Abstract
Dual-assignment of codons as termination and elongation codons is used to expand the genetic code. In mammals, UGA can be reassigned to selenocysteine during translation of selenoproteins by a mechanism involving a 3΄ untranslated region (UTR) selenocysteine insertion sequence (SECIS) and the SECIS-binding protein Secisbp2. Here, we present data from ribosome profiling, RNA-Seq and mRNA half-life measurements that support distinct roles for Secisbp2 in UGA-redefinition and mRNA stability. Conditional deletions of the Secisbp2 and Trsp (tRNASec) genes in mouse liver were compared to determine if the effects of Secisbp2 loss on selenoprotein synthesis could be attributed entirely to the inability to incorporate Sec. As expected, tRNASec depletion resulted in loss of ribosome density downstream of all UGA-Sec codons. In contrast, the absence of Secisbp2 resulted in variable effects on ribosome density downstream of UGA-Sec codons that demonstrate gene-specific differences in Sec incorporation. For several selenoproteins in which loss of Secisbp2 resulted in greatly diminished mRNA levels, translational activity and Sec incorporation efficiency were shown to be unaffected on the remaining RNA. Collectively, these results demonstrate that Secisbp2 is not strictly required for Sec incorporation and has a distinct role in stabilizing mRNAs that can be separated from its effects on UGA-redefinition.
INTRODUCTION
The prevailing view is that the genetic code is mainly fixed and its readout predictable. However, for a subset of genes, mRNA decoding is dynamic and may be altered to regulate expression or to produce new proteins not predicted by standard decoding rules. Both aspects are illustrated by selenoproteins, which are the fundamental paradigm for expansion of the genetic code by codon reassignment in mammals. Selenoproteins contain the rare amino acid selenocysteine (Sec) that is incorporated during translation in response to an in-frame UGA codon (1) and its insertion is regulated by selenium (Se) availability (2). In mammals, there are up to 25 genes encoding selenoproteins (3) with known Sec-dependent oxidoreductase activities involved in redox regulation, protection from oxidative damage, thyroid hormone metabolism, protein folding and cellular Ca2+ homeostasis (1).
The incorporation of Sec during translation of a mRNA in eukaryotes requires the presence of a selenocysteine insertion sequence (SECIS element) located in the 3΄ untranslated region (UTR). The SECIS element consists of a kink-turn secondary structure with a central tandem non-Watson–Crick G-A tandem pair (4) and conserved apical AAR (R = G or A) motif. At least two other trans-acting factors, the SECIS binding protein 2 (Secisbp2), which interacts specifically with SECIS elements (5), and a specialized elongation factor Eefsec are thought to be required to complete redefinition of UGA to Sec (Figure 1A) (6,7). In addition, Secisbp2 has also been shown to interact with the ribosome (8) and Eefsec (9) leading to models in which Secisbp2 plays a physical role in the recruitment of the Sec-tRNA[Ser]Sec to the ribosome. Layered on top of these core components of the Sec-incorporation machinery are other trans-acting factors, such as L30 (10), nucleolin (11,12) and Eif4a3 (13), that have been reported to modulate Sec incorporation efficiency.
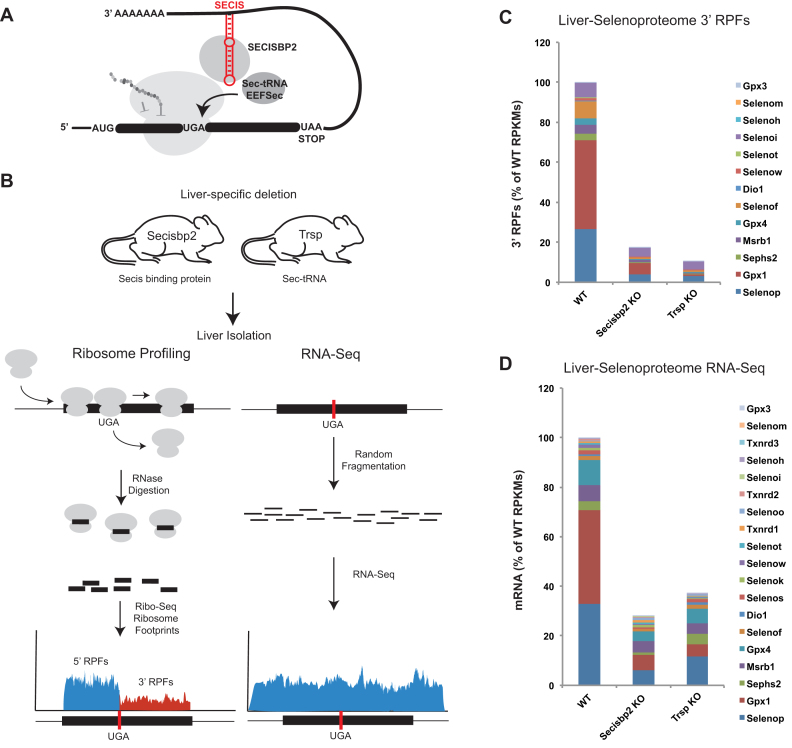
Figure 1.
Experimental design and selenoproteome analysis. (A) Illustration of current models for selenocysteine (Sec) incorporation. Included are components of the Sec insertion machinery, selenocysteine insertion sequence (SECIS) (red), SECIS binding protein (Secisbp2), Sec-tRNA[Ser]Sec (Sec-tRNA) and the elongation factor (Eefsec). (B) Schematic of ribosome profiling and RNA-Seq. For ribosome profiling, ribosome protected fragments (RPFs) are considered upstream (blue) and downstream (red) of UGA. (C) Overview of the effects of Secisbp2 and Trsp deletions on translation of all selenoproteins in mouse liver. RPFs were determined and normalized for total mapped reads and length (reads/kb/million mapped reads – RPKM) from UGA to the termination codon. RPKMs were then summed and expressed as a percentage of the wild type. (D) Same as C for randomly sheared RNA fragments (RNA-Seq) with the exception that RPKMs were determined across the entire length of selenoprotein mRNAs.
Selenoprotein expression is regulated at multiple levels and depends on the bioavailability of Se. A hierarchy among selenoproteins has been observed, which means that upon Se shortage, some selenoproteins are made at normal rates, while the biosynthesis of others is greatly diminished (14–16). One factor affecting selenoprotein expression is the efficiency of Sec incorporation. Both in vitro and in vivo experiments demonstrate that the process of Sec incorporation is an inherently inefficient process that is subject to gene-specific regulation in response to Se availability (2,17,18). Several models including differential affinity for binding of Secisbp2 to SECIS elements, competition with accessory factors (13), and Se-responsive modification of tRNA[Ser]Sec (19–21) have been proposed to account for the gene-specific translation efficiency and the effects of Se on UGA-redefinition efficiency.
Another factor implicated in selenoprotein hierarchy is differential mRNA stability (15,22). For example, glutathione peroxidase 1 (Gpx1) mRNA is sensitive to the nonsense-mediated decay (NMD) pathway (23). NMD initiates mRNA degradation if a termination codon is encountered >50 nts upstream of a splice junction complex. While the UGA-Sec codon in Gpx1 mRNA conforms to this rule, Gpx4 mRNA is relatively stable in Se-deficiency despite its homologous gene structure (24). Moreover, Sephs2 mRNA is labile, although it is produced from an intron-less gene, and many selenoprotein mRNAs carry UGA close to their termination signal within the last exon and thus cannot be substrates of NMD. These observations raise questions as to whether other cellular mechanisms are involved in determining mRNA stability and imply gene-specific effects that are not readily explained by current NMD models.
While numerous studies indicate that Secisbp2 is required for Sec incorporation in vitro, recent studies from our group have suggested that inactivation of the Secisbp2 gene in vivo is less detrimental than loss of tRNA[Ser]Sec and that Secisbp2 may have a role in stabilizing a subset of selenoprotein mRNAs in vivo (25). Here, we have developed an experimental model to examine in detail the mechanisms by which Secisbp2 affects expression of individual selenoproteins. Murine hepatocytes were chosen for conditional deletion of the Secisbp2 gene as they express a high number of selenoproteins and the organ tolerates the complete loss of selenoprotein expression. We assess UGA-redefinition efficiency by deep sequencing of ribosome protected mRNA footprints (ribosome profiling) and comparisons of the densities of ribosome protected fragments (RPFs) located 5΄ and 3΄ of UGA. In addition, effects on mRNA levels and stability were measured by RNA-Seq and half-life experiments. The results unexpectedly reveal that Secisbp2 is not absolutely required for UGA redefinition of all selenoprotein mRNAs, but rather has differential gene-specific effects on UGA-redefinition efficiency. We further propose that Secisbp2 stabilizes mRNAs on which UGA-redefinition has failed.
MATERIALS AND METHODS
This manuscript adopts the new systematic nomenclature of selenoprotein names (26).
Mice
Generation and maintenance of mice carrying hepatocyte-specific conditional alleles of Trsp and Secisbp2 were described previously (25,27). In brief, mice were kept in individually ventilated cages under a 12:12 h light-dark cycle and fed standard breeding chow according to local regulations. Whole livers were isolated from three young adult (5–8 weeks) male mice for each genetic background, Secisbp2 KO, Trsp KO and matched male littermates carrying wild-type alleles. Livers were rapidly frozen in liquid nitrogen, pulverized and stored at −80°C. To assess the completeness of Cre-mediated recombination, we crossed a Cre reporter mouse strain, ROSA26mT/mG into the conditional Secisbp2 knockout line and examined liver sections from Alb-Cre; Secisbp2fl/fl and respective control mice. Liver sections were analyzed by confocal microscopy and the fraction of non-recombined hepatocytes was 8/250 cells in control (3.2%) and 5/250 in Secisbp2 knockout (2%; Supplementary Figure S7).
Ribosome profiling and RNA-Seq
For ribosome profiling, ∼100 mg of material was suspended in 1.5 ml of lysis buffer (10 mM Tris-Cl (pH 7.5), 300 mM KCl, 10 mM MgCl2, 200 µg/ml cycloheximide, 1 mM DTT and 1% Triton X-100. Insoluble debris was removed by centrifugations at 12 000 × g at 4°C. Next 600 U of RNase1 (Ambion) were added and the sample was incubated at RT for 45 min. Monosomes were isolated by centrifugation through 50% sucrose. Ribosomes pellets were resuspended in Qiazol (Qiagen) and ribosome protected fragments were isolated using the miRNAeasy kit (Qiagen) with modification to retain small RNAs, as described by the manufacturer. For RNA-Seq, frozen pulverized liver was suspended in Qiazol, and total RNA was isolated as described by the manufacturer. PolyA mRNA was isolated using the Poly(A) Purist Mag kit (Ambion). Both RPFs and randomly fragmented PolyA enriched total RNA were electrophoresed on a 15% Tris-Borate-EDTA (TBE) Urea gel and RNA fragments between 20 and 40 nts in size were purified prior to the construction of libraries for deep-sequencing. Small RNA sequencing libraries were constructed using the Illumina TruSeq Small RNA Sample Prep kit (Illumina), according to the manufacturer's instructions. Libraries were subjected to 50-cycle single-end sequencing on the Illumina HiSeq 2000 Instrument. Data can be obtained from the NCBI GEO repository, entry GSE84112.
Bioinformatic analysis of deep-sequence data
Adapter sequences were trimmed from all sequences using the Hannon laboratory FastX toolkit. Sequences from ribosome profiling and polyA enriched libraries that aligned uniquely to selenoprotein mRNAs, allowing for two mismatches, were identified using bowtie. RefSeq entries used for this alignment are shown in Supplementary Table S2.
For RPFs, the 5΄ ends of RPFs were offset 15 nts to approximate the position of the A-site (see Supplementary Figure S1). Reads with the A-site mapping to the first and last 15 nts of the annotated coding sequence were excluded to avoid bias at the initiation and termination codon. Analysis of RPFs upstream of UGA-Sec codons excluded UGA and the five preceding codons. Total mapped reads used to derive RPKM calculations were determined by aligning sequences against RefSeq entries obtained from the UCSC genome browser in which all mRNAs derived from the same gene were reduced to a single entry corresponding to the longest isoform.
Western blot analysis
Liver protein lysates from wild-type, Secisbp2 and Trsp KO mice were extracted with RIPA buffer, electrophoresed in a 12% acrylamide SDS-PAGE gel and transferred to a nitrocellulose membrane (GE Healthcare). After blocking, membranes were probed with antibodies and detected as described (25).
Hepatocyte culture and treatment
Livers from Secisbp2, Trsp KO and wild-type mice were perfused with collagenase buffer via the vena cava. After mechanical disaggregation, cells were passed through a cell strainer, counted and plated on collagen-coated plates. After 4 h media was replaced and cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin (GIBCO).
The following day, hepatocytes were treated with 2.5 μg/ml actinomycin D or 2.5 μg/ml actinomycin D and 3 μg/ml cycloheximide (Panreac Applichem), respectively, for the times indicated in the Figure legends.
Northern blot of selenoproteins
Total RNA was extracted by Trizol (Invitrogen). A total of 5 μg of RNA sample were loaded on a denaturing agarose gel. 5S RNA and 18S RNA were used as loading controls. A 5S RNA probe was made as indicated in the tRNA Northern blot section. Membrane was exposed and radioactivity was detected by PhosphorImager (BAS-1800 II Fujifilm). Densitometry quantification was performed with AIDA Imager analyzer (raytest).
Northern blot of tRNA
Five micrograms of total RNA from mouse liver samples were electrophoresed using a 10% urea-acrylamide gel. Probes for tRNA[Ser]Sec (5΄- CGCCCGAAAGGTGGAATTGAA -3΄), tRNASer (5΄- CGTAGTCGGCAGGATTCGAA -3΄) and 5S RNA (5΄- TCTCCCATCCAAGTACTAACCA -3΄), were labeled with [γ-32P]ATP by T4 polynucleotide kinase (NEB). After washing, the membrane was exposed to a PhosphorImager screen.
Distinguishing tRNA[Ser]Sec isoforms
One gram of liver from wild type or Secisbp2 KO mice was used for total tRNA isolation. After aminoacylation with [3H]serine, seryl-charged tRNA was chromatographed twice on a RPC-5 column, initially in absence of Mg2+ and then in presence of Mg2+ as described previously (28). By two-step chromatography, tRNA[Ser]Sec can be separated and quantified from total tRNASer and distribution of two tRNA[Ser]Sec isoforms (containing either mcm5U or mcm5Um at position 34) determined (29).
RESULTS
Global analysis of selenoprotein mRNA and translation
To test how selenoprotein expression is altered when Secisbp2 and the tRNA[Ser]Sec gene (Trsp) are removed in vivo, we took advantage of two hepatocyte-specific knockout mouse models, Alb-Cre; Secisbp2fl/fl (25) and Alb-Cre; Trspfl/fl (27,30), which are deficient in Secisbp2 and tRNA[Ser]Sec, respectively. As selenoprotein synthesis is proportional to the amount of mRNA and the rate of translation, we first sought to assess these parameters and analyze to what extent they are affected by the lack of Secisbp2 or tRNA[Ser]Sec. To this end, livers were harvested and ribosome profiling and RNA-Seq experiments were performed as described under Materials and Methods. In ribosome profiling, which entails deep sequencing of ribosome protected mRNA fragments (RPFs), the number of ribosome footprints mapping to each mRNA provides a quantitative measure of translational activity across the mRNA (31). Assessment of RPFs aligned to all RefSeq mRNAs revealed that they were highly enriched in the coding sequences relative to the UTRs (Supplementary Figure S1), were ∼30 nts in size, and were positioned with a strong triplet phasing corresponding to the expected step size (3 nts) of actively translating ribosomes. For selenoproteins, it is of special interest whether, and at what efficiency, UGA is translated. While RPFs located 5΄ of UGA (Figure 1B, 5΄ RPFs) reflect the number of ribosomes that initiated translation, RPFs 3΄ of UGA (Figure 1B, 3΄ RPFs) are proportional to the number of ribosomes that have successfully redefined UGA and are translating downstream codons. Triplet phasing and ∼30 nt sizes were observed for the RPFs located both upstream and downstream of selenoprotein UGA-Sec codons (Supplementary Figure S2) demonstrating that the RPFs obtained from selenoprotein mRNAs have the expected features of footprints obtained from actively translating ribosomes. As UGA-redefinition efficiency is not 100%, we surmised that 3΄ RPFs, representing ribosomes that have successfully incorporated Sec, provide the most accurate measure of effective full-length selenoprotein synthesis rates.
To examine the aggregate effects of deleting Secisbp2 on selenoprotein translation, 3΄ RPFs for all selenoproteins (excluding selenoproteins with UGA-Sec codons near the termination codon) were normalized for gene length and total mapped sequence reads in each sample (reads/kilobase/million mapped reads – RPKMs) and shown as a percentage of selenoprotein 3΄ RPFs found in wild-type liver samples (Figure 1C). Secisbp2 inactivation results in an ∼80% reduction of 3΄ RPFs in liver. We then analyzed livers in which selenoprotein expression is abrogated by inactivation of tRNA[Ser]Sec (Figure 1C) and found a >90% reduction of 3΄ RPFs. In the absence of tRNA[Ser]Sec, the remaining 3΄ RPFs may derive from liver cells that are not hepatocytes (e.g. endothelial cells, Kupffer and Ito cells) and/or low-level near-cognate decoding of UGA and continued translation to the termination codon. Since 3΄ RPFs are less abundant in tRNA[Ser]Sec-deficient liver than in Secisbp2-deficient liver, our findings suggest that in-frame UGA-Sec codons within selenoprotein mRNAs can be successfully decoded in the absence of Secisbp2.
We then examined the abundance of selenoprotein mRNAs by RNA-Seq. RPKMs were calculated across the entire length of each selenoprotein, summed and shown as a percentage of wild type (Figure 1D). Most selenoprotein mRNAs were reduced in both mutants, however, in contrast to ribosome profiling data, deletion of Secisbp2 had a greater effect on total selenoprotein mRNA abundance than deletion of tRNA[Ser]Sec (70% and 60% reduction, respectively). Collectively, this global analysis of the selenoproteome suggests that UGA redefinition occurs at a reduced level in the absence of Secisbp2, and that Secisbp2 may play a previously under-appreciated role in maintaining selenoprotein mRNA levels.
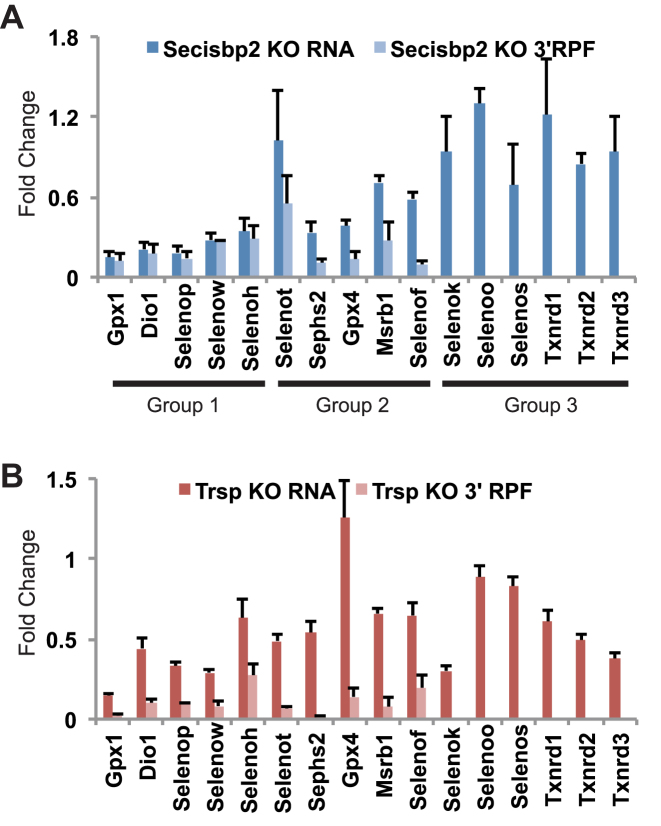
The effects of Secisbp2 and tRNA[Ser]Sec-depletion on 3΄ RPFs and mRNA levels are shown for each individual selenoprotein in Figure 2. The effects of Secisbp2-depletion on selenoprotein expression can be observed to fall into three categories (see Figure 2A), those in which mRNA abundance and 3΄ RPFs are reduced to approximately equivalent amounts (Gpx1, Dio1, Selenop, Selenow and Selenoh), those in which 3΄ RPFs are reduced to a greater degree than mRNA levels (Selenot, Sephs2, Gpx4, Msrb1 and Selenof), and the selenoproteins with UGA near the 3΄ end of the coding sequence that have relative preserved mRNA levels (Selenok, Selenoo, Selenos, Txnrd1, Txnrd2 and Txnrd3). In contrast, 3΄ RPFs are dramatically reduced for all selenoproteins in the tRNA[Ser]Sec-depleted liver although mRNAs are affected to varying degrees (Figure 2B). Notably, Gpx4 mRNA was preserved in the tRNA[Ser]Sec-deficient liver but strongly reduced by inactivation of Secisbp2. Further, several selenoprotein mRNAs with UGA codons near the end of the open reading frame were reduced in tRNA[Ser]Sec-deficient liver but somewhat less affected by the loss of Secisbp2.
Figure 2.
Comparison of the effects of Secisbp2-depletion on selenoprotein mRNA levels and translation. Fold change in selenoprotein mRNA levels and 3΄ RPFs between (A) Secisbp2-deficient, (B) tRNA[Ser]Sec-deficient and their corresponding wild-type samples were calculated. Three groups are indicated corresponding to selenoproteins in which mRNA levels and 3΄ RPFs were reduced to similar degrees (Group1), 3΄ RPFs were reduced more than mRNA levels (Group 2) and selenoproteins in which Sec is located near the C-terminus (Group 3).
Ribosome profiling measurements of Secisbp2 effects on translation initiation and UGA redefinition
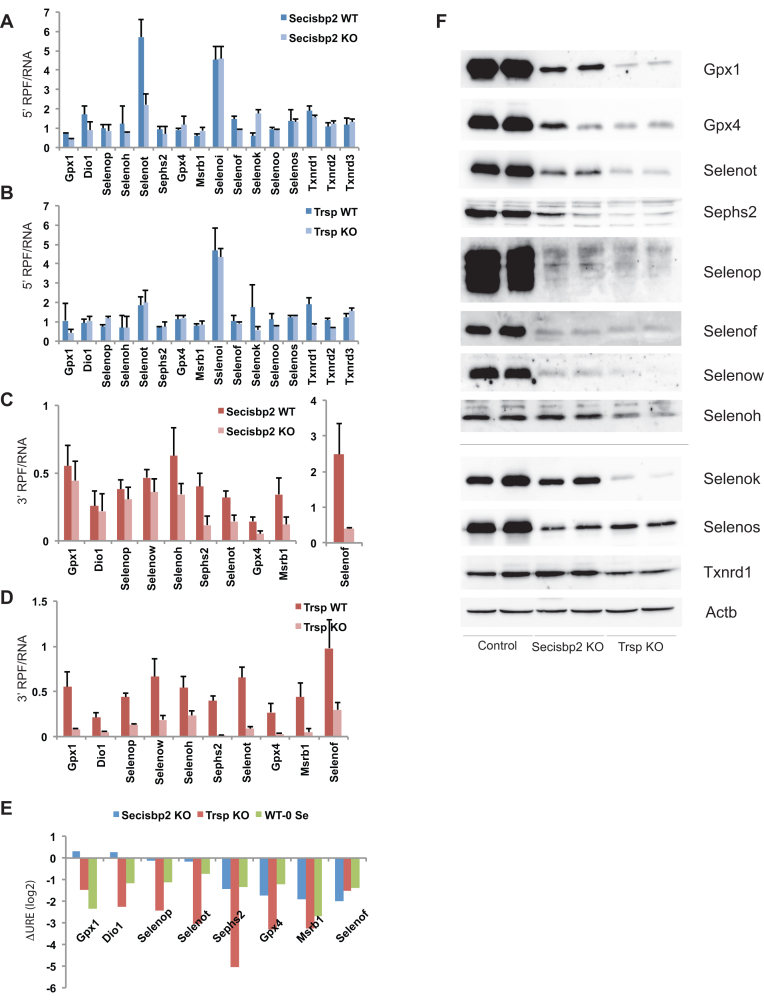
Assuming that selenoprotein translation downstream of UGA (3΄ RPFs) is proportional to not only the rate of UGA redefinition but also translation initiation, we sought to separately assess these parameters for each selenoprotein and analyze to what extent they are affected by the lack of Secisbp2 or tRNA[Ser]Sec. Changes in translational efficiency can be separated from changes in mRNA abundance by normalizing RPFs to mRNA levels (RPF density). First, we examined the RPFs upstream of UGA (5΄ RPFs) for individual selenoproteins. We surmised that an increase in translation initiation would result in an increase in 5΄ RPF density. Conversely, a reduction in translation initiation would reduce 5΄ RPF density. The 5΄ RPF densities are shown for hepatic selenoproteins in Figure 3A and B. A comparison of selenoprotein 5΄ RPF density between wild type, Secisbp2- and tRNA[Ser]Sec-deficient livers revealed that for most selenoproteins, translation 5΄ of UGA was unaffected by the loss of Secisbp2 or tRNA[Ser]Sec. Exceptions were observed for Selenok and Selenot in which 5΄ RPF density was altered approximately 2- to 3-fold upon deletion of Secisbp2, and for Txnrd1 and Txnrd2, where the loss of tRNA[Ser]Sec reduced 5΄ RPF density by approximately 1.5- to 2-fold.
Figure 3.
Ribosome profiling in Secisbp2- and tRNA[Ser]Sec-deficient mice. 5΄ and 3΄ RPFs were quantified (RPKMs) and normalized to the abundance of the corresponding mRNAs. Comparisons of 5΄ RPF density in Secisbp2- and tRNA[Ser]Sec-deficient liver relative to the wild-type control are shown in (A) and (B), respectively. Likewise, 3΄ RPF density is shown in (C) and (D). For Selenop, ribosome densities were determined upstream and downstream of the first UGA-Sec codon only. (E) UGA-redefinition efficiency (URE) is shown as the ratio between 3΄ RPF and 5΄ RPF density. The changes in URE log2 values are shown for selenoproteins in which both 5΄ and 3΄ RPF density could be determined. (F) Western blot analysis of selenoproteins from the liver of Secisbp2 KO and Trsp KO mice (Alb-Cre; Secisbp2fl/fl and Alb-Cre; Trspfl/fl, respectively).
To examine the effects of UGA redefinition and Sec incorporation on translation downstream of UGA, we compared selenoprotein 3΄ RPF density in Secisbp2-deficient, tRNA[Ser]Sec-deficient and wild-type livers (Figure 3C and D). In Secisbp2-deficient liver, 3΄ RPF density is reduced between 2- and 5-fold on Selenot, Msrb1, Gpx4 and Sephs2 mRNAs in agreement with the proposed role of Secisbp2 in mediating UGA redefinition (Figure 3C). In contrast, the density of ribosomes translating downstream of UGA is preserved in Secisbp2-deficient liver for Dio1, Selenop, Gpx1 and Selenow (see Discussion Figure 6). In tRNA[Ser]Sec-deficient liver, 3΄ RPF density was significantly reduced on all selenoprotein mRNAs, including those only moderately or not affected by Secisbp2-inactivation (Figure 3D).
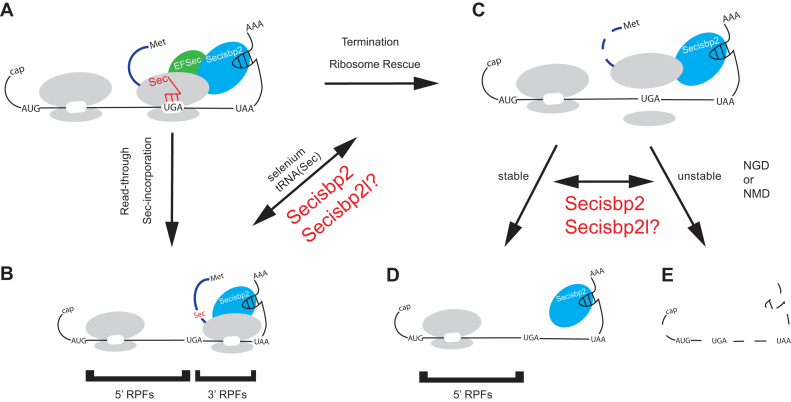
Figure 6.
A model of UGA-redefinition and selenoprotein mRNA stability. Schematic of the roles of Secisbp2 in determining gene-specific Sec incorporation rates and RNA stability when Sec incorporation fails. When the ribosome encounters a UGA during translation of (A) selenoprotein mRNAs, either (B) Sec is incorporated or (C) ribosomes are released from the mRNA. As indicated, the efficiency of Sec incorporation is affected by selenium availability, Sec-tRNA[Ser]Sec (tRNASec) modification status and abundance or Secisbp2. It is possible that the Secisbp2 homolog Secisbp2l may have Sec incorporation stimulatory activity on a subset of selenoprotein mRNAs when Secisbp2 is absent. When Sec incorporation fails, the data presented here indicates that (D and E) Secisbp2 has an additional role to play in stabilizing the mRNA, which is gene specific and separable from its role in UGA redefinition. Again, Secisbp2l may bind and thus stabilize selenoprotein mRNAs. Since inhibition of translation with cycloheximide (i.e. A-site bound to tRNA) blocks mRNA decay in Secisbp2 mutants, we propose that mRNA decay is translation-coupled.
We reasoned that the 5΄ and 3΄ RPF measurements from ribosome profiling data should allow an estimate of the impact of Secisbp2 on UGA-redefinition efficiency for each selenoprotein. The ratios of 3΄ to 5΄ RPFs were calculated excluding mRNAs where UGA was too close to the 5΄ or 3΄ end of the coding sequence or had low expression values (Table 1). These ratios ranged from 0.1 to 0.8. While the numbers are qualitatively consistent with previous findings of inefficient UGA redefinition, care should be taken not to interpret these numbers as absolute redefinition efficiencies since any differences in translation elongation rates or library construction bias between the 5΄ and 3΄ regions might impact these ratios. In addition, reduced stability of mRNAs on which termination occurs at the UGA further influences this ratio (see Discussion Figure 6). Nevertheless, it should be possible to estimate changes in UGA-redefinition efficiency on stable mRNAs by comparing the differences in 3΄ RPF:5΄ RPF ratios for each mRNA between wild-type and mutant samples. In this way, the confounding factors of translation rates and library bias should be roughly equivalent for each mRNA revealing only the change in UGA-redefinition. Changes in UGA-redefinition efficiency (ΔURE; log2) are shown in Figure 3E. The blue bars show that URE is unaffected for Gpx1, Dio1, Selenop and Selenot in Secisbp2-deficient liver, while Sephs2, Gpx4, Msrb1 and Selenof UREs are reduced. These results are in contrast to the observed reduction in UREs found for all selenoproteins in tRNA[Ser]Sec-deficient liver (red bars). For further comparison, we calculated ΔURE for the same selenoprotein mRNAs from livers of mice fed Se-deficient diets analyzed previously (2) and plotted them into the same diagram (green bars). These data show that selenoproteins with URE values that were relatively unaffected by Secisbp2-deficiency change their URE under conditions of nutritional Se deficiency, when both factors are present, but aminoacylated Sec-tRNA[Ser]Sec is limiting. The impact of RNA stability on the apparent changes in URE is considered in detail in the Discussion.
Table 1. Ratio of 3΄RPF/5΄RPF, in wild type control mice.
| Gene | Secisbp2 wild type | Trsp wild type |
|---|---|---|
| Gpx1 | 0.76 | 0.52 |
| Dio1 | 0.15 | 0.22 |
| Selenop | 0.39 | 0.59 |
| Selenoh | 0.51 | 0.78 |
| Sephs2 | 0.43 | 0.58 |
| Gpx4 | 0.16 | 0.24 |
| MsrB1 | 0.56 | 0.55 |
It should be noted that while there is some variation between wild-type mice in this table that mutants have been matched and compared to the corresponding wild-type littermates of the same genetic background, while the Secisbp2 and Trsp mouse lines are not congenic.
To confirm that the abundance of 3΄ RPFs is a good measure of selenoprotein synthesis, hepatic selenoproteins were visualized by Western blot (Figure 3F). Comparisons between the fold changes in protein abundance between wild type and Secisbp2- or tRNA[Ser]Sec-deficient livers calculated from Western blots and 3΄ RPFs had a positive correlation coefficient of R2 = 0.47 (data not shown), which is in agreement with the previously reported correlation between ribosome profiling and protein levels in yeast for the whole proteome of R2 = 0.42 (31). However, comparisons of 3΄ RPFs normalized to mRNA abundance (Figure 3C and D) and ΔURE calculations (Figure 3E) reveal that the selenoproteins in which 3΄ ribosome density was least affected by Secisbp2 deletion (e.g. Selenop, Gpx1 and Selenow), were in fact significantly reduced in protein abundance (Figure 3F and Supplementary Figure S3). For these selenoproteins, as shown in Figure 2, reduced mRNA abundance is the primary effect of Secisbp2 loss, with UGA redefinition remaining relatively unaffected during translation of the remaining mRNA. Finally, we performed 75Se metabolic labelling in hepatocytes isolated from wild type and Secisbp2 knockout mice as well as the human HAP1 cell line carrying a null mutation in Secisbp2 to illustrate that Sec is incorporated in the absence of Secisbp2 (Supplementary Figure S4). 75Se labeling can clearly be seen for several selenoproteins, including Txnrd1 and Gpx4, in cells lacking Secisbp2.
Ribosome coverage analysis
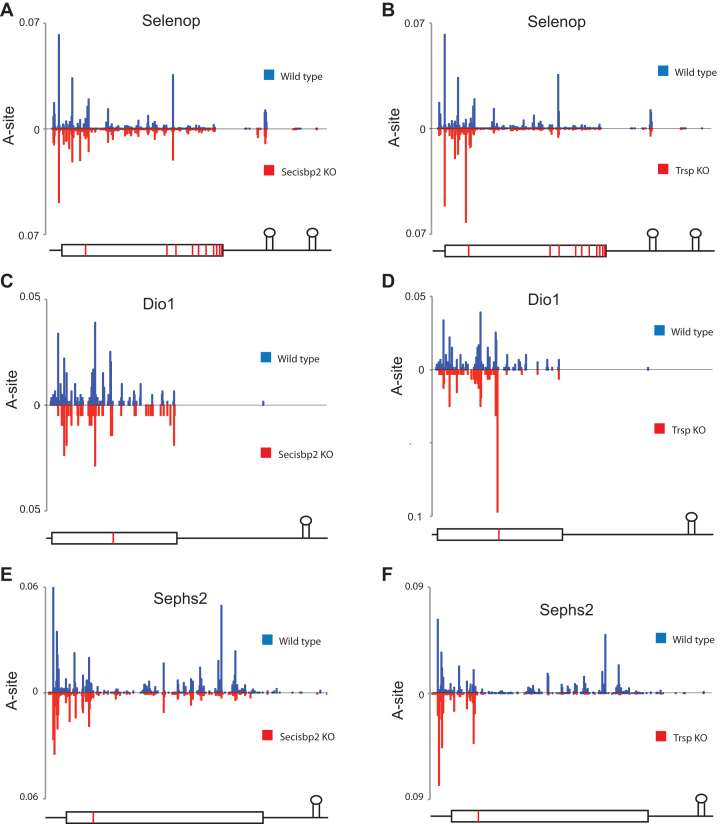
Ribosome A-site coverage corrected for mRNA abundance is shown in Figure 4. Selenop is the only mammalian selenoprotein with multiple UGA-Sec codons. As a Se transport protein, it contains up to 10 Sec residues in mice, which are encoded by 10 UGA codons in the mRNA. Consistent with the results of Figure 3, it can be seen in Figure 4A that the distribution of RPFs density across the Selenop coding sequence is similar in wild type and Secisbp2-deficient liver. In contrast, in tRNA[Ser]Sec-deficient liver (Figure 4B), ribosome density upstream of the first UGA-Sec is comparable to wild type with the exception that ribosome density is increased immediately 5΄ to the first UGA. Downstream of the first UGA-Sec codon, RPFs are significantly lower in Trsp compared to Secisbp2 mutants. In addition, the presence of two ribonuclease resistant fragments can be observed in Selenop that overlap with the first SECIS element (Figure 4A and B; see Supplementary Figure S5A for nucleotide level protection of SECIS 1). The size of the first ribonuclease resistant fragment (∼30 nts) is consistent with a ribosome footprint, whereas the second ribonuclease resistant fragment is larger (∼38 nts) (Supplementary Figure S5B and S5C).
Figure 4.
RPF A-site coverage map for Selenop, Dio1 and Sephs2. The A-site position of each RPF was determined in wild type (blue-plotted above the x-axis) and Secisbp2- or tRNA[Ser]Sec-deficient liver (red- plotted below the x-axis) for each gene, summed at each position and normalized to million mapped reads and RNA levels. (A and B) Selenop A-site plots. (C and D) Dio1 A-site plots. (E and F) Sephs2 A-site plots. The position of UTRs (thin lines), coding sequences (rectangular box), UGA-Sec codons (red vertical bars) and SECIS structures are indicated graphically below the plots for each gene.
In the case of Dio1, Secisbp2 deletion has little effect on downstream RPF density (Figure 4C), whereas Trsp deletion severely reduces 3΄ RPF density (Figure 4D). Similar to Selenop, RPFs immediately upstream of the Dio1 UGA-Sec codon show increased density when tRNA[Ser]Sec is deficient. In contrast to the above examples, Sephs2 reveals a loss of RPFs 3΄ of UGA in the Secisbp2 deletion (Figure 4E), although not to the same degree as is seen in the Trsp deletion (Figure 4F).
To further examine the effect of Secisbp2 deficiency on ribosome pausing near UGA, the position of ribosome A-sites for all RPFs that were positioned with the ribosomal A-site codon located either at UGA or in the 5 preceding codons was determined as a percentage of all RPFs on the same mRNA (Supplementary Table S1). As observed for changes in dietary Se levels, the stress-related selenoproteins Gpx1, Selenow, Selenoh and Msrb1 revealed the highest levels of ribosome pausing upstream of UGA with relatively few RPF A-sites residing on UGA. Pausing upstream of UGA was increased in the Trsp knockout and under conditions of Se deficiency for several of the stress related selenoproteins, compared with samples obtained from wild type or Secisbp2 deletion mice.
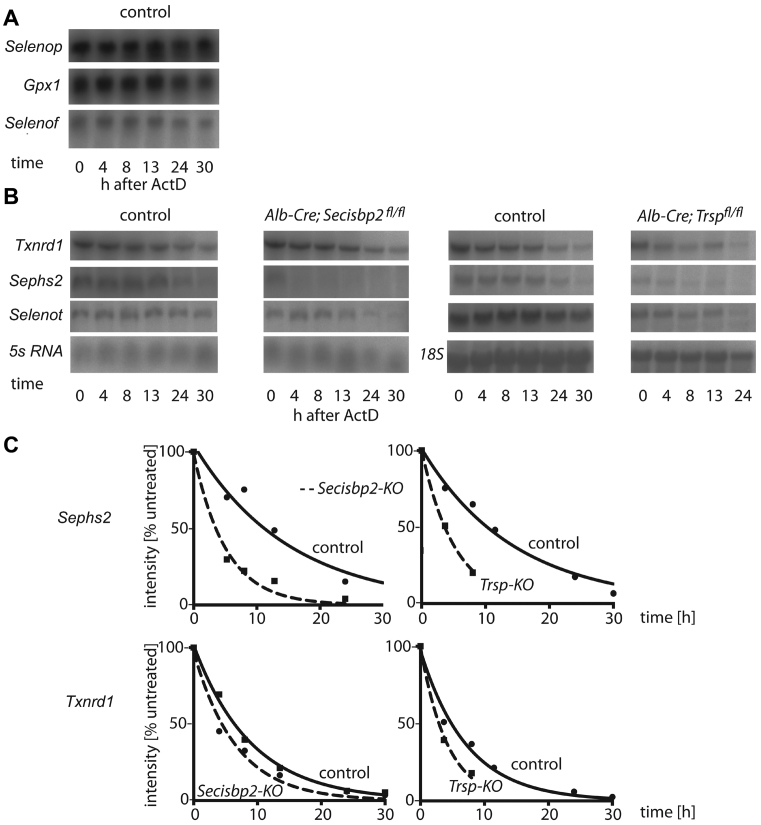
Effects on selenoprotein mRNA half-life
We assessed selenoprotein mRNA stability in primary hepatocytes isolated from mice. Cells were exposed to actinomycin D to inhibit RNA polymerase II activity and decay of mRNAs was followed by Northern blot analyses over a period of 30 h. Messenger RNA half-lives were calculated from auto-radiographic images for Secisbp2-deficient and wild-type hepatocytes and normalized to ribosomal RNA levels. Selenoproteins like Gpx1, Selenop and Selenof displayed reduced mRNA levels in Secisbp2 mutant cells, but their half-lives, in the range of days, were too long to be reasonably determined within 30 h of cell culture in the presence of actinomycin D (Figure 5A). Txnrd1, Sephs2 and Selenot mRNA stability could be reasonably assessed (Figure 5B and C). Txnrd1 a selenoprotein mRNA mildly affected by mRNA degradation displayed half-lives of 8.1 h and 7.2 h in wild-type control and Secisbp2-deficient cells, respectively. Sephs2 mRNA half-life was reduced from 10.5 h in controls to 3.6 h in Secisbp2-deficient cells. Likewise, Sephs2 mRNA half-life was reduced to 3.6 h in tRNA[Ser]Sec-deficient cells compared to 10.8 h in the wild-type control culture. The Txnrd1 mRNA half-life was 2.9 h in tRNA[Ser]Sec-deficient cells compared to 5.1 h in controls. Selenot was the only selenoprotein mRNA with relatively unaffected URE that could be assessed for half-life. Its half-life in Secisbp2-deficient cells was 19.6 h, while it was 20.1 h in wild type. In comparison, Selenot half-life in the absence of tRNA[Ser]Sec was 13.7 h versus 15.1 h in wild-type controls.
Figure 5.
Selenoprotein mRNA stability in primary hepatocytes. (A) Selenop, Gpx1 and Selenof mRNA levels were stable over 30 h of actinomycin D treatment such that half-lives could not be reliably estimated. (B) Txnrd1, Sephs2 and Selenot mRNAs half-lives were determined in the presence of actinomycin D in Secisbp2-deficient, tRNA[Ser]Sec-deficient and control cell cultures. (C) Graphs showing the degradation of Sephs2 and Txnrd1 mRNA in Secisbp2- and tRNA[Ser]Sec-deficient cells. 5s RNA and 18s RNA were used as loading controls.
To test whether selenoprotein mRNA destabilization is translation-dependent (e.g. NMD or No-go decay), we assessed the stability of Txnrd1 and Sephs2 mRNA in the presence of cycloheximide. A dose-response study on wild-type cells was performed to determine the optimal concentration of cycloheximide (Supplementary Figure S6). The incubation over 30 h in the presence of both actinomycin D and cycloheximide stabilized both Sephs2 and Txnrd1 mRNAs. Sephs2 showed half-lives of 7.9 h and 8.4 h in controls and Secisbp2-deficient cells, respectively (Supplementary Figure S6B). These data suggest that the shorter mRNA half-lives of Sephs2 mRNA in Secisbp2-deficient cells depend on a translation-dependent mRNA degradation pathway.
tRNA[Ser]Sec abundance and 2΄-O methylation status
2΄-O methylation of tRNA[Ser]Sec at the wobble base U34 is responsive to bioavailability of Se (32,33). It may be argued that the gene-specific response of selenoproteins to Secisbp2 deficiency reflects tRNA[Ser]Sec methylation status or abundance. To investigate this possibility, we isolated tRNA from Secisbp2-deficient and wild-type liver, enriched tRNA[Ser]Sec, charged the tRNAs with [3H]-Ser and assessed tRNA[Ser]Sec profiles by chromatography on a RPC-5 column. As shown in Supplementary Figure S7A, tRNA[Ser]Sec from both wild type and Secisbp2-deficient liver showed the characteristic double peak with the earlier eluting left peak representing U34 tRNA[Ser]Sec and the later eluting right peak representing Um34 tRNA[Ser]Sec. Thus, there is no indication that lack of Secisbp2 interferes with tRNA[Ser]Sec U34 2΄-O-methylation or affects the abundance of tRNA[Ser]Sec (Supplementary Figure S7B).
DISCUSSION
Global analysis of selenoprotein expression suggests a model describing dual roles for Secisbp2 in UGA redefinition and gene-specific rates of RNA decay
The redefinition of UGA codons to Sec illustrates that the readout of the genetic code need not be fixed across the genome and, in the case of selenoproteins, is required for the insertion of the uncommon amino acid Sec that is utilized for selenoprotein mediated redox reactions. Since UGA redefinition is thought to depend on the SECIS-Secisbp2 interaction, we analyzed liver-specific Secisbp2-deficient mice in order to probe the role of Secisbp2 in UGA redefinition and selenoprotein biosynthesis. tRNA[Ser]Sec-deficient mice were also used here as a negative control, because UGA redefinition and thus, selenoprotein expression is dependent on Sec-tRNA[Ser]Sec. Ratios of RPFs upstream and downstream of UGA codons were utilized as a proxy for UGA-redefinition efficiency. The results demonstrate that UGA redefinition occurs with low efficiency, as had been suggested by studies with model and in vitro experimental systems (18,34–37).
By comparing changes in selenoprotein mRNA levels directly with 3΄ RPFs (Figure 2) in Secisbp2-deficient tissue, we find that the effects of loss of Secisbp2 can be categorized into three groups: (i) Selenoproteins in which RNA and 3΄ RPFs are reduced to equivalent levels (Gpx1, Dio1, Selenop, Selenow and Selenoh) suggesting that mRNAs on which UGA redefinition fails are rapidly degraded and that Sec incorporation on the remaining mRNA occurs with near wild-type efficiency. (ii) Selenoproteins in which 3΄ RPFs are reduced more than the mRNA levels (Selenot, Sephs2, Gpx4, Msrb1 and Selenof), which is consistent with a partial reduction in mRNA stability and translation, but clearly shows that Secisbp2 affects the efficiency of UGA-redefinition; and (iii) selenoproteins with near terminal UGA codons in which RNA levels are mostly preserved (Selenok, Selenoo, Selenos, Txnrd1, Txnrd2 and Txnrd3). The observation that a subset of selenoproteins are strongly reduced in mRNA abundance, but that 3΄ RPFs are preserved on the remaining RNAs in the absence of Secisbp2 demonstrates that, in contrast to the prevailing view, Secisbp2 is not absolutely essential for UGA recoding. In addition, the RNA levels for several mRNAs are reduced to a greater degree by the loss of Secisbp2 than by loss of Sec-tRNA[Ser]Sec indicating that Secisbp2 protects these mRNAs from degradation in a manner that is not simply due to defects in Sec incorporation.
We introduce ΔURE (Figure 3) as a new parameter to assess the combined impacts of a biological condition or genetic mutation on both RNA stability and UGA-redefinition efficiency (Figure 6). As shown schematically in Figure 6, upon encountering a UGA codon (Figure 6A), the ribosome either redefines the UGA codon (Figure 6B) and continues translation or fails to incorporate Sec (Figure 6C) and is released from the mRNA. In the instances when redefinition fails, ribosomes may either terminate or perhaps be released by the No-go decay pathway, either of these outcomes may lead to RNA decay (Figure 6D and E). The ability to monitor 5΄ RPFs when Sec incorporation fails is dependent on the extent of RNA degradation. For example, when RNAs are stable, the 5΄ RPFs are fully accounted for and ΔURE is proportional to changes in UGA redefinition efficiency (URE). However, if the mRNA is rapidly degraded following failure to redefine UGA, then ribosomes 5΄ of the UGA will be lost and the corresponding 5΄ RPFs will be underestimated leading to an apparent increase in ΔURE. This observation is most clearly reflected in the group 1 selenoproteins (Gpx1, Dio1, Selenot and Selenop) where ΔURE is unchanged by Secisbp2 deletion. We interpret these results to indicate that the failure to redefine UGA in these mRNAs leads to rapid RNA decay such that 5΄ and 3΄ RPFs are only measured for the subpopulation of mRNAs on which UGA redefinition was successful. Conversely, those mRNAs showing reductions in ΔURE have more stable mRNAs that escape RNA decay even when UGA redefinition fails. Due to the confounding effects of changes in RNA decay, ΔURE does not reflect actual changes in UGA redefinition, and thus, should only be used as a measure of changing URE if mRNA levels are unchanged between the conditions compared. Regardless, these findings, as well as the observation that many selenoprotein mRNAs are more stable in the absence of Sec-tRNA[Ser]Sec than Secisbp2, indicate that Secisbp2 stimulates, but is not required for UGA redefinition, and may additionally affect the stability of mRNAs on which UGA redefinition failed.
Ribosomal profiling of selenoproteins as a tool to assess redefinition
We assume that UGA-redefinition is reasonably reflected by the abundance of 3΄ RPFs, since these correlate well with protein levels as judged by western blot (R2 = 0.47). It may be assumed that read-through of UGA might be achieved by recruitment of near-cognate tRNAs (Cys in response to UGY codons, Trp in response to UGG codon) or even tRNAs misreading the second base (Ser for UCA, Leu for UUA). Insertion of any amino acid except Cys is expected to result in catalytically inactive selenoenzymes and Cys incorporation will reduce activity by orders of magnitude, e.g. as observed for the hepatocyte-specific selenoenzyme Dio1 (34,38). Dio1 activity is greatly reduced in primary hepatocytes isolated from liver-specific Secisbp2 KO mice, but in proportion to its mRNA level (25). If Sec was replaced by Cys or any other amino acid in this enzyme, its residual activity would have been even lower. In addition, the mRNA level of Txnrd1 was only marginally reduced in livers from these mice and activity in the Sec-specific Txnrd1 assay of insulin reduction was almost 20% of wild type (25). Thus, enzymatic activity of two enzymes is supported by Sec incorporation in Secisbp2 KO cells. In order to investigate whether Sec is still incorporated into selenoproteins, we have metabolically labeled primary hepatocytes derived from Secisbp2 KO mice and found that 75Se incorporation is greatly reduced, yet detectable, in particular in the more abundant selenoproteins Txnrd1, Gpx4 and Selenof (Supplementary Figure S4A). The same has recently been observed in a human haploid HAP1 cell line engineered deficient in Secisbp2. Dubey and Copeland have metabolically labeled these cells with 75Se and showed that 75Se-selenoprotein labeling is reduced, but clearly present (39). Similarly, we found significant 75Se incorporation into TXNRD1 in SECISBP2-deficient HAP1 cells. Western blot analysis of TXNRD1, GPX1 and GPX4 showed that GPX4 is less affected than GPX1 in HAP1 SECISBP2-deficient cells (Supplementary Figure S4B).
While translation initiation rate estimated from 5΄ RPF density varies significantly between selenoproteins, it remains relatively unchanged upon Secisbp2 inactivation for most selenoproteins, with the notable exception that loss of Secisbp2 appears to reduce 5΄ RPFs of Selenot, which is not regulated on the level of URE or mRNA abundance. Overall, it is evident from the data that 3΄ RPF density is higher in Secisbp2-deficient livers than in tRNA[Ser]Sec-deficient livers. These findings support our earlier results that suggested limited Secisbp2-independent translation of several selenoprotein mRNAs (25).
Coverage plots along the length of selenoprotein mRNAs show, at high resolution and for individual selenoproteins, how the lack of Secisbp2 or tRNA[Ser]Sec impacts the read-through of UGA-Sec codons. In the case of Selenop, the largest drop of coverage is seen after the first UGA, while at later UGAs the URE is relatively unchanged (Figure 4A). This is consistent with earlier findings (36,40,41) and suggests that a ribosome having successfully incorporated Sec can more efficiently read through subsequent UGA-Sec codons. While such a model explains the ability of ribosomes to produce full-length Selenop with its multiple Sec residues, the data clearly indicate that incorporation of Sec during translation of selenoprotein mRNAs, including at least the first UGA-Sec of Selenop, is a rate-limiting step in selenoprotein translation that is subject to regulation.
Upon deeper analysis of the profiling data, two new ribonuclease resistant fragments were detected in the 3΄ UTR of Selenop mRNA overlapping SECIS1. As shown in Supplementary Figure S5, the two ribonuclease resistant fragments cover an extended region of SECIS1 outside the region known to be bound by Secisbp2 (5). The experimental procedure involving purification of ribosomes suggests that the ribonuclease resistant fragments were either protected by a ribosome or by a complex that associates with ribosomes. Although ribosomes are greatly depleted in 3΄ UTRs, a recent study demonstrates that ribosomes do access the 3΄ UTR in certain circumstances and that components of the No-Go decay pathway, which can clear ribosomes that are unable to terminate normally, are involved in release of those ribosomes (42). The protected fragments cannot be due to protection by Secisbp2, because they were also found in Secisbp2-deficient livers.
For selenoproteins involved in stress-related responses, such as Gpx1, Selenow, Selenoh and Msrb1, we see significant accumulation of ribosome footprints upstream of UGA. These results imply that a delay in translation occurs prior to the ribosome encountering UGA that is either unique or particularly pronounced for these mRNAs. Interestingly, the lack of Secisbp2 does not lead to increased ribosomal stacking upstream of UGA-Sec codons compared to the wild type, while increased ribosome density is observed in livers lacking Sec-tRNA[Ser]Sec (Figure 4 and Supplementary Table S1). Perhaps this delay is required in order for the ribosome to receive signals for Sec insertion that is dependent upon the Sec-tRNA[Ser]Sec prior to encountering UGA. Whether a lack of ribosome stacking reflects rapid termination at the UGA codon when Sec incorporation is reduced, or perhaps clearing of paused ribosomes by translational quality control mechanisms such as No-go decay (43) deserves further study.
Secisbp2 stabilizes selenoprotein mRNAs
Selenoproteins Dio1, Selenot, Selenop and Gpx1 illustrate that regulation of selenoprotein expression can operate through different mechanisms. While their UREs remained unchanged on a subpopulation of mRNAs in Secisbp2-deficient liver, their mRNA levels were reduced to an extent that largely explained their reduced protein amounts. Reduced Gpx1 and Dio1 mRNA levels are consistent with earlier reports on dietary Se deficiency in rats, where these mRNAs were decreased and mRNA half-lives were reduced (44). Similar results have been found in Secisbp2-knockdown cells (45). One surprising observation was the finding that Sephs2 and Gpx4 mRNA levels were decreased in the Secisbp2 mutant, and notably Gpx4 was unaffected in the Trsp mutant. This implies that Gpx4 and Sephs2 mRNAs, which are resistant to changes in Se levels, are stabilized by a mechanism involving Secisbp2.
We have analyzed the role of Secisbp2 in regulating mRNA stability by determining the half-lives of selenoprotein mRNAs in primary hepatocytes in the presence of actinomycin D. The half-lives of Txnrd1 (9 h) and Selenot (15 h) are near the median of 9 h found for mRNAs in murine 3T3 cells (46). Gpx1, Selenop and Selenof mRNAs had very long half-lives that were too long to be determined reliably (20–50 h). This finding is consistent with two of these transcripts being among the most abundant in liver. The half-life of Sephs2 mRNA is clearly reduced from 10.5 to 3.6 h in Secisbp2-deficient hepatocytes. However, as inactivation of Trsp leads to the same change in mRNA stability, it remains unclear whether this effect is specific for Secisbp2 or related to the general inability to translate Sephs2 mRNA. Similarly Txnrd1 mRNA is destabilized by both mutations (half-lives reduced from 6–7 to 3–5 h).
NMD was suggested as a mechanism of Gpx1 mRNA destabilization and UPF1 identified as a factor involved (23,47), but neither Sephs2 nor Txnrd1 are canonical NMD targets (48). By incubation of primary hepatocytes from the Secisbp2 mutant and controls with actinomycin D and cycloheximide, we found that Sephs2 and Txnrd1 mRNAs are stabilized by inhibition of translation. These observations combined with evidence from ribosome profiling that ribosome pausing occurs prior to the ribosome encountering UGA in a subset of selenoproteins raises the question as to whether the No-Go decay translational control pathway (43) may, in addition to NMD, be involved in regulating selenoprotein mRNA levels. Consistent with this view is the observation that a mutation within the selenocysteine redefinition element of Selenon that decreases read-through of UGA also destabilizes the respective mRNA in the muscle of a patient (49). It is also intriguing that ribosome pausing as well as the effects on RNA levels are more pronounced in the tRNA[Ser]Sec-deficient tissue when Secisbp2 is present. Based on this, it is tempting to speculate that in the presence of Secisbp2, information is conveyed to the ribosome to decode UGA as Sec rather than terminate and that consequently, when Sec-tRNA[Ser]Sec is absent, the ribosome stalls waiting for the Sec-tRNA[Ser]Sec that never arrives, thus activating the No-Go Decay pathway.
Can Secisbp2l compensate for the lack of Secisbp2?
The low level of Sec redefinition in the absence of Secisbp2 in hepatocytes raises the question whether its paralog Secisbp2l may function at low level in selenoprotein expression. This question may be best answered in a Secisbp2/Secisbp2l double mutant mouse model, provided that inactivation of Secisbp2 is not lethal to the animal. The physiological role of Secisbp2l, however, is not known, albeit Secisbp2l genes are not present in plants and fungi, organisms that do not possess the selenoprotein biosynthesis machinery (50). The human gene was cloned owing to its sequence homology to Secisbp2, which is greatest in the RNA-binding domain and the selenocysteine insertion domain (50). Human Secisbp2l did not support Sec incorporation in an in vitro system of selenoprotein biosynthesis despite its ability to bind human SECIS elements with lower affinities (51). It is possible that Secisbp2l and Secisbp2 have diverged in structure and function during evolution, since Secisbp2l from the worm Capitella, that does not contain a Secisbp2 gene, is able to substitute for rat Secisbp2 in the in vitro system (51). It is also possible, but has not been demonstrated, that in mouse, Secisbp2l is able to bind and protect selenoprotein mRNAs from degradation without promoting UGA redefinition. There may as well exist signals in some selenoprotein mRNAs, as e.g. selenocysteine redefinition elements (52), which might be able to protect certain selenoprotein mRNAs by increasing redefinition or by recruiting other general RNA-binding proteins.
Multiple mechanisms coordinate to achieve gene-specific regulation of selenoprotein expression
Based on the data presented here, selenoprotein synthesis (S) can be approximated by measuring mRNA abundance (R), translation initiation efficiency (T) and URE, or S α R x T x URE. Herein, we show that changes in each of these parameters can be measured using a combination of RNA-Seq (R = RNA RPKM) and ribosome profiling (T = (5΄ RPF RPKM/RNA RPKM); URE = (3΄ RPF RPKM)/(5΄ RPF RPKM)). As discussed above, variations in RNA decay rates should be considered carefully in applying this model. For analyses concerned only with estimating selenoprotein synthesis rates irrespective of mechanism, mathematical reduction of this formula leads to a simplified equation, S α 3΄ RPF RPKM. Using all these parameters, we find that the abundance of selenoprotein mRNAs is regulated in a gene-specific manner. For example, Gpx1 is strongly regulated by mRNA decay. Selenot in contrast does not change its mRNA abundance or redefinition efficiency in Secisbp2-deficient hepatocytes, but down-regulates translation initiation. Gpx4 mRNA and UGA-redefinition are dependent on Secisbp2, but mRNA stability is remarkably unaffected by Se availability or tRNA[Ser]Sec inactivation. Thus, in contrast to a widely held notion, there may not be one overarching mechanism that governs selenoprotein expression, but each selenoprotein has evolved a coordinated set of mechanisms that respond specifically in the context of its biological function within a given organ to determine expression levels. The power of the methods presented here is that the different parameters involved can now be directly addressed in a gene-specific manner and on a global scale.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Brian Dalley of the University of Utah's High Throughput Genomics Core Facility for helpful discussions and Simon Bohleber, Bonn, for preparing Supplementary Figure S3. U.S. and N.F.-V. acknowledge skilful technical support by Uschi Reuter and Tobias Lindenberg. The corresponding authors would like to especially acknowledge D.L.H.'s contribution to the origin of the present study. While his many scientific contributions to the field of selenium research are well known, his role in stimulating collaborations and supporting the development of younger scientists from many countries and across several decades is equally deserving of recognition.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R01GM114291 and R21ES022716 to M.T.H.]; Intramural Research Program of the National Institutes of Health [to D.L.H. and B.A.C.]; Deutsche Forschungsgemeinschaft [DFG SCHW914/2-2, SCHW914/5-1]; Rheinische Friedrich-Wilhelms-Universität Bonn [U.S.]. Funding for open access charge: National Institutes of Health [R01GM114291 and R21ES022716 to M.T.H.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Labunskyy V.M., Hatfield D.L., Gladyshev V.N.. Selenoproteins: molecular pathways and physiological roles. Physiol. Rev. 2014; 94:739–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Howard M.T., Carlson B.A., Anderson C.B., Hatfield D.L.. Translational redefinition of UGA codons is regulated by selenium availability. J. Biol. Chem. 2013; 288:19401–19413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kryukov G.V., Castellano S., Novoselov S.V., Lobanov A.V., Zehtab O., Guigo R., Gladyshev V.N.. Characterization of mammalian selenoproteomes. Science. 2003; 300:1439–1443. [DOI] [PubMed] [Google Scholar]
- 4. Walczak R., Westhof E., Carbon P., Krol A.. A novel RNA structural motif in the selenocysteine insertion element of eukaryotic selenoprotein mRNAs. RNA. 1996; 2:367–379. [PMC free article] [PubMed] [Google Scholar]
- 5. Fletcher J.E., Copeland P.R., Driscoll D.M., Krol A.. The selenocysteine incorporation machinery: interactions between the SECIS RNA and the SECIS-binding protein SBP2. RNA. 2001; 7:1442–1453. [PMC free article] [PubMed] [Google Scholar]
- 6. Copeland P.R., Fletcher J.E., Carlson B.A., Hatfield D.L., Driscoll D.M.. A novel RNA binding protein, SBP2, is required for the translation of mammalian selenoprotein mRNAs. EMBO J. 2000; 19:306–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fagegaltier D., Hubert N., Yamada K., Mizutani T., Carbon P., Krol A.. Characterization of mSelB, a novel mammalian elongation factor for selenoprotein translation. EMBO J. 2000; 19:4796–4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Copeland P.R., Stepanik V.A., Driscoll D.M.. Insight into mammalian selenocysteine insertion: domain structure and ribosome binding properties of Sec insertion sequence binding protein 2. Mol. Cell. Biol. 2001; 21:1491–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zavacki A.M., Mansell J.B., Chung M., Klimovitsky B., Harney J.W., Berry M.J.. Coupled tRNA(Sec)-dependent assembly of the selenocysteine decoding apparatus. Mol. Cell. 2003; 11:773–781. [DOI] [PubMed] [Google Scholar]
- 10. Chavatte L., Brown B.A., Driscoll D.M.. Ribosomal protein L30 is a component of the UGA-selenocysteine recoding machinery in eukaryotes. Nat. Struct. Mol. Biol. 2005; 12:408–416. [DOI] [PubMed] [Google Scholar]
- 11. Miniard A.C., Middleton L.M., Budiman M.E., Gerber C.A., Driscoll D.M.. Nucleolin binds to a subset of selenoprotein mRNAs and regulates their expression. Nucleic Acids Res. 2010; 38:4807–4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wu R., Shen Q., Newburger P.E.. Recognition and binding of the human selenocysteine insertion sequence by nucleolin. J. Cell. Biochem. 2000; 77:507–516. [DOI] [PubMed] [Google Scholar]
- 13. Budiman M.E., Bubenik J.L., Miniard A.C., Middleton L.M., Gerber C.A., Cash A., Driscoll D.M.. Eukaryotic initiation factor 4a3 is a selenium-regulated RNA-binding protein that selectively inhibits selenocysteine incorporation. Mol. Cell. 2009; 35:479–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Behne D., Hilmert H., Scheid S., Gessner H., Elger W.. Evidence for specific selenium target tissues and new biologically important selenoproteins. Biochim. Biophys. Acta. 1988; 966:12–21. [DOI] [PubMed] [Google Scholar]
- 15. Bermano G., Nicol F., Dyer J.A., Sunde R.A., Beckett G.J., Arthur J.R., Hesketh J.E.. Tissue-specific regulation of selenoenzyme gene expression during selenium deficiency in rats. Biochem. J. 1995; 311:425–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sunde R.A., Thompson B.M., Palm M.D., Weiss S.L., Thompson K.M., Evenson J.K.. Selenium regulation of selenium-dependent glutathione peroxidases in animals and transfected CHO cells. Biomed. Environ. Sci. 1997; 10:346–355. [PubMed] [Google Scholar]
- 17. Berry M.J., Harney J.W., Ohama T., Hatfield D.L.. Selenocysteine insertion or termination: factors affecting UGA codon fate and complementary anticodon:codon mutations. Nucleic Acids Res. 1994; 22:3753–3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Suppmann S., Persson B.C., Bock A.. Dynamics and efficiency in vivo of UGA-directed selenocysteine insertion at the ribosome. EMBO J. 1999; 18:2284–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim L.K., Matsufuji T., Matsufuji S., Carlson B.A., Kim S.S., Hatfield D.L., Lee B.J.. Methylation of the ribosyl moiety at position 34 of selenocysteine tRNA[Ser]Sec is governed by both primary and tertiary structure. RNA. 2000; 6:1306–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chittum H.S., Hill K.E., Carlson B.A., Lee B.J., Burk R.F., Hatfield D.L.. Replenishment of selenium deficient rats with selenium results in redistribution of the selenocysteine tRNA population in a tissue specific manner. Biochim. Biophys. Acta. 1997; 1359:25–34. [DOI] [PubMed] [Google Scholar]
- 21. Carlson B.A., Moustafa M.E., Sengupta A., Schweizer U., Shrimali R., Rao M., Zhong N., Wang S., Feigenbaum L., Lee B.J. et al. Selective restoration of the selenoprotein population in a mouse hepatocyte selenoproteinless background with different mutant selenocysteine tRNAs lacking Um34. J. Biol. Chem. 2007; 282:32591–32602. [DOI] [PubMed] [Google Scholar]
- 22. Weiss S.L., Evenson J.K., Thompson K.M., Sunde R.A.. The selenium requirement for glutathione peroxidase mRNA level is half of the selenium requirement for glutathione peroxidase activity in female rats. J. Nutr. 1996; 126:2260–2267. [DOI] [PubMed] [Google Scholar]
- 23. Moriarty P.M., Reddy C.C., Maquat L.E.. Selenium deficiency reduces the abundance of mRNA for Se-dependent glutathione peroxidase 1 by a UGA-dependent mechanism likely to be nonsense codon-mediated decay of cytoplasmic mRNA. Mol. Cell. Biol. 1998; 18:2932–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sun X., Li X., Moriarty P.M., Henics T., LaDuca J.P., Maquat L.E.. Nonsense-mediated decay of mRNA for the selenoprotein phospholipid hydroperoxide glutathione peroxidase is detectable in cultured cells but masked or inhibited in rat tissues. Mol. Biol. Cell. 2001; 12:1009–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Seeher S., Atassi T., Mahdi Y., Carlson B.A., Braun D., Wirth E.K., Klein M.O., Reix N., Miniard A.C., Schomburg L. et al. Secisbp2 is essential for embryonic development and enhances selenoprotein expression. Antioxid. Redox Signal. 2014; 21:835–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gladyshev V.N., Arner E.S., Berry M.J., Brigelius-Flohe R., Bruford E.A., Burk R.F., Carlson B.A., Castellano S., Chavatte L., Conrad M. et al. Selenoprotein Gene Nomenclature. J. Biol. Chem. 2016; 291:24036–24040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schweizer U., Streckfuss F., Pelt P., Carlson B.A., Hatfield D.L., Kohrle J., Schomburg L.. Hepatically derived selenoprotein P is a key factor for kidney but not for brain selenium supply. Biochem. J. 2005; 386:221–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hatfield D., Matthews C.R., Rice M.. Aminoacyl-transfer RNA populations in mammalian cells chromatographic profiles and patterns of codon recognition. Biochim. Biophys. Acta. 1979; 564:414–423. [DOI] [PubMed] [Google Scholar]
- 29. Moustafa M.E., Carlson B.A., El-Saadani M.A., Kryukov G.V., Sun Q.A., Harney J.W., Hill K.E., Combs G.F., Feigenbaum L., Mansur D.B. et al. Selective inhibition of selenocysteine tRNA maturation and selenoprotein synthesis in transgenic mice expressing isopentenyladenosine-deficient selenocysteine tRNA. Mol. Cell. Biol. 2001; 21:3840–3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Carlson B.A., Novoselov S.V., Kumaraswamy E., Lee B.J., Anver M.R., Gladyshev V.N., Hatfield D.L.. Specific excision of the selenocysteine tRNA[Ser]Sec (Trsp) gene in mouse liver demonstrates an essential role of selenoproteins in liver function. J. Biol. Chem. 2004; 279:8011–8017. [DOI] [PubMed] [Google Scholar]
- 31. Ingolia N.T., Ghaemmaghami S., Newman J.R., Weissman J.S.. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009; 324:218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Diamond A.M., Choi I.S., Crain P.F., Hashizume T., Pomerantz S.C., Cruz R., Steer C.J., Hill K.E., Burk R.F., McCloskey J.A. et al. Dietary selenium affects methylation of the wobble nucleoside in the anticodon of selenocysteine tRNA([Ser]Sec). J. Biol. Chem. 1993; 268:14215–14223. [PubMed] [Google Scholar]
- 33. Hatfield D., Lee B.J., Hampton L., Diamond A.M.. Selenium induces changes in the selenocysteine tRNA[Ser]Sec population in mammalian cells. Nucleic Acids Res. 1991; 19:939–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Berry M.J., Maia A.L., Kieffer J.D., Harney J.W., Larsen P.R.. Substitution of cysteine for selenocysteine in type I iodothyronine deiodinase reduces the catalytic efficiency of the protein but enhances its translation. Endocrinology. 1992; 131:1848–1852. [DOI] [PubMed] [Google Scholar]
- 35. Mehta A., Rebsch C.M., Kinzy S.A., Fletcher J.E., Copeland P.R.. Efficiency of mammalian selenocysteine incorporation. J. Biol. Chem. 2004; 279:37852–37859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fixsen S.M., Howard M.T.. Processive Selenocysteine Incorporation during Synthesis of Eukaryotic Selenoproteins. J. Mol. Biol. 2010; 399:385–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gupta N., DeMong L.W., Banda S., Copeland P.R.. Reconstitution of selenocysteine incorporation reveals intrinsic regulation by SECIS elements. J. Mol. Biol. 2013; 425:2415–2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lu J., Zhong L., Lonn M.E., Burk R.F., Hill K.E., Holmgren A.. Penultimate selenocysteine residue replaced by cysteine in thioredoxin reductase from selenium-deficient rat liver. FASEB J. 2009; 23:2394–2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dubey A., Copeland P.R.. The selenocysteine-specific elongation factor contains unique sequences that are required for both nuclear export and selenocysteine incorporation. PLoS One. 2016; 11:e0165642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shetty S.P., Shah R., Copeland P.R.. Regulation of selenocysteine incorporation into the selenium transport protein, selenoprotein P. J. Biol. Chem. 2014; 289:25317–25326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stoytcheva Z., Tujebajeva R.M., Harney J.W., Berry M.J.. Efficient incorporation of multiple selenocysteines involves an inefficient decoding step serving as a potential translational checkpoint and ribosome bottleneck. Mol. Cell. Biol. 2006; 26:9177–9184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Guydosh N.R., Green R.. Dom34 rescues ribosomes in 3΄ untranslated regions. Cell. 2014; 156:950–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lykke-Andersen J., Bennett E.J.. Protecting the proteome: eukaryotic cotranslational quality control pathways. J. Cell Biol. 2014; 204:467–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bermano G., Arthur J.R., Hesketh J.E.. Selective control of cytosolic glutathione peroxidase and phospholipid hydroperoxide glutathione peroxidase mRNA stability by selenium supply. FEBS Lett. 1996; 387:157–160. [DOI] [PubMed] [Google Scholar]
- 45. Squires J.E., Stoytchev I., Forry E.P., Berry M.J.. SBP2 binding affinity is a major determinant in differential selenoprotein mRNA translation and sensitivity to nonsense-mediated decay. Mol. Cell. Biol. 2007; 27:7848–7855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schwanhausser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M.. Global quantification of mammalian gene expression control. Nature. 2011; 473:337–342. [DOI] [PubMed] [Google Scholar]
- 47. Seyedali A., Berry M.J.. Nonsense-mediated decay factors are involved in the regulation of selenoprotein mRNA levels during selenium deficiency. RNA. 2014; 20:1248–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shetty S.P., Copeland P.R.. Selenocysteine incorporation: a trump card in the game of mRNA decay. Biochimie. 2015; 114:97–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Maiti B., Arbogast S., Allamand V., Moyle M.W., Anderson C.B., Richard P., Guicheney P., Ferreiro A., Flanigan K.M., Howard M.T.. A mutation in the SEPN1 selenocysteine redefinition element (SRE) reduces selenocysteine incorporation and leads to SEPN1-related myopathy. Hum. Mutat. 2009; 30:411–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Donovan J., Copeland P.R.. Evolutionary history of selenocysteine incorporation from the perspective of SECIS binding proteins. BMC Evol. Biol. 2009; 9:229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Donovan J., Copeland P.R.. Selenocysteine insertion sequence binding protein 2L is implicated as a novel post-transcriptional regulator of selenoprotein expression. Plos One. 2012; 7:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Howard M.T., Aggarwal G., Anderson C.B., Khatri S., Flanigan K.M., Atkins J.F.. Recoding elements located adjacent to a subset of eukaryal selenocysteine-specifying UGA codons. EMBO J. 2005; 24:1596–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.