant, repressed LexA led to a lower basal expression of lexA in Mu50ΔrecA strain (0.05-fold). Exposure to MMC upregulated the expression of ccrA, recA and lexA in the wild-type Mu50 and Mu50ΔrecA::precA strains. Little effect of MMC on the expression of ccrA was observed in the in recA deletion strain. The excision frequency of SCCmec was 6.8 × 10−5 and increased to 4.7 × 10−4 upon MMC treatment (Figure 6D). No significant difference was observed in the recA-deleted Mu50 with or without MMC exposure. These results show that the expression of ccrAB and excision of SCCmec in Mu50 is dependent on the RecA-LexA mediated SOS pathway.
Figure 6.
The SOS response promotes the expression of ccrA and SCCmec excision in Mu50. (A) Alignment of the promoter regions of ccrA genes from Staphylococcus aureus Mu50 (accession number BA000017), S. aureus COL (CP000046), S. aureus N315 (BA000018), Staphylococcus pseudintermedius KM241 (AM904731), S. aureus 85/3907 (AB047089) and S. aureus HDE288 (AF411935). The putative LexA-binding sequences are underlined. Promoter elements (−35 and −10) are boxed. The 5΄ ATG of ccrA gene is indicated by a black arrow. (B) EMSA experiments performed with the LexA protein and ccrA promoter. (C) Relative expression of ccrA, recA and lexA in wild-type Mu50, the recA-deleted mutant (ΔrecA) and recA-complemented mutant (ΔrecA::precA) compared to the control (wild-type without MMC). *, not detectable. (D) SCCmec excision frequency detection in different strains with and without MMC treatment (0.5 mg/l). Each sample was determined in triplicate. Data are presented as the average ± SEM.
DISCUSSION
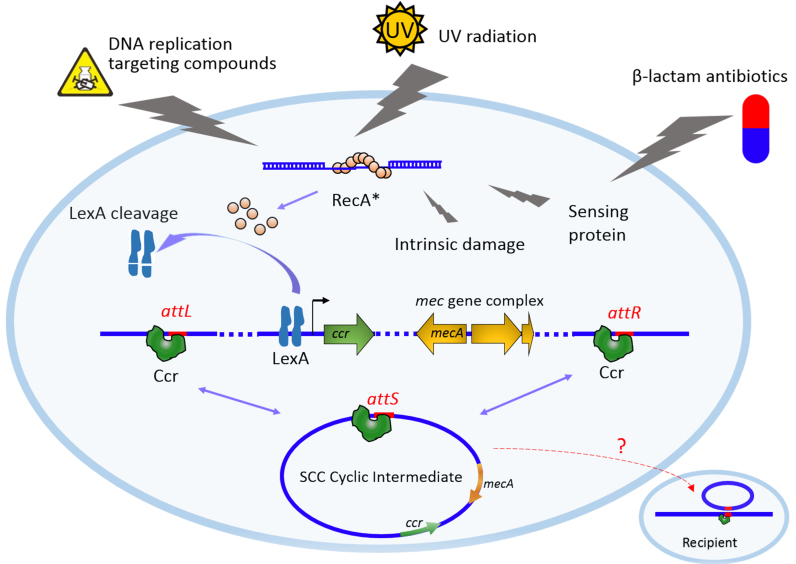
In this study, we propose a model of the regulatory pathway by which antibiotics promote SCCmec transfer (Figure 7). Bactericidal drugs inducing DNA and protein damage are usually involved in triggering the SOS response (20,35). LexA is initially bound to its binding sites (SOS box) on the promoters of ccrC1 and ccrAB, thus hindering the transcription of ccrC1 and ccrAB by blocking RNA polymerase activity. The RecA protein becomes activated (RecA*) during the SOS response, which in turn facilitates LexA autocleavage. Inactivation of LexA alleviates the repression of CcrC1 and CcrAB expression. The increased levels of Ccr proteins, which can bind recombination sites (attL and attR) on both ends of SCCmec, lead to the integration and excision of SCCmec on the chromosome. HGT of the SCCmec cyclic intermediate may lead to transfer and spread of β-lactam resistance among staphylococci.
Figure 7.
Proposed model of the regulatory pathway by which the SOS response promotes SCCmec transfer. Intermediary molecules involved in SOS induction are shown. The mechanism by which SCCmec transfers among staphylococci remains elusive.
The frequency of SCCmec excision was dependent on the type and the concentration of antibiotics in this study. Low levels of the β-lactam antibiotic (OXA at 1/8 MIC and 1/4 MIC) slightly affected expression of SOS proteins in S. haemolyticus NW19 (Figure 1 and Supplementary Figure S1), while OXA at the concentration of 1/100 MIC induced the SOS response in various Gram negative bacteria (39–41). The difference between S. haemolyticus NW19 and Gram-negative bacteria could be due to their different responses to OXA. S. haemolyticus NW19 became highly resistant to OXA (OXA MIC > 256 μg/ml) with OXA induction (36), due to increased expression of mecA (Supplementary Figure S1). In E. coli, cell wall stress induced by β-lactams induces the SOS response through a sensor protein DpiB and a replication site binding protein DpiA (18). Putative proteins (Figure 7) responsible for the cell wall stress response by β-lactams in staphylococcus remain to be identified. In our study, Ccr expression and SCCmec excision were also observed in non-stressed staphylococci, as reported in a previous study (42). This observation can be explained by the basal expression of ccr genes in the absence of SOS induction. In Stojanov's study, the ccr promoter activity was observed in the absence of antibiotic exposure, even though only in a small percentage of cell populations (∼3 and 1% among logarithmic and stationary growth phases respectively).
SCCmec is only part of the composite island in strain NW19; this island encodes not only methicillin resistance but also heavy metal resistance for cadmium, arsenic and copper. Many other composite islands in staphylococci have been reviewed by Shore and Coleman (43). Some SCCs that do not contain the mec gene complex are considered non-mec SCC. Ten non-mec SCC types have been identified, and four (SCCATCC12228, SCCpbp4, SCCHg and SCC476) carry additional antibiotic resistance or virulence genes (43). Because both the SCC composite island and non-mec SCC have ccr gene complexes, our results suggest that the excision and transfer of such SCCs could also be induced by antibiotics through the SOS response. Furthermore, we observed that the SCCmec excision frequency mediated by ccrC1 was much lower than that mediated by ccrAB, even upon induction by MMC, which might explain why more SCCs carry the ccrAB gene complex than ccrC1.
The overuse and improper use of antibiotics have contributed to the rise of multidrug-resistant MRSA. In our study, β-lactams and antibiotics targeting DNA were observed to promote the SCCmec excision. Antibiotics were also reported to be responsible for maintaining antibiotic resistance in bacteria (44,45). The findings from this study and other studies may explain why MRS is widely distributed worldwide. An individual mecA positive staphylococcal bacterium with the SCCmec excised would become methicillin-susceptible and be destroyed under lactam pressure. It has been suggested that these few individuals commit this suicide, thus sacrificing themselves to transfer SCCmec into new recipient strains (42). However, we favor an alternative model in which the excision of SCCmec is an active process and is initiated when MRS is exposed to damage or environmental stress (Figure 7). Excision allows SCCmec to hitchhike from a feeble individual or species to a vigorous one. This ‘hitchhike model’ ensures that SCCmec is not eliminated in the long-term process of evolution.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Foundation of China [31372282]; University Scientific Research Fund project [Z111021305]. Natural Science and Engineering Research Council of Canada discovery grant (in part). Funding for open access charge: University Scientific Research Fund project [Z111021305].
Conflict of interest statement. None declared.
REFERENCES
- 1. Hanssen A.M., Ericson S.J.. SCCmec in staphylococci: genes on the move. FEMS Immunol. Med. Microbiol. 2006; 46:8–20. [DOI] [PubMed] [Google Scholar]
- 2. Otto M. Coagulase-negative staphylococci as reservoirs of genes facilitating MRSA infection: Staphylococcal commensal species such as Staphylococcus epidermidis are being recognized as important sources of genes promoting MRSA colonization and virulence. Bioessays. 2013; 35:4–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wu S., Piscitelli C., de Lencastre H., Tomasz A.. Tracking the evolutionary origin of the methicillin resistance gene: cloning and sequencing of a homologue of mecA from a methicillin susceptible strain of Staphylococcus sciuri. Microb. Drug. Resist. 1996; 2:435–441. [DOI] [PubMed] [Google Scholar]
- 4. Tsubakishita S., Kuwahara-Arai K., Sasaki T., Hiramatsu K.. Origin and molecular evolution of the determinant of methicillin resistance in staphylococci. Antimicrob. Agents Chemother. 2010; 54:4352–4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Archer G.L., Thanassi J.A., Niemeyer D.M., Pucci M.J.. Characterization of IS1272, an insertion sequence-like element from Staphylococcus haemolyticus. Antimicrob. Agents Chemother. 1996; 40:924–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jones R.N., Barry A.L., Gardiner R.V., Packer R.R.. The prevalence of staphylococcal resistance to penicillinase-resistant penicillins. A retrospective and prospective national surveillance trial of isolates from 40 medical centers. Diagn. Microbiol. Infect. Dis. 1989; 12:385–394. [DOI] [PubMed] [Google Scholar]
- 7. Schmitz F.J., Verhoef J., Fluit A.C.. Prevalence of resistance to MLS antibiotics in 20 European university hospitals participating in the european SENTRY surveillance programme. sentry participants group. J. Antimicrob. Chemother. 1999; 43:783–792. [DOI] [PubMed] [Google Scholar]
- 8. Ito T., Kuwahara-Arai K., Katayama Y., Uehara Y., Han X., Kondo Y., Hiramatsu K.. Staphylococcal cassette chromosome mec (SCCmec) analysis of MRSA. Methods Mol. Biol. 2014; 1085:131–148. [DOI] [PubMed] [Google Scholar]
- 9. Misiura A., Pigli Y.Z., Boyle-Vavra S., Daum R.S., Boocock M.R., Rice P.A.. Roles of two large serine recombinases in mobilizing the methicillin-resistance cassette SCCmec. Mol. Microbiol. 2013; 88:1218–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boundy S., Safo M.K., Wang L., Musayev F.N., O’Farrell H.C., Rife J.P., Archer G.L.. Characterization of the Staphylococcus aureus rRNA methyltransferase encoded by orfX, the gene containing the staphylococcal chromosome Cassette mec (SCCmec) insertion site. J. Biol. Chem. 2013; 288:132–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ito T., Ma X.X., Takeuchi F., Okuma K., Yuzawa H., Hiramatsu K.. Novel type V staphylococcal cassette chromosome mec driven by a novel cassette chromosome recombinase, ccrC. Antimicrob. Agents Chemother. 2004; 48:2637–2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Katayama Y., Ito T., Hiramatsu K.. A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 2000; 44:1549–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wu Z., Li F., Liu D., Xue H., Zhao X.. Novel type XII Staphylococcal cassette chromosome mec harboring a new cassette chromosome recombinase, CcrC2. Antimicrob. Agents Chemother. 2015; 59:7597–7601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kinnevey P.M., Shore A.C., Brennan G.I., Sullivan D.J., Ehricht R., Monecke S., Slickers P., Coleman D.C.. Emergence of sequence type 779 methicillin-resistant Staphylococcus aureus harboring a novel pseudo staphylococcal cassette chromosome mec (SCCmec)-SCC-SCCCRISPR composite element in Irish hospitals. Antimicrob. Agents Chemother. 2013; 57:524–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Higgins P.G., Rosato A.E., Seifert H., Archer G.L., Wisplinghoff H.. Differential expression of ccrA in methicillin-resistant Staphylococcus aureus strains carrying staphylococcal cassette chromosome mec type II and IVa elements. Antimicrob. Agents Chemother. 2009; 53:4556–4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cuirolo A., Plata K., Rosato A.E.. Development of homogeneous expression of resistance in methicillin-resistant Staphylococcus aureus clinical strains is functionally associated with a beta-lactam-mediated SOS response. J. Antimicrob. Chemother. 2009; 64:37–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Plata K.B., Riosa S., Singh C.R., Rosato R.R., Rosato A.E.. Targeting of PBP1 by beta-lactams determines recA/SOS response activation in heterogeneous MRSA clinical strains. PLoS One. 2013; 8:e61083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Miller C., Thomsen L.E., Gaggero C., Mosseri R., Ingmer H., Cohen S.N.. SOS response induction by beta-lactams and bacterial defense against antibiotic lethality. Science. 2004; 305:1629–1631. [DOI] [PubMed] [Google Scholar]
- 19. Kohanski M.A., Dwyer D.J., Hayete B., Lawrence C.A., Collins J.J.. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007; 130:797–810. [DOI] [PubMed] [Google Scholar]
- 20. Erill I., Campoy S., Barbe J.. Aeons of distress: an evolutionary perspective on the bacterial SOS response. FEMS Microbiol. Rev. 2007; 31:637–656. [DOI] [PubMed] [Google Scholar]
- 21. Al M.A., Lombardo M.J., Shee C., Lisewski A.M., Gonzalez C., Lin D., Nehring R.B., Saint-Ruf C., Gibson J.L., Frisch R.L. et al. Identity and function of a large gene network underlying mutagenic repair of DNA breaks. Science. 2012; 338:1344–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schlacher K., Goodman M.F.. Lessons from 50 years of SOS DNA-damage-induced mutagenesis. Nat. Rev. Mol. Cell Biol. 2007; 8:587–594. [DOI] [PubMed] [Google Scholar]
- 23. Janion C. Some aspects of the SOS response system–a critical survey. Acta Biochim. Pol. 2001; 48:599–610. [PubMed] [Google Scholar]
- 24. Fornelos N., Browning D.F., Butala M.. The use and abuse of LexA by mobile genetic elements. Trends Microbiol. 2016; 24:391–401. [DOI] [PubMed] [Google Scholar]
- 25. Ubeda C., Maiques E., Knecht E., Lasa I., Novick R.P., Penades J.R.. Antibiotic-induced SOS response promotes horizontal dissemination of pathogenicity island-encoded virulence factors in staphylococci. Mol. Microbiol. 2005; 56:836–844. [DOI] [PubMed] [Google Scholar]
- 26. Maiques E., Ubeda C., Campoy S., Salvador N., Lasa I., Novick R.P., Barbe J., Penades J.R.. Beta-lactam antibiotics induce the SOS response and horizontal transfer of virulence factors in Staphylococcus aureus. J. Bacteriol. 2006; 188:2726–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Beaber J.W., Hochhut B., Waldor M.K.. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature. 2004; 427:72–74. [DOI] [PubMed] [Google Scholar]
- 28. Guerin E., Cambray G., Sanchez-Alberola N., Campoy S., Erill I., Da R.S., Gonzalez-Zorn B., Barbe J., Ploy M.C., Mazel D.. The SOS response controls integron recombination. Science. 2009; 324:1034. [DOI] [PubMed] [Google Scholar]
- 29. Cambray G., Sanchez-Alberola N., Campoy S., Guerin E., Da R.S., Gonzalez-Zorn B., Ploy M.C., Barbe J., Mazel D., Erill I.. Prevalence of SOS-mediated control of integron integrase expression as an adaptive trait of chromosomal and mobile integrons. Mob. DNA. 2011; 2:20–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hocquet D., Llanes C., Thouverez M., Kulasekara H.D., Bertrand X., Plesiat P., Mazel D., Miller S.I.. Evidence for induction of integron-based antibiotic resistance by the SOS response in a clinical setting. PLoS Pathog. 2012; 8:e1002778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Clinical and Laboratory Standards Institute Performance Standards for Antimicrobial Susceptibility Testing: Twenty-fourth Informational Supplement M100-S24. 2014; Wayne: CLSI. [Google Scholar]
- 32. Noto M.J., Archer G.L.. A subset of Staphylococcus aureus strains harboring staphylococcal cassette chromosome mec (SCCmec) type IV is deficient in CcrAB-mediated SCCmec excision. Antimicrob. Agents Chemother. 2006; 50:2782–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bae T., Schneewind O.. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid. 2006; 55:58–63. [DOI] [PubMed] [Google Scholar]
- 34. Xue H., Wu Z., Li L., Li F., Wang Y., Zhao X.. Coexistence of heavy metal and antibiotic resistance within a novel composite staphylococcal cassette chromosome in a Staphylococcus haemolyticus isolate from bovine mastitis milk. Antimicrob. Agents Chemother. 2015; 59:5788–5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Baharoglu Z., Mazel D.. SOS, the formidable strategy of bacteria against aggressions. FEMS Microbiol. Rev. 2014; 38:1126–1145. [DOI] [PubMed] [Google Scholar]
- 36. Liu P., Xue H., Wu Z., Ma J., Zhao X.. Effect of bla regulators on the susceptible phenotype and phenotypic conversion for oxacillin-susceptible mecA-positive staphylococcal isolates. J. Antimicrob. Chemother. 2016; 71:2105–2112. [DOI] [PubMed] [Google Scholar]
- 37. Bisognano C., Kelley W.L., Estoppty T., Francois P., Schrenzel J., Li D., Lew D.P., Hooper D.C., Cheung A.L., Vaudaux P.. A RecA-LexA-dependent pathway mediates ciprofloxacin-induced fibronectin binding in Staphylococcus aureus. J. Biol. Chem. 2004; 279:9064–9071. [DOI] [PubMed] [Google Scholar]
- 38. Cirz R.T., Jones M.B., Gingles N.A., Minogue T.D., Jarrahi B, Peterson S.N., Romesberg F.E.. Complete and SOS-mediated response of Staphylococcus aureusto the antibioticciprofloxacin. J. Bacteriol. 2007; 189:531–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gutierrez A., Laureti L., Crussard S., Abida H., Rodriguez-Rojas A., Blazquez J., Baharoglu Z., Mazel D., Darfeuille F., Vogel J. et al. β-lactam antibiotics promote bacterial mutagenesis via an RpoS-mediated reductionin replication fidelity. Nat. Commun. 2013; 4:1610–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Baharoglu Z., Krin E., Mazel D.. RpoS plays a central role in the SOS induction by sub-lethal aminoglycoside concentrations in Vibrio cholerae. PLoS Genet. 2013; 9:e1003421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Baharoglu Z., Mazel D.. Vibrio cholerae triggers SOS and mutagenesis in response to a wide range of antibiotics: a route towards multiresistance. Antimicrob. Agents Chemother. 2011; 55:2438–2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stojanov M., Sakwinska O., Moreillon P.. Expression of SCCmec cassette chromosome recombinases in methicillin-resistant Staphylococcus aureus and Staphylococcus epidermidis. J. Antimicrob. Chemother. 2013; 68:749–757. [DOI] [PubMed] [Google Scholar]
- 43. Shore A.C., Coleman D.C.. Staphylococcal cassette chromosome mec: recent advances and new insights. Int. J. Med. Microbiol. 2013; 303:350–359. [DOI] [PubMed] [Google Scholar]
- 44. Bottery M.J., Wood A.J., Brockhurst M.A.. Selective conditions for a multidrug resistance plasmid depend on the sociality of antibiotic resistance. Antimicrob. Agents Chemother. 2016; 60:2524–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhu Y.G., Johnson T.A., Su J.Q., Qiao M., Guo G.X., Stedtfeld R.D., Hashsham S.A., Tiedje J.M.. Diverse and abundant antibiotic resistance genes in chinese swine farms. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:3435–3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.