Abstract
The conserved INO80 chromatin remodeling complex is involved in regulation of DNA damage repair, replication and transcription. It is commonly recruited to the transcription start region and contributes to the establishment of promoter-proximal nucleosomes. We find a substantial influence of INO80 on nucleosome dynamics and gene expression during stress induced transcription. Transcription induced by osmotic stress leads to genome-wide remodeling of promoter proximal nucleosomes. INO80 function is required for timely return of evicted nucleosomes to the 5΄ end of induced genes. Reduced INO80 function in Arp8-deficient cells leads to correlated prolonged transcription and nucleosome eviction. INO80 and the related complex SWR1 regulate incorporation of the H2A.Z isoform at promoter proximal nucleosomes. However, H2A.Z seems not to influence osmotic stress induced gene regulation. Furthermore, we show that high rates of transcription promote INO80 recruitment to promoter regions, suggesting a connection between active transcription and promoter proximal nucleosome remodeling. In addition, we find that absence of INO80 enhances bidirectional promoter activity at highly induced genes and expression of a number of stress induced transcripts. We suggest that INO80 has a direct repressive role via promoter proximal nucleosome remodeling to limit high levels of transcription in yeast.
INTRODUCTION
The chromatin structure of eukaryotic cells has a large impact on the accessibility and interpretation of the genetic information. Despite disrupting activities such as replication, repair and transcription, the chromatin structure is largely ordered and stable. Yeast RNA polymerase II promoters enable access of regulatory factors via a nucleosome depleted region (5΄ NDR) framed by boundary nucleosomes commonly designated as −1 and +1 (1,2). In higher eukaryotes active RNA Pol II genes are at least partially depleted of nucleosomes around transcription start and termination sites (3,4). In the course of initiation and elongation of transcription, these nucleosomes become reversibly evicted (5). The question how their specific positioning is established and maintained is subject of debate (reviewed by 6,7).
In yeast, the 5΄ NDR is maintained by combined action of sequence specific transcription factors such as Rap1, Reb1 and Abf1, chromatin remodeling complexes (CRCs) and sequence determinants (8–14). The +1 nucleosome, downstream of the 5΄ NDR, is located in close proximity to the transcription start site (TSS) (2,15–17). The histone isoform H2A.Z is enriched in +1 nucleosomes and reduces their stability (18,19). The location of the +1 nucleosome, and its high propensity for eviction suggests functional participation in transcription (reviewed by 1,20,21). However, the interdependence between insertion and eviction of the +1 nucleosome and regulation of transcription is not fully understood. The anchoring and maintenance of the +1 nucleosome is controlled by several activities. In vitro and in vivo studies showed that the underlying DNA sequence is involved but is not the major determinant (11,22). Moreover, in vitro chromatin reconstitution studies failed to recapitulate the precise +1 positioning, suggesting a complex and perhaps dynamic interplay of factors in vivo (6). Furthermore, it has been suggested that the transcription machinery is involved in positioning of the +1 nucleosome. The pre-initiation complex (PIC) and the +1 nucleosome are in close proximity and components of the transcription machinery interact with +1 and are proposed to recruit chromatin remodelers (7,23).
Chromatin remodeling is facilitated by a variety of factors which act as monomers or in large complexes. INO80 has a conserved role in several distinct chromatin-related processes such as nucleosome sliding, repair of double strand breaks, replication, sense and antisense transcription and silencing (24–34). Mammalian INO80 subunit composition shares a core set of subunits with yeast but includes a number of metazoan-specific subunits suggesting further specific functions (35). INO80-dependent replacement of the histone variant H2A.Z against conventional H2A is important to cope with replication stress and DNA damage (30,36–38). Removal of H2A.Z by INO80 promotes homologous recombination (39). The related SWR-C catalyzes the exchange of H2A for the variant H2A.Z close to NDRs (40–43). Recently, H2A.Z has been shown to have a positive role for expression of antisense transcripts and non-coding RNAs (44,45). INO80 is present within genic regions and enriched genome-wide at the site of the +1 nucleosome (17,31,34,46). Specific subunits of CRCs bind to nucleosomes and recognize certain covalent histone modifications (47). The INO80 complex comprises 15 subunits organized in the Nhp10, Arp5 and Arp8 modules and the head module containing the Rvb1/2 helicases plus the Ino80 ATPase (48–50). INO80 mutants lacking one of the Actin-related proteins Arp4, Arp5 and Arp8, Nhp10 or the ATPase subunit Ino80 have enhanced expression of stress-induced genes and genes involved in respiration (32,51,52).
In yeast, the change of external physical parameters such as temperature and osmolarity or exposure to chemicals causing oxidative stress, leads to rapid (within minutes) and often transient change of expression of a substantial fraction of genes (e.g. for hyperosmolarity stress (53)). Stress-triggered activation causes transient disequilibrium of the chromatin structure and enables observation of otherwise masked effects. At stress-induced loci alterations of chromatin structure may happen rapidly. For example, recent surveys of a compendium of mutants identified chromatin regulators with pronounced effects on gene expression during oxidative (diamide) stress (54,55).
Here we examined the role of INO80 for the chromatin structure and transcription of osmostress-inducible genes. Using high resolution tiling microarrays we found that INO80 facilitates re-insertion of promoter proximal nucleosomes following transcription induced eviction. In addition, we identified cryptic transcripts that are repressed by INO80. Furthermore, INO80-dependent change of the +1 nucleosome level is correlated with transcription and suggests a repressive role during dynamic changes of gene expression.
MATERIALS AND METHODS
Strains and growth conditions
Strains used in this study are listed in Supplementary Table S1. For stress treatment Saccharomyces cerevisiae strains were grown in YPD (yeastextract, peptone dextrose) (2% Glucose) to early exponential growth phase (OD600 0.8–1) followed by exposure to hyperosmolarity stress by addition of 5M NaCl solution to a final concentration of 0.4M. Plasmid carrying strains were pre-grown in minimal SC-medium then grown in YPD before treatment. Anchor-Away strains HHY168 (wild-type), HHY154 (Tbp1-Frb) and EKYH154 (Tbp1-Frb arp8Δ) were grown to early exponential phase, then 1 μg/ml rapamycin (LC-Laboratories) was added for 45 min, followed by treatment with hyperosmotic stress as above.
Nucleosome scanning assay
NuSA was performed essentially as described (56). Cells were grown to an OD600 0.8–1.0 and aliquots of 45ml were treated with 0.4M NaCl followed by crosslinking with 1% formaldehyde for 10 min and washing with ice-cold 1× tris-buffered salineTBS. Pellets were resuspended in 8 ml Buffer Z2 (1M Sorbitol, 50 mM Tris–HCl pH 7.4, 10 mM freshly added Mercaptoethanol) with Zymolyase (10 mg/ml Seikagaku Corp, 200 μl per sample) and incubated on 30°C for 20–30 min. Spheroplasts were harvested by centrifugation (1500 g) for 10 min and resuspended in 1.5 ml NPS buffer (0.075% NP-40, 50 mM NaCl, 10 mM Tris pH 7.4, 5 mM MgCl2 1 mM CaCl2, freshly added 0.5 mM Spermidine and 1 mM Mercaptoethanol). Aliquots of 600 μl were digested with 50U of Micrococcus Nuclease (15U/μl in 10 mM Tris–HCL pH 7.5, 10 mM NaCl, 100 mg/μl bovine serum albumin, Fermentas). The reaction was stopped by addition of 12 μl 0.5M ethylenediaminetetraacetic acid pH 8.0 followed by reversal of crosslink in presence of 10 μl Proteinase K (10 mg/ml) and 60 μl 10% sodium dodecyl sulphate and incubation at 65°C over night. DNA fragments were phenol-chloroform purified and separated on a 1% Agarose gel. Mono nucleosome DNA bands were purified (Wizard® SV Gel and polymerase chain reaction (PCR) Clean-Up System, Promega). Samples were analyzed by qPCR with three technical replicates. The CTT1 locus was covered with overlapping amplicons of about 100 bp length. Oligos are listed in Supplementary Table S4. As a reference the stable +1 nucleosome of VCX1 was used as described earlier (57). At least three biological replicates were analyzed (except Supplementary Figure S1C and D).
RNA extraction and analysis
Cells were grown to early exponential phase and treated with hyperosmotic stress. Isolation of RNA by phenol–chloroform extraction was performed essentially as described (51). RNA concentration was determined using Nanodrop 2000 (ThermoScientific) and 2 μg were used for synthesizing cDNA by Revert Aid Reverse Transcriptase (Thermo Scientific, 200U) and oligo-dT primers (0.5 μg) at 42°C for 60 min. Samples were analyzed by qPCR (Biorad CFX 96) using amplicons within the coding regions of CTT1 (centered around +275, see Supplementary Table S4) and the stress unresponsive VCX1 locus which served as an internal standard. Samples were diluted 1:40 and 5μl were analyzed in a total reaction volume of 25 μl. Data were normalized to the maximum expression of the wild-type strain (10 min). Samples were analyzed at least two times.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed as described (51). After crosslinking and digestion, spheroplasts were resuspended in 600 µl Lysis Buffer and Chromatin was sonicated to an average fragment size of 200 bp (Bioruptor, Diagenode). For immunoprecipitation we used antibodies against Rpb1 (8WG16, Covance Inc.) or Histone H3 (gift from E. Ogris, MedUni Vienna) and Dynabeads panMouse IgG (Invitrogen) for tandem affinity purification (TAP) tags and Dynabeads Protein G for Rpb1 and H3 antibodies. Purified ChIP DNA was diluted 1:10 and 5 μl were analyzed in a total reaction volume of 25 μl by qPCR. qPCR amplified amplicons centered at CTT1 −91 and +275 (for details see Supplementary Table S4) were used. As a reference we used VCX1 as described (51). All ChIP experiments were quantified by three technical replicas of at least three biological replicas. Data analysis was carried out using R packages (R Core Team 2016) through the wessa.net framework (http://www.wessa.net/ version 1.1.23-r7.).
Microarrays
wild-type and htz1Δ mutants were grown to exponential phase and treated with 0.4M NaCl for 20 min. Total RNAs from two biological replicates were isolated. Agilent's Low Input Quick Amp Labeling Kit, one-color was used to generate fluorescent cRNA. The amplified cyanine three-labeled cRNA samples were then purified using SV Total RNA Isolation System (Promega) and hybridized to Agilent Yeast (V2) Gene Expression Microarrays 8 × 15K. Microarray slides were washed and scanned with an Agilent Scanner, according to the standard protocol of the manufacturer. Information from probe features was extracted from microarray scan images using the Agilent Feature Extraction software v10.7.3. Further analyses were performed using with the Bioconductor limma package and Quality Controls were performed using the arrayQualityMetrics package. Microarray data have been deposited at GEO (GSE78766).
Tiling microarrays
Exponentially growing cells were treated with 0.4M NaCl and mononucleosomal DNA (as described for NuSA) and RNA were prepared. Hybridization and analysis followed described protocols (58). Mononucleosomal DNA samples were amplified using Sequenase 2.0 and random Primer A: GTTTCCCAGTCACGGTC(N)9 followed by purification with Illustra MicroSpin S-300 HR columns (GE Healthcare) and another amplification round using Dream Taq polymerase (Fermentas) and Primer B: GTTTCCCAGTCACGGTC in presence of 2 mM UTP. Samples were purified by MinElute PCR cleanup columns (Qiagen) before fragmentation and labeling with Affymetrix GeneChIP WT Terminal Labeling Kit. For transcription profiling total RNA was treated with RNase-free DNaseI using Turbo DNA-free kit (Ambion). For first-strand cDNA synthesis, 20 μg of total RNA was mixed with 1.72 μg of random hexamers, 0.034 μg of oligo(dT) primer and incubated at 70°C for 10 min followed by 10 min at 25°C, then transferred on ice. The synthesis included 2000 units of SuperScript II Reverse Transcriptase, 50 mM Tris–HCl, 75 mM KCl, 3 mM MgCl2, 0.01 M DTT, dNTP + dUTP mix (0.5 mM for dCTP, dATP and dGTP; 0.4 mM for dTTP and 0.1 mM for dUTP, Invitrogen), 20 μg/ml actinomycin D in a total volume of 105 μl. The reaction was carried out in 0.2 ml tubes in a thermal cycler with the following thermal profile: 25°C for 10 min, 37°C for 30 min, 42°C for 30 min followed by 10 min at 70° for heat inactivation and 4°C on hold. Samples were then subjected to RNase treatment of 20 min at 37°C (30 units RNase H, Epicentre, 60 units of RNase Cocktail, Ambion). First-strand cDNA was purified using the MinElute PCR purification kit (Qiagen) and 5 μg were fragmented and labeled using the GeneChip WT Terminal labeling kit (Affymetrix) according to manufacturer's protocol. The labeled cDNA samples were denatured in a volume of 300 μl containing 50 pM control oligonucleotide B2 (Affymetrix) and Hybridization mix (GeneChip Hybridization, Wash and Stain kit, Affymetrix) of which 250 μl were hybridized per array (S. cerevisiae yeast tiling array, Affymetrix, PN 520055). Hybridizations were carried out at 45°C for 16 h with 60 rpm rotation. The staining was carried out using the GeneChip Hybridization, Wash and Stain kit with fluidics protocol FS450_0001 in an Affymetrix Fluidics station.
Data analysis
Sequences and gene annotations were obtained from the Saccharomyces Genome Database (release 56; www.yeastgenome.org). Data handling and extraction was done in R (http://www.R-project.org. (59). Heat maps were generated using Java TreeView (60). The RNA tiling array data was normalized with R and Bioconductor using the package tilingArray as described earlier (61). The nucleosome tilingArray data were normalized by first dividing the quantile normalized genomic hybridizations.The divided values were then quantile normalized separately. Quantile normalization used the function normalizeQuantile from the R and Bioconductor package limma (62). The tilling array data derived from RNA and nucleosome samples were visualized as a heatmap (Xu et al. (85)). Expression values were obtained as the median expression at the respective strand within the annotated gene regions. Changes of gene expression were calculated as linear differences of median-normalized RNA values (Supplementary Table S2). Chromatin data of both strands were analyzed jointly. The missing data along the genome were imputed using running medians with window size 65. Chromatin levels of differentially expressed genes were visually inspected, and +1 nucleosome positions annotated in cases where it did not coincide with the annotated TSS. Chromatin plots were aligned at the +1 nucleosome position or the defined gene start and median centered over all values. The contrasts among time points or between wild-type and mutant were calculated from normalized log intensity values. Stress inducible cryptic transcripts (abbreviated as SITs) and other non-annotated transcripts were selected manually by screening the genome wide expression data in a heat map representation available at http://steinmetzlab.embl.de/schueller_ino80/index.html. Datasets have been submitted to arrayExpress with accession number E-MTAB-1810.
RESULTS
INO80 restores the promoter proximal nucleosome under stress conditions
The INO80 complex has a repressive role for heat and osmotic stress regulated gene expression (51). Previously we used ChIP to determine nucleosome content of promoter and open reading frame (ORF) regions. To increase the resolution we analyzed the chromatin structure of the highly stress inducible gene CTT1 by Nucleosome Scanning Assays (NuSA). We identified three nucleosomes (designated here as −3, −2 and −1) in the promoter region (Supplementary Figure S1). The −1 nucleosome covers the TSS and the TATA box. The transcription initiation site of the CTT1 promoter is near position −43 (relative to the ORF). Thus, the nucleosome with a dyad at approximately −50 relative to the TSS corresponds to the −1 nucleosome (20,63). We found the +1 nucleosome to be located approximately at position +130 relative to the TSS (Supplementary Figure S1A). During activation by osmotic stress (0.4M NaCl) the promoter region becomes transiently depleted of nucleosomes (Supplementary Figure S1A). We analyzed the CTT1 promoter region under stress conditions in cells lacking the INO80 subunits Arp8, Arp5 and the Ino80 core subunit (Supplementary Figure S1A, C and D). Promoter nucleosomes in cells lacking Arp8 were almost similar to wild-type (Supplementary Figure S1A), whereas in arp5Δ and ino80Δ cells nucleosome levels were reduced (Supplementary Figure S1C and D). Nucleosome eviction 10 min after exposure to hyperosmotic stress was similar in wild-type and mutants. In the wild-type the pre-exposure levels were reestablished rapidly. However, in the mutants, promoter nucleosome levels remained diminished for up to 45 min after induction. The level of the CTT1 -3 nucleosome showed a strong dependency on INO80 (Supplementary Figure S1A, C and D).
To explore a general function of INO80 for stress activated transcription and promotor chromatin structure we globally analyzed changes of nucleosome structure and transcripts. We chose to investigate arp8Δ mutants because they do not have the retarded growth of the arp5Δ and ino80Δ mutants but have reduced nucleosome remodeling activity (31,50,64). Furthermore, in the arp8Δ mutant the chromatin structure of CTT1 was similar to the wild-type (Figure 1D, time point 0, Supplementary Figure S1A). Exponentially growing cultures of wild-type and arp8Δ strains were exposed to 0.4M NaCl and samples were collected during a time course (0–45min). A graphical representation of the experimental outline is shown in Supplementary Figure S2A. Both RNA and micrococcal nuclease digested chromatin, were analyzed with genome wide tiling microarrays with 8 bp resolution. The results represented as searchable heatmaps are available online (http://steinmetzlab.embl.de/schueller_ino80/index.html). The nucleosome density data were processed as described in materials and methods and resulted in the well-established phased nucleosome pattern (Figure 1A).
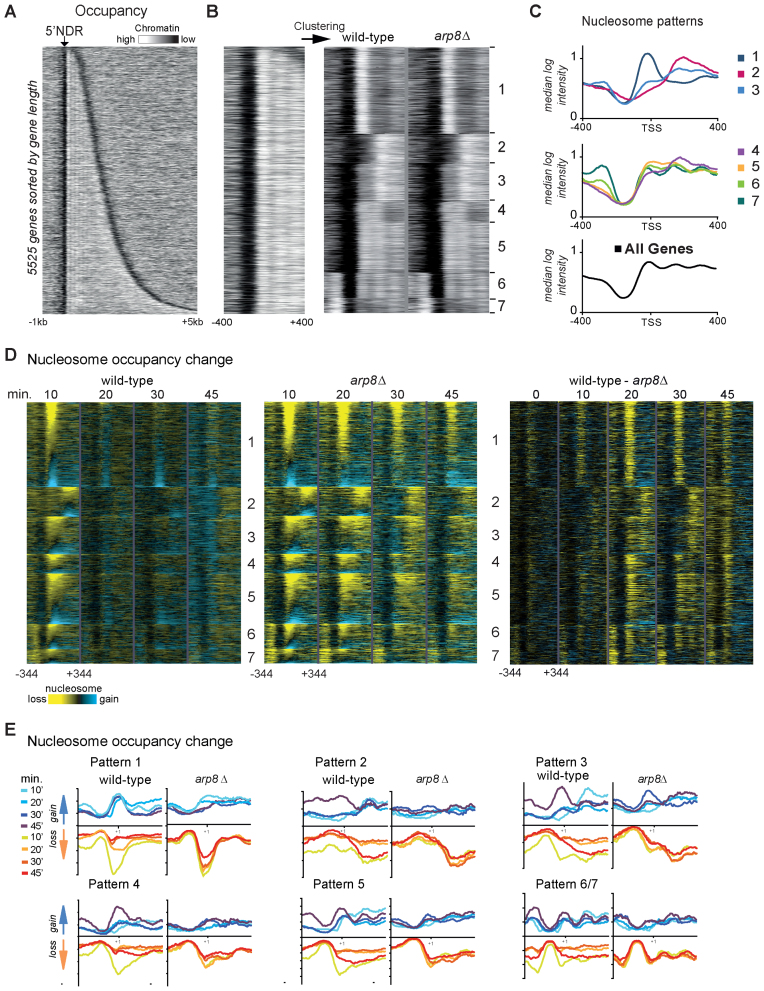
Figure 1.
INO80 alleviates osmotic stress induced chromatin remodeling. (A) Nucleosome occupancy map in wild-type cells of 5525 genes sorted by their ORF length. Each row represents 1 kb upstream and 5 kb downstream of the TSS. Data are derived from 8 bp tiling array data of MNase digested chromatin. (white: high nucleosome occupancy; black: low nucleosome occupancy). The heat map shows the typical pattern of nucleosome depleted regions flanked by positioned nucleosomes. (B) Seven different promoter chromatin patterns resulting from k-means clustering of nucleosome occupancy around the 5΄ NDR (−400 to +400 bp of the 5΄ border of +1 nucleosome) are shown. The arp8Δ mutant has a very similar pattern. Heatmaps were generated with identical settings. (C) Graphical illustration of the median nucleosome intensity of the respective promoter chromatin patterns shown in (B). (D) Nucleosome loss during hyperosmotic stress is more persistent in arp8Δ. Changes of nucleosome occupancy (reduction yellow, gain blue) in wild-type and arp8Δ (left and middle panel) are shown as difference between treated and untreated at the respective time points. The differences between wild-type and arp8Δ at every time point are also shown (right panel). Genes were ranked within the groups according to the change of the nucleosome intensity at the 10 min time point of the wild-type and adjusted to the position of the +1 nucleosome. (E) Graphs of nucleosome occupancy change (15th and 85th percentile) of patterns 1–6 show the persistence of nucleosome loss in the arp8Δ mutant.
The nucleosome density array data were qualitatively similar to the NuSA profiles (Supplementary Figure S1A and B). For the CTT1 locus, the array data of the arp8Δ mutant showed hyperinduced transcripts and prolonged eviction of nucleosomes flanking the 5΄ NDR after stress and, thus, consistency with earlier results (Supplementary Figure S1B). Genes that are repressed during hyperosmotic stress show a transient increase of promoter proximal nucleosomes to a varying degree as shown for ZEO1, URA5 and RPL3 (Supplementary Figure S1E). The transcript repression and a parallel increase of nucleosome level is slightly enhanced and sometimes prolonged in arp8Δ mutants compared to wild-type. These differences are small compared to changes at induced genes.
Alignment of the nucleosome data of all genes according to ascending gene length resulted in the typical pattern of canonical spaced nucleosomes and NDRs (Figure 1A and B). However, the same data rearranged by k-means clustering of the region near the start site (400 bp upstream and downstream) revealed distinct patterns of promoter nucleosomes in wild-type and mutant and thus, substructures within the nucleosome density data (Figure 1B). By varying this approach we could distinguish between seven characteristic patterns of promoter architecture shown here as heat maps and median intensities (Figure 1B and C). These patterns display differences in 5΄ NDR size, position and levels of the promoter proximal nucleosomes. For example, pattern 1 has a prominent +1 nucleosome covering the TSS, while pattern 2 has a +1 nucleosome located downstream of the TSS and a broader NDR (Figure 1B and C). Pattern 3–5 have positioned +1, +2 and +3 nucleosomes with different intensities, whereas 6 and 7 are characterized by a closely positioned −1 nucleosome (Figure 1B and C).
Next, we visualized the stress induced changes of nucleosome levels. We calculated the local differences of the nucleosome occupancy between treated and untreated samples of wild-type and arp8Δ and visualized them as heat maps (Figure 1D). This approach reveals that depending on the local nucleosome structure, eviction might reach beyond the first boundary (+1) nucleosome (e.g. pattern 3, 4 and 5). Hyperosmotic stress induced nucleosome gain (blue) and nucleosome loss (in yellow) does not seem to be influenced by the local pattern. In the wild-type eviction was transient and changes were largely alleviated at the 20 min time point, while in the arp8Δ mutant nucleosomes remained evicted for up to 45 min (Figure 1D and E). The largest difference between wild-type and mutant could be observed at the 20 min time point (Figure 1D). Nucleosome loss (in blue) during the stress time course seems largely not to be different in wild-type and arp8Δ. In contrast to the start site, the chromatin structure surrounding the transcription end sites was not visibly influenced by hyperosmotic stress (Supplementary Figure S2B–E). The +1 nucleosome level differs in the arp8Δ mutant at a number of genes in the unstressed state, however, osmotic stress gives rise to a different and expression specific pattern of changes (Supplementary Figure S2F). In summary, stress-induced eviction of promoter proximal nucleosomes occurs according to local patterns and re-association is delayed in the arp8Δ mutant.
Promoter proximal nucleosome dynamics correlates with stress-induced transcription
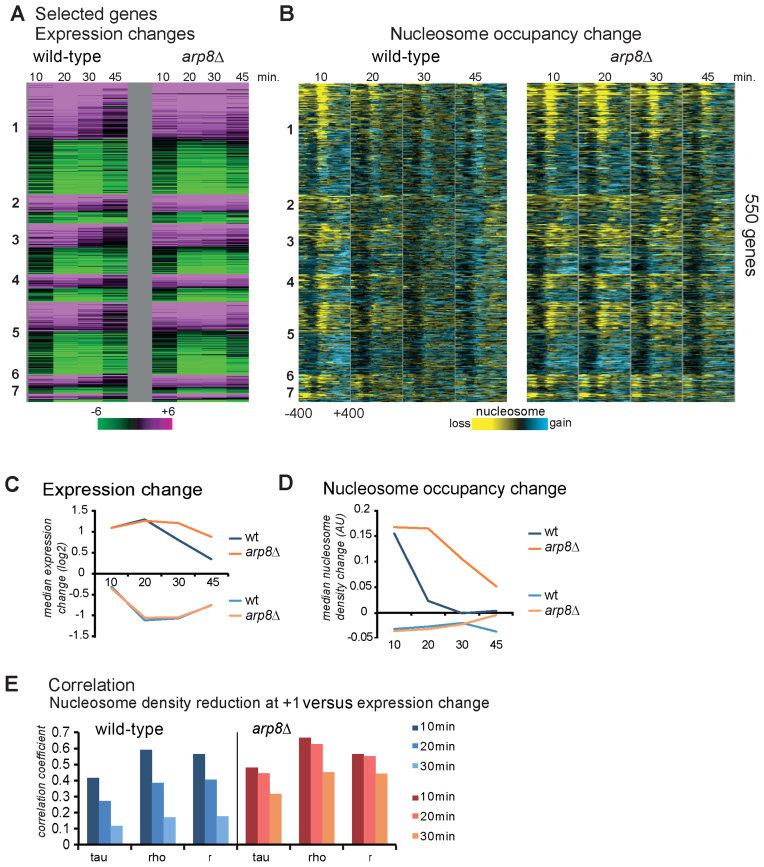
To analyze the effect of nucleosome eviction on transcripts in wild-type and arp8Δ we compared the changes of expression with the corresponding nucleosome density differences (Supplementary Figure S3). The resulting heatmaps suggested an overall trend, however, the Pearson's correlation of 0.35 (arp8Δ 20 min) for the expression change of all genes versus their nucleosome density change indicated correlation for only a subset. Therefore, we focused on the dataset of genes that are highly expressed and regulated. We selected genes from the time points with largest expression changes (20 min for arp8Δ and 10 min for wild-type) falling into the top and bottom 5th percentile according to their fold change of expression and absolute expression difference. The rationale for the inclusion of the absolute expression difference was to select only for genes with substantial transcription activity. This resulted in a selection of 550 genes. Their expression change and corresponding nucleosome density change data are shown here as heatmaps (Figure 2A and B; Supplementary Table S2). We did not find overrepresentation of the selected genes in specific promoter nucleosome patterns. Induced genes had prolonged median gene expression and nucleosome eviction in the arp8Δ mutant (Figure 2C and D). In contrast, repressed genes had similar median expression and nucleosome levels in mutant and wild-type (Figure 2C and D). For the selection we found a robust correlation (0.7) according to Spearman (rho), Kendall (tau) and Pearson (r) of promoter proximal nucleosome change and gene expression (Figure 2E). In the arp8Δ mutant the correlation persisted to later time points (Figure 2E).
Figure 2.
Nucleosome eviction and expression levels show correlation during hyperosmotic stress. (A) Gene expression changes of the top 550 expressed genes after treatment with hyperosmotic stress in wild-type (left) and arp8Δ (right) cells at the corresponding time points are shown (Relative to time point 0 at each strain). Genes were selected as highly regulated and highly expressed (top 5%) and sorted according to expression levels. Green represents downregulated and purple upregulated transcripts. (B) Differences of nucleosome levels (−400 to +400 bp around the TSS) sorted by the same criteria as in (A) are shown as heatmap (reduction yellow, gain blue). (C) Median expression levels of upregulated and downregulated genes in wild-type and arp8Δ. (D) Median nucleosome occupancy change in wild-type and arp8Δ. (E) Correlation coefficients of nucleosome density change at +1 and gene expression according to Spearman's rho, Kendall's tau and Pearson's r.
INO80 represses stress-induced activation of cryptic and bidirectional promoters
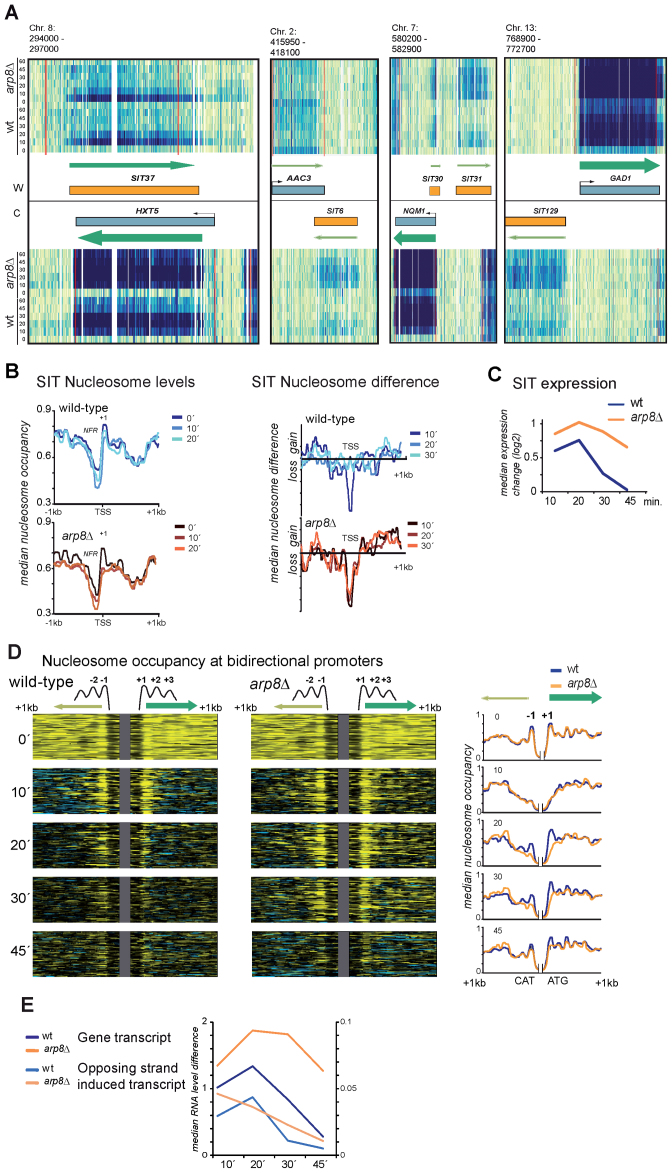
Histone modifiers and CRCs are crucial factors for the repression of transcription from cryptic promoters and divergent transcription (65–69). We manually scanned the genome-wide expression data (Figure 3A) for anomalies caused by stress treatment or absence of INO80 function and found 138 previously undetected SITs. Fifty-six of these originate from cryptic promoters owning an NDR and a positioned promoter-proximal +1 nucleosome (Supplementary Table S3). The remaining SITs were caused by bidirectional promoter activity of stress-induced genes on the opposite strand (Figure 3D and E; Supplementary Table S3). Some SITs are antisense to expressed genes as exemplified by the HXT5 locus (Figure 3A). The majority of SIT RNAs was only detectable under stress and their +1 (and sometimes +2) nucleosome became transiently evicted during stress (Figure 3A–C). SITs behaved similar to common stress-induced genes and were hyper-induced in the arp8Δ mutant accompanied by prolonged +1 nucleosome eviction (Figure 3B and C).
Figure 3.
Stress-induced cryptic transcripts are repressed by INO80. Properties of stress induced cryptic transcripts induced by osmotic stress (0.4M NaCl) ientified by genome survey. (A) Example transcript maps for SITs. RNA signal intensities for both strands (W and C) are shown for the different profiled samples (y-axis): annotated ORFs are indicated as blue boxes and SITs as orange boxes. Transcription orientation and intensity are indicated by green arrows. SIT37 and SIT30 are antisense oriented, SIT6 is downstream of AAC3, and SIT31 and SIT129 are originating from bidirectional promoters. (B) Nucleosome eviction at SITs is similar to canonical genes. Median nucleosome occupancy is reduced at SIT transcript start sites after treatment with hyperosmotic stress in wild-type cells and arp8Δ cells but nucleosome eviction (shown as nucleosome difference, right panel) persists in arp8Δ cells. (C) Mutants lacking Arp8 have prolonged SIT expression shown here as median expression change (log2) wild-type (blue) and arp8Δ (orange). (D) Eviction at strong promoters is bidirectional and enhanced in arp8Δ cells. The left panel shows nucleosome density maps of the bidirectional transcripts of the 56 most highly activated stress-induced loci. The right panel shows the median nucleosome occupancy in wild-type and arp8Δ of the selected genes. (E) Median expression differences of 5΄ opposing strand transcripts compared to gene transcripts.
Bidirectional activity of promoters is a common feature (70–72). We found that highly induced stress genes frequently contain bidirectional promoters directing induced expression of opposing strand SITs (Figure 3A, SIT31, SIT129). Activation of these promoters caused eviction of the −1 simultaneously to the +1 nucleosome (Figure 3E). In the arp8Δ mutant we observed enhanced expression of these transcripts and prolonged eviction of their promoter proximal nucleosome (Figure 3D and E). Hence, eviction of the −1 nucleosome of strongly induced genes is in fact due to bidirectional activity of the promoter and correlates with the sustained activation of the corresponding opposing transcript. These data demonstrate that INO80 represses the activity of a number of cryptic stress-inducible promoters and bidirectional promoter activity.
Stress gene regulation appears independent from the histone variant H2A.Z
Enhanced eviction of the promoter proximal nucleosomes of stress-induced genes in cells lacking Arp8 points to a possible role of H2A.Z (1,20). INO80 activity is opposing the SWR-C driven H2A.Z incorporation (31,36). Moreover, lacking of the subunit Arp5 might lead to enhanced incorporation of H2A.Z at the +1 nucleosome (31). Therefore, if INO80 is absent or has reduced activity increased levels of H2A.Z might destabilize the promoter proximal nucleosomes leading to prolonged eviction and changed expression dynamics. Moreover, H2A.Z could also influence recruitment of INO80 to stress induced genes.
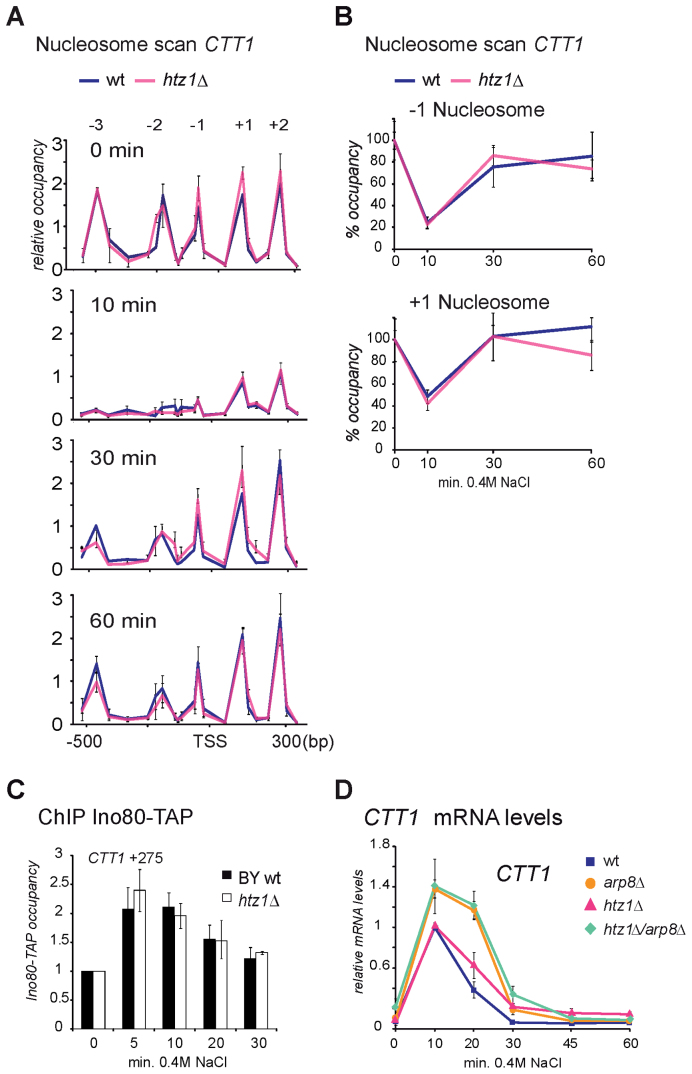
To investigate a possible role of H2A.Z (encoded by HTZ1) for eviction dynamics we analyzed the nucleosome structure of the CTT1 locus in wild-type and htz1Δ cells by nucleosome scanning. Nucleosome eviction following activation by osmotic stress and re-insertion of all detected nucleosomes including the +1 nucleosome were similar in wild-type and htz1Δ mutant (Figure 4A and B). Furthermore, a possible role of H2A.Z for recruitment of INO80 to the +1 region of CTT1 was ruled out by a ChIP experiment detecting INO80-TAP recruitment after stress exposure (Figure 4C).
Figure 4.
The H2A variant H2A.Z is not involved in INO80 related effects at stress genes. H2A.Z is not involved in delayed promoter nucleosome repopulation in arp8Δ. (A and B) Nucleosome scanning assay of the CTT1 locus (−600 to +350 bp). Wild-type and htz1Δ cells were treated with 0.4M NaCl for 10, 30 and 60 min or left untreated. NuSA are shown in (A) and quantification of NuSA of the CTT1 −1 and +1 nucleosomes in wild-type and htz1Δ are shown in (B). Graphs in (B) are normalized to the untreated wild-type sample. (C) The stress induced transient recruitment of INO80 to the 5΄ end of the CTT1 locus is similar in wild-type and htz1Δ mutant. Recruitment to the CTT1 5΄ region (+275) including the +1 nucleosome during osmotic stress (for indicated time points) was measured by ChIP of Ino80-TAP. (D) Expression of the CTT1 locus during osmotic stress is not influenced by absence of H2A.Z. Wild-type cells (BY4741), arp8Δ, htz1Δ and htz1Δarp8Δ mutants were treated with 0.4M NaCl for indicated time points. CTT1 expression levels were quantified relative to VCX1 by qRT-PCR. Expression levels were normalized to the maximum expression levels in wild-type (10 min).
Next, we analyzed whether expression dynamics of the CTT1 locus was dependent on H2A.Z. Transcript levels of CTT1 were not changed in cells lacking H2A.Z (htz1Δ) compared to wild-type and hyper-induction was similar in the arp8Δ and arp8Δ htz1Δ double mutant (Figure 4D).
To analyze a possible global impact of H2A.Z on stress gene expression we used microarrays (Supplementary Figure S4). We could not detect significant differences between wild-type and htz1Δ cells during standard (unstressed) growth conditions. The expression pattern after osmotic stress (20 min 0.4M NaCl) was not influenced by absence of HTZ1. These results suggest that H2A.Z is of minor importance for global rapid changes of stress dependent transcription.
Dynamics of promoter activity correlates with INO80 association
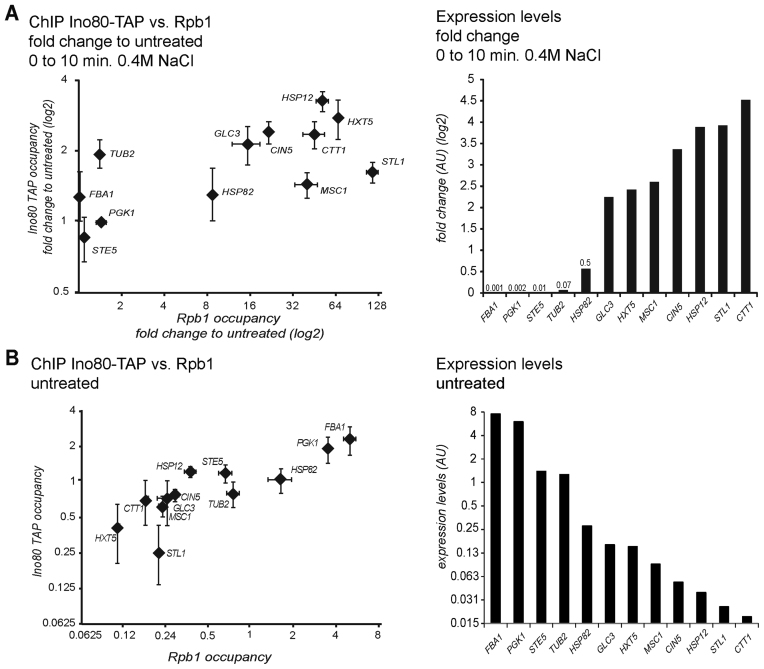
The requirement of INO80 for repression of highly inducible stress genes might suggest a general coupling of promoter activity and Ino80 recruitment. Thus, we measured by ChIP the association of both, RNA Pol II (Rpb1) and Ino80 to selected targets. We chose genes with constant expression after stress exposure (FBA1, PGK1, STE5), with small (TUB2, HSP82), medium (GLC3, HXT5, MSC1) and strong increase (CIN5, HSP12, STL1, CTT1) (Figure 5A). We found correlation between the change of RNA Pol II and Ino80 occupancy to regions close to the TSS (Figure 5A, left). This suggests a significant correlation between induced promoter activity and Ino80 recruitment (Spearman's rho = 0.69, P = 0.013; Kendall's tau = 0.55, P = 0.017; Supplementary Table S5). In addition, we compared Rpb1 and Ino80 occupancy in unstressed cells for this set of genes. The expression of the chosen genes span about three orders of magnitude. We detected a substantial amount of Ino80 at highly expressed genes and a robust correlation (Spearman's rho = 0.88, P = 0.0002; Kendall's tau = 0.72, P = 0.0012; Pearson's r = 0.91 P = 0.0003) (Figure 5B and Supplementary Table S5). Therefore, based on the small number of genes analyzed here, we suspect that association of INO80 tends to increase with promoter activity.
Figure 5.
INO80 recruitment to transcribed genes. ChIP analysis of the association of Ino80-TAP and the large Pol II subunit Rpb1 to the transcription start region for a range of stress inducible and constitutively transcribed genes. (A) Stress induction (0.4M NaCl for 10 min) leads to increased Ino80-TAP levels at the TSS region. Right panel shows mRNA levels extracted from tiling array data. ChIP signals of Ino80-TAP and Rpb1 are expressed in relation to VCX1 as reference. (B) Expression levels are correlated to Ino80 presence at the 5΄ end of genes. ChIP signals were quantified with VCX1 as reference. Standard deviation of three biological replicates is indicated.
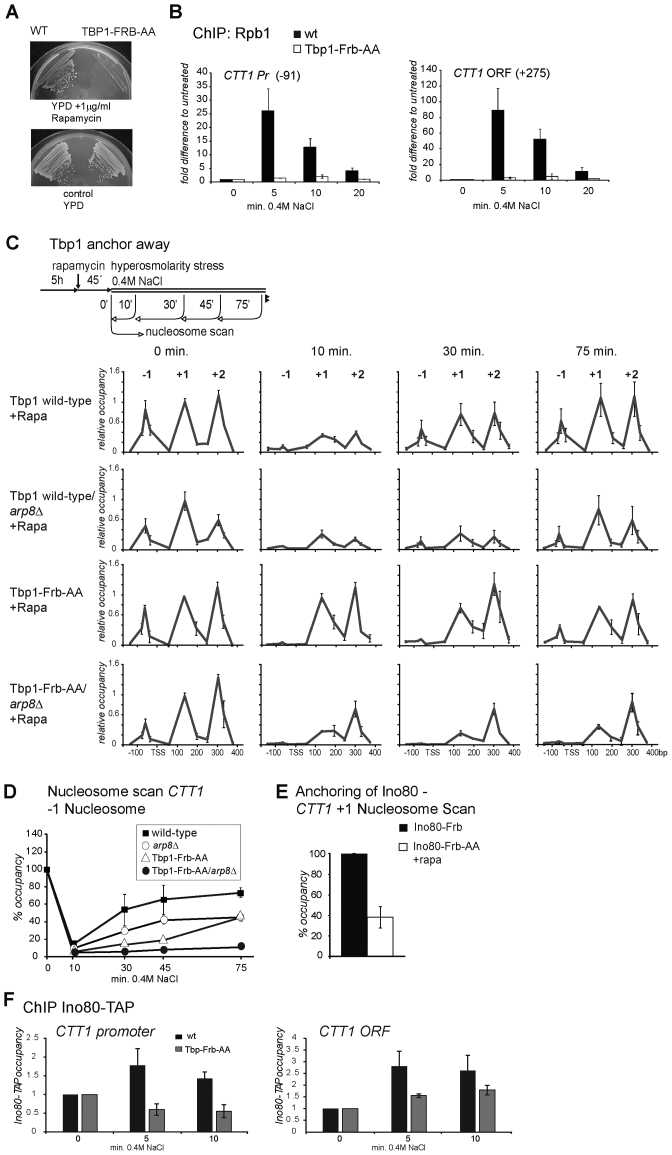
INO80 is necessary for transcription-dependent installing of promoter boundary nucleosomes
Genome-wide approaches showed the relevance of transcription for positioning of promoter proximal (+1) nucleosomes (7,23,73). Thus, INO80-facilitated insertion of these nucleosomes might be supported by interaction with components of the transcription machinery. To test this hypothesis in vivo we inhibited the formation of the PIC by conditionally removing the TATA binding protein Tbp1/Spt15 from the nucleus. We applied the Anchor-Away technique by using a Tbp1-FRB fusion protein which, in presence of rapamycin, forms a ternary complex with the ribosomal protein Rpl13A fused to FKBP12 (74). Nuclear export of the assembled ribosomal large subunit leads to rapid depletion of the tethered nuclear protein from the nucleus (Figure 6A). As expected, the rapamycin insensitive allele tor1-1 prevented any effect of rapamycin on transcripts, nucleosomes and physiology of yeast cells. Anchoring of Tbp1 reduced the transient stress induced signal of Rpb1 at the CTT1 promoter and coding region to approximately 3% of the wild-type (Figure 6B). Nevertheless, the −1 nucleosomes were depleted after exposure to hyperosmotic stress similar to wild-type (Figure 6C, third panel and 6D). The −1 nucleosome covers the CTT1 TATA-Box and is therefore likely a ‘fragile nucleosome’ that is evicted by the action of factors preceding PIC assembly, such as transcription factors and chromatin remodelers (23,75,76). Surprisingly, presence of Tbp1 was necessary for efficient increase of the −1 after stress induced eviction (Figure 6C third panel, 30 and 75 min). In contrast, the +1 nucleosome remained stably associated in the Tbp1 anchored state throughout all analyzed time points. (Figure 6C, third panel). This result shows that a basal transcription or PIC formation is necessary for re-association of the −1 and eviction of the +1 nucleosome. Prolonged depletion of the −1 nucleosome did not cause constitutive activation of stress responses (Supplementary Figure S5C and D). To distinguish nucleosome delocalization from eviction under stress in the Tbp1 anchored condition we determined histone H3 by ChIP covering the regions of CTT1 −1 and the +2 nucleosome and obtained similar results as from the Nucleosome Scan (Supplementary Figure S5B). To address the role of INO80 in conditions with low levels of PIC assembly we used a Tbp1 anchor strain lacking Arp8 (Tbp1-FRB arp8Δ). Surprisingly, deletion of Arp8 in Tbp1 anchored conditions caused almost complete and lasting reduction of both, the −1 in parallel to the +1 nucleosome (Figure 6C, lower panel and Supplementary Figure S5A). Therefore, INO80 and Tbp1 are both necessary for re-association of the −1 nucleosome after eviction but have antagonizing effects on the levels of the +1 nucleosome. Changes of the −1 nucleosome might be caused by remodeling activities triggered by the upstream (−325) binding of Msn2 and Msn4 as well as Hog1 due to osmotic stress (77).
Figure 6.
INO80 and transcription stabilize the +1 nucleosome of CTT1. (A) Depletion of the TATA binding protein 1 (Tbp1/Spt15) from the nucleus by anchoring to the cytosolic ribosomes is rapid and prevents gene transcription. The strain HHY154 (TBP1-FRB) does not grow on medium containing 1 μg/ml rapamycin while HHY168 (TBP1 wild-type) does. (B) Nuclear depletion of Tbp1 by anchoring inhibits association of RNA Pol II to promoter and ORF of the CTT1 locus. Strains HHY168 (wild-type) and HHY154 (TBP1-FRB) were treated with rapamycin (1 μg/μl, 45 min) to anchor Tbp1-FRB to the cytosol followed by treatment with 0.4M NaCl for the indicated time points. RNA Pol II was determined by ChIP of Rpb1. (C) Tbp1/Spt15 is essential for reconstitution of promoter chromatin of the CTT1 locus after osmotic stress activation. Panels show nucleosome scan data of the CTT1 promoter and a part of the coding region (−200 to +350) of wild-type (upper panel), arp8Δ (upper middle panel), TBP1-FRB (lower middle panel) and TBP1-FRB arp8Δ (lower panel). (D) Occupancy of the CTT1 −1 nucleosome after stress treatment as determined by nucleosome scan (see Figure 4B). (E) Anchoring of Ino80 to the cytoplasm leads to reduction of CTT1 +1 nucleosome levels in unstressed cells.
A prediction following the above result and previous mechanistic insights (31) would be a dynamic effect of INO80 on the stabilization of the +1 nucleosome during non-stress conditions. In fact, depletion of INO80 increases the fuzziness of nucleosomes (76). We anchored Ino80 from the nucleus and determined CTT1 +1 nucleosome levels by nucleosome scanning. After 1 h of treatment with rapamycin we measured a 60% reduction of the +1 nucleosome in the anchored strain compared to non-anchored (Figure 6E). Finally, we find that INO80 recruitment is reduced at the CTT1 locus in ORF and promoter region in the Tbp1 anchored condition under stress conditions (Figure 6F). These results show that INO80 is attracted by active transcription and required for dynamic insertion of promoter proximal nucleosomes.
DISCUSSION
Stable nucleosome positions at the 5΄ end of genes in the yeast genome are partly encoded and partly determined by a large number of remodeling factors (10,13,14,16,17,46,67,78). Stress-induced loci undergo a rapid and transient activation and allow assessment of the direct contributions of factors to the chromatin structure during the transition from silent to the induced and then to the adapted state. Here, we explore the function of INO80 during stress-induced transcriptional changes and provide evidence for its role in re-association of promoter proximal nucleosomes and for repression of transcription.
It has been suggested that chromatin remodeling driven by the INO80 ATPase may be connected to transcription as well as DNA damage repair (79). INO80 has been shown to be involved in regulation of gene expression in a number of cases (17,26,32,51,80–82). The parallel analysis of the dynamic change of nucleosome and transcript patterns under stress conditions allowed us to explore specific chromatin rearrangements connected to local changes of transcription. Classification of promoter chromatin structure using k-means clustering revealed distinct nucleosome patterns of promoter proximal nucleosomes (−1, +1, +2 and +3). These nucleosomes displayed characteristic transient eviction after hyperosmotic stress treatment. The most dramatic changes were observed in the region of the +1 nucleosome, but also at position 2 and 3. Importantly, genes with substantial expression changes had a corresponding change of nucleosome levels. The correlation of promoter nucleosomes with gene expression is interesting since promoter regions seem to be quite permissive for nucleosome changes caused by impairment of the related remodelers RSC (remodel the structure of chromatin) and SWI/SNF (83). We further demonstrate a role for INO80 as an activity largely contributing to establish promoter proximal nucleosomes after transcription induced eviction. In the arp8Δ mutant, activated genes became hyper-induced coinciding with a pronounced delay of nucleosome reappearance.
We screened the genome data for peculiarities caused by the absence of Arp8 and found several novel transcripts. Cryptic initiation of transcription within genes is prevented by histone methylation by Set2 in conjunction with Isw1B and Chd1 (67) and surveillance mechanisms degrade products of cryptic initiation events (70,72,84,85). However, in wild-type cells, specific and relatively stable transcripts emerge depending on environmental conditions. We identified a number of initiation events triggered by hyperosmolarity stress leading to transient expression of SITs. Most of these become hyper-induced in cells lacking Arp8. Antisense ncRNAs are involved in positive and negative regulation of transcription (71,86,87). Yeast species closely related to S. cerevisiae, such as Saccharomyces castellii, have an RNAi system (88). Therefore, conserved SITs could function as regulatory RNAs in other Saccharomyces species. In addition, we observed consistent bidirectional activity of promoters of highly stress-induced genes. INO80 promotes efficient downregulation of these bidirectional promoters in both directions by facilitating re-association of the +1 and −1 nucleosomes. Since INO80 is recruited to flanking nucleosomes (31) these remodeling events are presumably independent (68,69). While stress-induced genes display transient association of RNA Pol II, constantly transcribed genes were transiently reduced. The cause for the latter is perhaps due to competition for RNA Pol II or for other components required for initiation such as for example the mediator complex. Transient cessation of transcription of housekeeping genes such as ribosomal protein genes upon stress exposure was reflected by a transient increase of promoter proximal nucleosome levels.
We were interested in a possible role of H2A.Z for INO80 recruitment or promoter nucleosome turnover. The histone variant H2A.Z is predominantly present in +1 nucleosomes (reviewed in 1,19,20). SWR-C incorporates H2A.Z and INO80 catalyses the reverse reaction (36). H2A.Z has been reported to contribute to transcription of several loci and to heat shock dependent gene activation (19,89). H2A.Z is preferentially enriched at nucleosomes covering the promoters of inactive genes and becomes depleted when those genes are activated (19,90). This observation might be explained by eviction of H2A.Z containing nucleosomes from active genes followed by the rapid re-establishment of H2A nucleosomes by INO80. These are subsequently converted to H2A.Z-containing nucleosomes by SWR-C. INO80 and H2A.Z also have distinct roles for genome stability which is regulated by H2A.Z acetylation. H2A.Z-K3,8,10,14Q, a variant lacking acetylation sites, is a potent suppressor of the genomic instability phenotypes of strains lacking INO80 function. However, it does not change the gene expression pattern (36). Gene expression in htz1Δ mutants is similar to the wild-type in unstressed cells (19,90–92) and under osmostress (shown here). Furthermore, we could not detect a function of H2A.Z for the local chromatin structure dynamics of the stress-inducible CTT1 gene. Effects of the histone variant H2A.Z on promoter nucleosome dynamics are either minor or stress overridden by eviction at highly induced genes.
We addressed the interdependence of transcription initiation and INO80 activity. To test the contribution of active transcription on +1 nucleosome level changes in a dynamic system we reduced the assembly of the transcription initiation machinery by conditionally removing the TATA-binding protein Tbp1 from the nucleus. This approach reduced the stress induced association of RNA Pol II to about 3% of the wild-type. Diminished PIC assembly prevented eviction of the +1 and +2 nucleosome within the CTT1 coding region, which is consistent with previous observations (5,73,93). The CTT1 -1 nucleosome was efficiently evicted in the absence of Tbp1. This is likely caused by chromatin remodelers connected to the activity of the transcription factors Msn2/4 which are major activators of CTT1 and bind upstream of the −1 at position −325. Osmotic stress leads to recruitment of these factors and of Hog1 to the CTT1 promoter (77). The −1 nucleosome of the CTT1 promoter was not efficiently reintroduced in the Tbp1 anchored condition suggesting requirement of events coupled to transcription. In the arp8Δ mutant we find lasting eviction of the −1 and the +1 nucleosome in the Tbp1 anchored condition. Assuming a very low initiation rate in the Tbp1 anchored condition we conclude that re-installation of the evicted +1 nucleosome is dependent on INO80.
INO80 recognizes and remodels nucleosomes adjacent to non-chromatinized DNA (29) and is commonly associated with promoter regions close to the position of the +1 nucleosome (31). Our data provide an in vivo example for the dynamic aspect of INO80 activity. The function of INO80 for maintenance during steady state is supported by our analysis of a number of selected genes for which we show that association of INO80 increases proportionally to RNA Pol II and promoter activity and is therefore possibly coupled to initiation dependent eviction events. Recruitment of INO80 might also be coupled to the depletion of promoter nucleosomes. The repressive role of INO80 for transcription is suggested to be due to insertion of promoter proximal nucleosomes.
ACCESSION NUMBER
Datasets have been submitted to arrayExpress with accession number E-MTAB-1810.
Supplementary Material
ACKNOWLEDGEMENTS
We thank F. Klein, P. Kovarik and G. Adam and U. Wintersberger for critically reading the manuscript and G. Ammerer for his substantial support.
Author contributions: E.K. conceived and designed experiments, performed experiments and wrote the manuscript. H.A.S. analyzed the data and wrote the manuscript. S.C.M. performed experiments. L.M.S. conceived and designed experiments and contributed reagents and materials. C.S. conceived and performed experiments, analyzed data, contributed materials and reagents and wrote the manuscript.
Footnotes
Present Address: Eva Klopf, Max F. Perutz Laboratories (MFPL), University of Vienna, Dr Bohrgasse 9 1030 Vienna, Austria.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Research Council under the European Union's 7th Framework Programme [FP7/2007-2013, ERC Grant agreement n° AdG-294542 to L.M.S.]; Herzfelder Foundation (to C.S.); Austrian Science Fund [B23355, P19966 to C.S.]; University of Vienna [I031-B to C.S.]; Verein zur Förderung der Genomforschung (to C.S.). Funding for open access charge: Austrian Science Fund.
Conflict of interest statement. None declared.
REFERENCES
- 1. Jiang C., Pugh B.F.. Nucleosome positioning and gene regulation: advances through genomics. Nat. Rev. Genet. 2009; 10:161–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brogaard K., Xi L., Wang J.P., Widom J.. A map of nucleosome positions in yeast at base-pair resolution. Nature. 2012; 486:496–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Teif V.B., Vainshtein Y., Caudron-Herger M., Mallm J.P., Marth C., Hofer T., Rippe K.. Genome-wide nucleosome positioning during embryonic stem cell development. Nat. Struct. Mol. Biol. 2012; 19:1185–1192. [DOI] [PubMed] [Google Scholar]
- 4. Schones D.E., Cui K., Cuddapah S., Roh T.Y., Barski A., Wang Z., Wei G., Zhao K.. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008; 132:887–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schwabish M.A., Struhl K.. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 2004; 24:10111–10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Struhl K., Segal E.. Determinants of nucleosome positioning. Nat. Struct. Mol. Biol. 2013; 20:267–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hughes A.L., Jin Y., Rando O.J., Struhl K.. A functional evolutionary approach to identify determinants of nucleosome positioning: a unifying model for establishing the genome-wide pattern. Mol. Cell. 2012; 48:5–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bernstein B.E., Liu C.L., Humphrey E.L., Perlstein E.O., Schreiber S.L.. Global nucleosome occupancy in yeast. Genome Biol. 2004; 5:R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gavin A.C., Aloy P., Grandi P., Krause R., Boesche M., Marzioch M., Rau C., Jensen L.J., Bastuck S., Dumpelfeld B. et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006; 440:631–636. [DOI] [PubMed] [Google Scholar]
- 10. Bai L., Ondracka A., Cross F.R.. Multiple sequence-specific factors generate the nucleosome-depleted region on CLN2 promoter. Mol. Cell. 2011; 42:465–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kaplan N., Moore I.K., Fondufe-Mittendorf Y., Gossett A.J., Tillo D., Field Y., LeProust E.M., Hughes T.R., Lieb J.D., Widom J. et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009; 458:362–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Segal E., Fondufe-Mittendorf Y., Chen L., Thastrom A., Field Y., Moore I.K., Wang J.P., Widom J.. A genomic code for nucleosome positioning. Nature. 2006; 442:772–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hartley P.D., Madhani H.D.. Mechanisms that specify promoter nucleosome location and identity. Cell. 2009; 137:445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cairns B.R. The logic of chromatin architecture and remodelling at promoters. Nature. 2009; 461:193–198. [DOI] [PubMed] [Google Scholar]
- 15. Yuan G.C., Liu Y.J., Dion M.F., Slack M.D., Wu L.F., Altschuler S.J., Rando O.J.. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005; 309:626–630. [DOI] [PubMed] [Google Scholar]
- 16. Lee W., Tillo D., Bray N., Morse R.H., Davis R.W., Hughes T.R., Nislow C.. A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 2007; 39:1235–1244. [DOI] [PubMed] [Google Scholar]
- 17. van Bakel H., Tsui K., Gebbia M., Mnaimneh S., Hughes T.R., Nislow C.. A compendium of nucleosome and transcript profiles reveals determinants of chromatin architecture and transcription. PLoS Genet. 2013; 9:e1003479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Santisteban M.S., Kalashnikova T., Smith M.M.. Histone H2A.Z regulats transcription and is partially redundant with nucleosome remodeling complexes. Cell. 2000; 103:411–422. [DOI] [PubMed] [Google Scholar]
- 19. Zhang H., Roberts D.N., Cairns B.R.. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell. 2005; 123:219–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Albert I., Mavrich T.N., Tomsho L.P., Qi J., Zanton S.J., Schuster S.C., Pugh B.F.. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007; 446:572–576. [DOI] [PubMed] [Google Scholar]
- 21. Liu C.L., Kaplan T., Kim M., Buratowski S., Schreiber S.L., Friedman N., Rando O.J.. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 2005; 3:e328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang Y., Moqtaderi Z., Rattner B.P., Euskirchen G., Snyder M., Kadonaga J.T., Liu X.S., Struhl K.. Intrinsic histone-DNA interactions are not the major determinant of nucleosome positions in vivo. Nat. Struct. Mol. Biol. 2009; 16:847–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rhee H.S., Pugh B.F.. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature. 2012; 483:295–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. van Attikum H., Fritsch O., Hohn B., Gasser S.M.. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell. 2004; 119:777–788. [DOI] [PubMed] [Google Scholar]
- 25. Shimada K., Oma Y., Schleker T., Kugou K., Ohta K., Harata M., Gasser S.M.. Ino80 chromatin remodeling complex promotes recovery of stalled replication forks. Curr. Biol. 2008; 18:566–575. [DOI] [PubMed] [Google Scholar]
- 26. Barbaric S., Luckenbach T., Schmid A., Blaschke D., Hörz W., Korber P.. Redundancy of chromatin remodeling pathways for the induction of the yeast PHO5 promoter in vivo. J. Biol. Chem. 2007; 282:27610–27621. [DOI] [PubMed] [Google Scholar]
- 27. Conaway R.C., Conaway J.W.. The INO80 chromatin remodeling complex in transcription, replication and repair. Trends Biochem. Sci. 2009; 34:71–77. [DOI] [PubMed] [Google Scholar]
- 28. Bao Y., Shen X.. INO80 subfamily of chromatin remodeling complexes. Mutat. Res. 2007; 618:18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Udugama M., Sabri A., Bartholomew B.. The INO80 ATP-dependent chromatin remodeling complex is a nucleosome spacing factor. Mol. Cell. Biol. 2011; 31:662–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chambers A.L., Ormerod G., Durley S.C., Sing T.L., Brown G.W., Kent N.A., Downs J.A.. The INO80 chromatin remodeling complex prevents polyploidy and maintains normal chromatin structure at centromeres. Genes Dev. 2012; 26:2590–2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yen K., Vinayachandran V., Pugh B.F.. SWR-C and INO80 chromatin remodelers recognize nucleosome-free regions near +1 nucleosomes. Cell. 2013; 154:1246–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yao W., King D.A., Beckwith S.L., Gowans G.J., Yen K., Zhou C., Morrison A.J.. The INO80 complex requires the Arp5-Ies6 subcomplex for chromatin-remodeling and metabolic regulation. Mol. Cell. Biol. 2016; 6:979–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lafon A., Taranum S., Pietrocola F., Dingli F., Loew D., Brahma S., Bartholomew B., Papamichos-Chronakis M.. INO80 chromatin remodeler facilitates release of RNA polymerase II from chromatin for ubiquitin-mediated proteasomal degradation. Mol. Cell. 2015; 60:784–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xue Y., Van C., Pradhan S.K., Su T., Gehrke J., Kuryan B.G., Kitada T., Vashisht A., Tran N., Wohlschlegel J. et al. The Ino80 complex prevents invasion of euchromatin into silent chromatin. Genes Dev. 2015; 29:350–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Seeber A., Dion V., Gasser S.M.. Checkpoint kinases and the INO80 nucleosome remodeling complex enhance global chromatin mobility in response to DNA damage. Genes Dev. 2013; 27:1999–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Papamichos-Chronakis M., Watanabe S., Rando O.J., Peterson C.L.. Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell. 2011; 144:200–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hur S.K., Park E.J., Han J.E., Kim Y.A., Kim J.D., Kang D., Kwon J.. Roles of human INO80 chromatin remodeling enzyme in DNA replication and chromosome segregation suppress genome instability. Cell Mol. Life Sci. 2010; 67:2283–2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Papamichos-Chronakis M., Peterson C.L.. Chromatin and the genome integrity network. Nat. Rev. Genet. 2013; 14:62–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Alatwi H.E., Downs J.A.. Removal of H2A.Z by INO80 promotes homologous recombination. EMBO Rep. 2015; 16:986–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wu W.H., Alami S., Luk E., Wu C.H., Sen S., Mizuguchi G., Wei D., Wu C.. Swc2 is a widely conserved H2AZ-binding module essential for ATP-dependent histone exchange. Nat. Struct. Mol. Biol. 2005; 12:1064–1071. [DOI] [PubMed] [Google Scholar]
- 41. Morrison A.J., Shen X.. Chromatin remodelling beyond transcription: the INO80 and SWR1 complexes. Nat. Rev. Mol. Cell Biol. 2009; 10:373–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Watanabe S., Peterson C.L.. Chromatin dynamics: flipping the switch on a chromatin remodeling machine. Cell Cycle. 2013; 12:2338–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nguyen V.Q., Ranjan A., Stengel F., Wei D., Aebersold R., Wu C., Leschziner A.E.. Molecular architecture of the ATP-dependent chromatin-remodeling complex SWR1. Cell. 2013; 154:1220–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gu M., Naiyachit Y., Wood T.J., Millar C.B.. H2A.Z marks antisense promoters and has positive effects on antisense transcript levels in budding yeast. BMC Genomics. 2015; 16:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rege M., Subramanian V., Zhu C., Hsieh T.H., Weiner A., Friedman N., Clauder-Munster S., Steinmetz L.M., Rando O.J., Boyer L.A. et al. Chromatin dynamics and the RNA exosome function in concert to regulate transcriptional homeostasis. Cell Rep. 2015; 13:1610–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yen K., Vinayachandran V., Batta K., Koerber R.T., Pugh B.F.. Genome-wide nucleosome specificity and directionality of chromatin remodelers. Cell. 2012; 149:1461–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Clapier C.R., Cairns B.R.. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 2009; 78:273–304. [DOI] [PubMed] [Google Scholar]
- 48. Fenn S., Breitsprecher D., Gerhold C.B., Witte G., Faix J., Hopfner K.P.. Structural biochemistry of nuclear actin-related proteins 4 and 8 reveals their interaction with actin. EMBO J. 2012; 30:2153–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tosi A., Haas C., Herzog F., Gilmozzi A., Berninghausen O., Ungewickell C., Gerhold C.B., Lakomek K., Aebersold R., Beckmann R. et al. Structure and subunit topology of the INO80 chromatin remodeler and Its nucleosome complex. Cell. 2013; 154:1207–1219. [DOI] [PubMed] [Google Scholar]
- 50. Watanabe S., Tan D., Lakshminarasimhan M., Washburn M.P., Hong E.J., Walz T., Peterson C.L.. Structural analyses of the chromatin remodelling enzymes INO80-C and SWR-C. Nat. Commun. 2015; 6:7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Klopf E., Paskova L., Sole C., Mas G., Petryshyn A., Posas F., Wintersberger U., Ammerer G., Schüller C.. Cooperation between the INO80 complex and histone chaperones determines adaptation of stress gene transcription in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 2009; 29:4994–5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Görzer I., Schüller C., Heidenreich E., Krupanska L., Kuchler K., Wintersberger U.. The nuclear actin-related protein Act3p/Arp4p of Saccharomyces cerevisiae is involved in transcription regulation of stress genes. Mol. Microbiol. 2003; 50:1155–1171. [DOI] [PubMed] [Google Scholar]
- 53. Miller C., Schwalb B., Maier K., Schulz D., Dumcke S., Zacher B., Mayer A., Sydow J., Marcinowski L., Dolken L. et al. Dynamic transcriptome analysis measures rates of mRNA synthesis and decay in yeast. Mol. Syst. Biol. 2011; 7:458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Weiner A., Chen H.V., Liu C.L., Rahat A., Klien A., Soares L., Gudipati M., Pfeffner J., Regev A., Buratowski S. et al. Systematic dissection of roles for chromatin regulators in a yeast stress response. PLoS Biol. 2012; 10:e1001369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Weiner A., Hsieh T.H., Appleboim A., Chen H.V., Rahat A., Amit I., Rando O.J., Friedman N.. High-resolution chromatin dynamics during a yeast stress response. Mol. Cell. 2015; 58:371–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Biddick R.K., Law G.L., Young E.T.. Adr1 and Cat8 mediate coactivator recruitment and chromatin remodeling at glucose-regulated genes. PLoS One. 2008; 3:e1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Reiter W., Klopf E., De Wever V., Anrather D., Petryshyn A., Roetzer A., Niederacher G., Roitinger E., Dohnal I., Gorner W. et al. Yeast protein phosphatase 2A-Cdc55 regulates the transcriptional response to hyperosmolarity stress by regulating Msn2 and Msn4 chromatin recruitment. Mol. Cell. Biol. 2013; 33:1057–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. David L., Clauder-Munster S., Steinmetz L.M.. Genome-wide transcriptome analysis in yeast using high-density tiling arrays. Methods Mol. Biol. 2011; 759:107–123. [DOI] [PubMed] [Google Scholar]
- 59. R Core Team R: A Language and Environment for Statistical Computing. 2016; Vienna, Austria: R Foundation for Statistical Computing; http://www.R-project.org. [Google Scholar]
- 60. Saldanha A.J. Java Treeview–extensible visualization of microarray data. Bioinformatics. 2004; 20:3246–3248. [DOI] [PubMed] [Google Scholar]
- 61. Huber W., Toedling J., Steinmetz L.M.. Transcript mapping with high-density oligonucleotide tiling arrays. Bioinformatics. 2006; 22:1963–1970. [DOI] [PubMed] [Google Scholar]
- 62. Bolstad B.M., Irizarry R.A., Astrand M., Speed T.P.. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003; 19:185–193. [DOI] [PubMed] [Google Scholar]
- 63. Basehoar A.D., Zanton S.J., Pugh B.F.. Identification and distinct regulation of yeast TATA box-containing genes. Cell. 2004; 116:699–709. [DOI] [PubMed] [Google Scholar]
- 64. Shen X., Ranallo R., Choi E., Wu C.. Involvement of actin-related proteins in ATP-dependent chromatin remodeling. Mol. Cell. 2003; 12:147–155. [DOI] [PubMed] [Google Scholar]
- 65. Carrozza M.J., Li B., Florens L., Suganuma T., Swanson S.K., Lee K.K., Shia W.J., Anderson S., Yates J., Washburn M.P. et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005; 123:581–592. [DOI] [PubMed] [Google Scholar]
- 66. Kaplan C.D., Laprade L., Winston F.. Transcription elongation factors repress transcription initiation from cryptic sites. Science. 2003; 301:1096–1099. [DOI] [PubMed] [Google Scholar]
- 67. Smolle M., Venkatesh S., Gogol M.M., Li H., Zhang Y., Florens L., Washburn M.P., Workman J.L.. Chromatin remodelers Isw1 and Chd1 maintain chromatin structure during transcription by preventing histone exchange. Nat. Struct. Mol. Biol. 2012; 19:884–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Marquardt S., Escalante-Chong R., Pho N., Wang J., Churchman L.S., Springer M., Buratowski S.. A chromatin-based mechanism for limiting divergent noncoding transcription. Cell. 2014; 157:1712–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Alcid E.A., Tsukiyama T.. ATP-dependent chromatin remodeling shapes the long noncoding RNA landscape. Genes Dev. 2014; 28:2348–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Neil H., Malabat C., d’Aubenton-Carafa Y., Xu Z., Steinmetz L.M., Jacquier A.. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature. 2009; 457:1038–1042. [DOI] [PubMed] [Google Scholar]
- 71. Xu Z., Wei W., Gagneur J., Clauder-Munster S., Smolik M., Huber W., Steinmetz L.M.. Antisense expression increases gene expression variability and locus interdependency. Mol. Syst. Biol. 2011; 7:468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Schulz D., Schwalb B., Kiesel A., Baejen C., Torkler P., Gagneur J., Soeding J., Cramer P.. Transcriptome surveillance by selective termination of noncoding RNA synthesis. Cell. 2013; 155:1075–1087. [DOI] [PubMed] [Google Scholar]
- 73. Weiner A., Hughes A., Yassour M., Rando O.J., Friedman N.. High-resolution nucleosome mapping reveals transcription-dependent promoter packaging. Genome Res. 2010; 20:90–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Haruki H., Nishikawa J., Laemmli U.K.. The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Mol. Cell. 2008; 31:925–932. [DOI] [PubMed] [Google Scholar]
- 75. Kubik S., Bruzzone M.J., Jacquet P., Falcone J.L., Rougemont J., Shore D.. Nucleosome stability distinguishes two different promoter types at all protein-coding genes in yeast. Mol. Cell. 2015; 60:422–434. [DOI] [PubMed] [Google Scholar]
- 76. Tramantano M., Sun L., Au C., Labuz D., Liu Z., Chou M., Shen C., Luk E.. Constitutive turnover of histone H2A.Z at yeast promoters requires the preinitiation complex. Elife. 2016; 5:e14243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Alepuz P.M., Jovanovic A., Reiser V., Ammerer G.. Stress-induced map kinase Hog1 is part of transcription activation complexes. Mol. Cell. 2001; 7:767–777. [DOI] [PubMed] [Google Scholar]
- 78. Zhang Z., Pugh B.F.. High-resolution genome-wide mapping of the primary structure of chromatin. Cell. 2011; 144:175–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Shen X., Mizuguchi G., Hamiche A., Wu C.. A chromatin remodelling complex involved in transcription and DNA processing. Nature. 2000; 406:541–544. [DOI] [PubMed] [Google Scholar]
- 80. Neuman S.D., Ihry R.J., Gruetzmacher K.M., Bashirullah A.. INO80-dependent regression of ecdysone-induced transcriptional responses regulates developmental timing in Drosophila. Dev. Biol. 2014; 387:229–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Konarzewska P., Esposito M., Shen C.H.. INO1 induction requires chromatin remodelers Ino80p and Snf2p but not the histone acetylases. Biochem. Biophys. Res. Commun. 2012; 418:483–488. [DOI] [PubMed] [Google Scholar]
- 82. Ebbert R., Birkmann A., Schüller H.J.. The product of the SNF2/SWI2 paralogue INO80 of Saccharomyces cerevisiae required for efficient expression of various yeast structural genes is part of a high-molecular-weight protein complex. Mol. Microbiol. 1999; 32:741–751. [DOI] [PubMed] [Google Scholar]
- 83. Tolkunov D., Zawadzki K.A., Singer C., Elfving N., Morozov A.V., Broach J.R.. Chromatin remodelers clear nucleosomes from intrinsically unfavorable sites to establish nucleosome-depleted regions at promoters. Mol. Biol. Cell. 2011; 22:2106–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Davis C.A., Ares M. Jr. Accumulation of unstable promoter-associated transcripts upon loss of the nuclear exosome subunit Rrp6p in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:3262–3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Xu Z., Wei W., Gagneur J., Perocchi F., Clauder-Munster S., Camblong J., Guffanti E., Stutz F., Huber W., Steinmetz L.M.. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009; 457:1033–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Uhler J.P., Hertel C., Svejstrup J.Q.. A role for noncoding transcription in activation of the yeast PHO5 gene. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:8011–8016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Camblong J., Beyrouthy N., Guffanti E., Schlaepfer G., Steinmetz L.M., Stutz F.. Trans-acting antisense RNAs mediate transcriptional gene cosuppression in S. cerevisiae. Genes Dev. 2009; 23:1534–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Drinnenberg I.A., Fink G.R., Bartel D.P.. Compatibility with killer explains the rise of RNAi-deficient fungi. Science. 2012; 333:1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Talbert P.B., Henikoff S.. Environmental responses mediated by histone variants. Trends Cell Biol. 2014; 24:642–650. [DOI] [PubMed] [Google Scholar]
- 90. Wan Y., Saleem R.A., Ratushny A.V., Roda O., Smith J.J., Lin C.H., Chiang J.H., Aitchison J.D.. Role of the histone variant H2A.Z/Htz1p in TBP recruitment, chromatin dynamics, and regulated expression of oleate-responsive genes. Mol. Cell. Biol. 2009; 29:2346–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Halley J.E., Kaplan T., Wang A.Y., Kobor M.S., Rine J.. Roles for H2A.Z and its acetylation in GAL1 transcription and gene induction, but not GAL1-transcriptional memory. PLoS Biol. 2010; 8:e1000401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Santisteban M.S., Hang M., Smith M.M.. Histone variant H2A.Z and RNA polymerase II transcription elongation. Mol. Cell. Biol. 2011; 31:1848–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Lee C.K., Shibata Y., Rao B., Strahl B.D., Lieb J.D.. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 2004; 36:900–905. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.