Abstract
Single cell RNA sequencing approaches are instrumental in studies of cell-to-cell variability. 5΄ selective transcriptome profiling approaches allow simultaneous definition of the transcription start size and have advantages over 3΄ selective approaches which just provide internal sequences close to the 3΄ end. The only currently existing 5΄ selective approach requires costly and labor intensive fragmentation and cell barcoding after cDNA amplification. We developed an optimized 5΄ selective workflow where all the cell indexing is done prior to fragmentation. With our protocol, cell indexing can be performed in the Fluidigm C1 microfluidic device, resulting in a significant reduction of cost and labor. We also designed optimized unique molecular identifiers that show less sequence bias and vulnerability towards sequencing errors resulting in an improved accuracy of molecule counting. We provide comprehensive experimental workflows for Illumina and Ion Proton sequencers that allow single cell sequencing in a cost range comparable to qPCR assays.
INTRODUCTION
The cell is the minimal building block of any living organism. Investigating the properties of individual cells rather than the average of a group of seemingly identical cells provided important insights in various domains such as cancer (1), development (2,3), immunology (4) and neurobiology (5–7). Single cell transcriptome sequencing is a key technology to address this cellular heterogeneity.
Since the first sequencing of a single cell transcriptome (8), advances in library preparation techniques greatly improved both efficiency and throughput (9).
Most mammalian cells contain just a few hundred thousand mRNA molecules (10). In consequence, efficient conversion of mRNA into cDNA is crucial and was the focus of several recent studies (1,10,11). Most current single cell mRNA cloning techniques exploit the template switching activity of reverse transcriptases (STRT-seq (12), Smart-seq (1)) to efficiently clone full length cDNA, which is subsequently amplified by PCR. The approach was further refined by Picelli et al. (11) (Smart-Seq2) and Islam et al. (10). Alternate approaches that use isothermal cRNA amplification were also developed (Mars-seq (13), CEL-seq (14,15)).
Early single cell profiling approaches processed single cells in tubes or in plates. Performing cDNA synthesis in tiny volumes either in microfluidic devices such as the Fluidigm C1 (2,3,10) or in microdroplets (16,17) was an important further development which increased throughput and reduced both reagent cost and labor. Single cell transcriptome profiling in microfluidic devices was also shown to yield increased transcript discovery rates and thus mRNA cloning efficiencies when compared to manual processing in tubes (18).
Amplification bias and library complexity are clearly issues that need to be considered since single cell library preparation requires huge amplification of tiny amounts (< 1pg) of cDNA. To address those issues, Islam et al. (10) and Jaitin et al. (13) stochastically tagged cDNA molecules during reverse transcription with short random nucleotide sequences (unique molecular identifiers, UMIs). The use of UMIs largely improved data quality since it allows counting of the initial unamplified cDNA molecules what is much less biased than counting transcript read numbers in heavily amplified cDNA.
UMIs are introduced during reverse transcription either at the 5΄ or 3΄ end of the cDNA. In consequence, only the UMI tagged extremity of the transcript is recovered and sequenced after cDNA fragmentation. The vast majority of currently used approaches introduce the UMI via the oligo-dT reverse transcription primer and sequence the 3΄ terminal, UMI tagged, fragment of the cDNA (14–17). Sequencing the actual 3΄ end of a transcript would require sequencing through the poly-dT stretch of the reverse transcription primer. Sequencing through such long repeats typically yields poor read qualities due to phasing issues and homopolymer length heterogeneity within flow cell clusters generated by polymerase slipping during amplification. In consequence, 3΄ selective single cell sequencing approaches don’t sequence the actual 3΄ end of a transcript but rather the 5΄ end of the most 3΄ fragment of the cDNA obtained after cDNA fragmentation.
Conversely, 5΄ selective approaches do not have this limitation and allow sequencing of the actual 5΄ end of the cDNA and thus a simultaneous definition of mRNA expression levels and transcription start sites. However, the only currently published 5΄ selective approach (10) has essentially two drawbacks: First, the cell index is introduced during cDNA fragmentation with indexed transposons independently for each cell, what is labor intensive and costly when commercial transposase is used. Secondly, only short UMIs of 5 nucleotides were used, since extension of the template switching oligonucleotide is thought to adversely affect mRNA capture efficiency. However such short UMIs get saturated for highly expressed transcripts and efficient UMI sequencing error correction strategies (16) cannot be used, since they would further decrease an already low UMI complexity.
Another limitation of all current efficient single cell sequencing approaches is that they are only available for Illumina sequencers and not for other low cost benchtop sequencers such as the Ion Proton.
We addressed those issues and developed a highly efficient cost- and labor effective 5΄ selective single cell transcriptome profiling approach for both Ion Torrent and Illumina sequencers. Our method introduces cell barcodes by PCR prior to cDNA fragmentation and requires just one fragmentation and library preparation for the pooled cDNAs from the individual cells. We also present a novel UMI design that allows better error correction and thus more reliable molecule counting. We show that our barcoding strategy and UMI design allows robust and efficient single cell transcriptome profiling with the Fluidigm C1 at just a fraction of the cost of currently available commercial approaches.
MATERIALS AND METHODS
Cell culture
HEK293 cells were cultured in DMEM medium supplemented with glutamine and 10% fetal calf serum. Human airway epithelial cells were isolated and cultured as described by Marcet et al. (19).
cDNA synthesis, PCR amplification—tube controls
Concentrations of reagents and enzymes as well as the reagent volume per cell were identical for tube controls and the Fluidigm C1 microfluidic device. Primer sequences are listed in Supplementary Data. A schematic workflow is provided in Supplementary Data.
Cell lysis
1000 HEK293 cells in 4.5 μl C1 wash buffer (Fluidigm) were lysed with 9 μl of lysis buffer (0.2% w/v Tween 20, 1 U/μl Promega RNAsin RNAse inhibitor, 2 μM reverse transcription primer, 2.5 mM dNTPs, 1× C1 loading reagent (Fluidigm), ERCC Spike-In Mix 1 at 20 000 molecules/cell (Life Technologies). The sample was incubated for 10 min at room temperature followed by 3 min at 70°C and 3 min at 10°C.
Reverse transcription
Reverse transcription was adapted from (20). 18 μl of 1.75× reverse transcription buffer (Thermo), 8.75 mM DTT, 1.75 M betaine, 10.5 mM MgCl2, 1.75 μM template switching oligonucleotide, 0.5 U/μl Promega RNAsin, 5.5 U/μl Life Technologies Superscript II reverse transcriptase, 1× C1 loading reagent were added to the lysed cells and the sample was incubated for 10 min at 25°C, 90 min at 42°C, 15 min at 70°C and kept <10°C until PCR amplification.
PCR amplification
One tenth of the reverse transcription (3.15 μl, 100 cells) was mixed with 27 μl 1.15× KAPA HiFi HotStart Ready Mix, 55 nM barcode primer and 1.1 μM biotinylated PCR primer. For Illumina sequencing forward and reverse primers for this PCR are distinct and an additional reverse PCR primer was added (1.1 μM). For PCR amplification samples were incubated 3 min at 98°C followed by 18 cycles at 98°C for 20 s, 64°C for 15 s, 72°C for 6 min, and a final extension at 72°C for 5 min. Primers and small fragments were removed by cleanup with 1 vol. SpriSelect beads. Typical cDNA size distributions are shown in Supplementary Data.
cDNA synthesis, PCR amplification – microfluidic device
Lysis, reverse transcription and PCR mixes were the same as for the tube controls. Lysis (7 μl) and reverse transcription mix (8 μl) were added to the wells of the microfluidic device specified in the script for the C1. 6.5 μl of PCR mix with one of the 96 barcodes was added to each of the outlet wells. The PCR mixes were backloaded from the outlet wells into the reaction chambers resulting in cell specific barcoding on the microfluidic chip during the PCR amplification. The script for the Fluidigm C1 has been submitted to the Fluidigm C1 OpenApp script repository (https://www.fluidigm.com/c1openapp).
Library preparation
Since Ion Torrent requires smaller fragment sizes than Illumina sequencers and no suitable commercial kits were available, we adapted a tagmentation protocol of Picelli et al. (21) for the Ion Proton. Assembly of transposomes was performed following Wang et al. (22). Two reverse complementary oligonucleotides containing the Tn5 mosaic end sequence (upper: 5΄-AGA TGT GTA TAA GAG ACA-G 3΄, lower: 5΄-PhosCTG TCT CTT ATA CAC ATC T-3΄) were annealed at a concentration of 50 μM each in TE (95°C 3 min, 70°C 3 min, cooling at 2°C min−1 to 26°C) and subsequently diluted to 10 μM in 50% glycerol. Annealed oligonucleotides and Ez-Tn5 transposase (1 U/μl, Epicentre) were mixed in a 4:1 ratio and incubated 30 min at room temperature for transposon assembly. The transposons were used immediately or stored at −20°C.
For the Fluidigm microfluidic chip, cDNA from the 96 output wells were pooled without quantification of the individual samples.
10 ng of cDNA in 9 μl water were mixed with 4 μl TAPS Buffer (50mM TAPS-NaOH, pH8.5 @ RT, 25 mM MgCl2), 2 μl dimethylformamide and 5 μl of transposon. After a 7 min incubation at 55°C, samples were cooled to 10°C and 4 μl of 0.1% SDS was added, and the tubes were incubated for 10 min at 65°C to detach the transposase, and then cooled to 4°C. For Illumina sequencing, tagmentation was done for 5 min at 55°C. Alternatively, a commercial Nextera tagmentation kit (Illumina) can be used for Illumina library preparation.
Terminal fragments that were biotinylated during the PCR amplification were captured with 24 μl Dynabeads® MyOne™ Streptavidin C1 beads (Life Technologies) and washed following the manufacturer supplied protocol. Beads were suspended in 10.5 μl water and 5΄ terminal fragments were amplified in 25 μl with KAPA HiFi HotStart Ready mix and 0.5 μM reverse library primer, 0.5 μM forward library primer and 0.125 μM extended forward library primer. For Illumina sequencing an extended reverse library primer was added (0.125 μM). A gap filing step was done at 72°C during 3 min followed by 98°C for 30 sec, 15 cycles of 98°C for 10 s, 55°C for 30 s and 72°c for 30 s, and 72°C for 2 min. Ion Torrent libraries were size selected (200–350 pb) with SPRIselect® beads (Beckman Coulter) following the manufacturer supplied protocol. 100 μl library in TE was incubated with 75 μl of SPRIselect® beads to deplete fragments >350 pb. Beads were discarded and an additional 15 μl of SPRIselect® beads were added to capture fragments >200 pb. Beads were washed with 85% EtOH and bound cDNA was eluted from the beads with 10 μl water.
Illumina libraries (100 μl) were just size selected for fragments > 200 bp with 90 μl SPRIselect beads. Beads were recovered, washed and eluted as described above.
Quality and yield was determined with an Agilent Bionalyzer (Supplementary Data).
Sequencing
Libraries were sequenced either on a Proton Ion PI™ Chip v3 (Thermo) or on a Nextseq 500 MID output flowcell (Illumina). For Illumina Nextseq sequencing, the custom sequencing primers listed in Supplementary Data were added to the reagent cartridge following the ‘NextSeq® System Custom Primers Guide’ (Illumina Part # 15057456). Instructions for the use of custom sequencing primers with Illumina HiSeq sequencers are in Illumina document # 15061846. Our protocol for Illumina sequencers uses single indexing and either single or paired end sequencing. For Illumina sequencers, sequencing more libraries from more than 96 cells is possible. This can be done by adding a plate index during the final library preparation using an indexed extended reverse library primer (see Supplementary Data) and sequencing the plate index as ‘index 2’.
Read alignments and gene-expression analysis
Ion Torrent sequencers generate just one read that contains the barcode and the insert sequence. In consequence, the read after barcode trimming starts with the TSO sequence (Supplementary Data). We first examined whether the TSO sequence including the UMI [(ATCG)4(ATC)4] and the three Guanines following the UMI were correctly formatted and free of substitutions or indels (Supplementary Data). Only reads with correctly formatted TSO sequences were processed further. UMI sequences were extracted and the TSO sequence was trimmed from the reads. Trimmed reads that were shorter than 26 nt were discarded. Typically 80 – 85% of the reads passed those filters.
In the case of Illumina sequencing, reads start with the UMI sequence and were just filtered for correct UMI formatting and the presence of three Guanines after the UMI.
Trimmed reads were mapped against the human genome (hg19) and ERCC sequences using STAR aligner (v2.4.0a), with default parameters. STAR indices were generated using Ensembl GTF file (release 75).
For molecule counting based on UMI counts, we used the Dropseq Core Computational Protocol version 1.0.1 (dropseq.jar) (16). Unless indicated otherwise, we used the uniq option (identical UMIs at two different transcript positions are only counted once) and edit distance = 1 (UMIs for a transcript that are potentially a substitution mutant of another UMI with higher read coverage of the same transcript are discarded).
Single cell quality filters
Capture sites were visually inspected for the presence and viability of cells after staining with the LIVE/DEAD® Viability/Cytotoxicity Kit for mammalian cells (Thermo) at 10× magnification. Only capture sites with one live cell were retained for analysis. Additional quality control of the remaining libraries was performed using the R package ‘Single-cell analysis toolkit for gene expression data in R’ (scater version 1.0.4, https://www.bioconductor.org). Briefly, a set of cell quality indicators, such as the total number of UMIs for the cell, the total number of detected genes and the percentage of UMIs corresponding to mitochondrial genes was computed for each cell (23). Then, all cells flagged as outlier in a principal component analysis based on these quality measures were excluded. Furthermore, we excluded cells with a percentage of counts on ERCC spike-ins greater than the median + 4 times the median absolute deviation for the batch (isOutlier function of the scater R package). Special cases, such as identification of rare quiescent cells with low transcript numbers in a heterogeneous cell population might require fine-tuning of those filters.
Statistical analysis
Statistical analysis was performed using the statistical package R version 3.3.1. ERCC capture efficiency was estimated as the intercept of a regression line with a constrained slope of 1 fitted between the expected number of ERCC molecules and the number of ERCC molecules counted. Only ERCC for which at least 10 molecules were spiked in were used for analysis. Correlation coefficients are calculated using Pearson's method. Single cell UMI count data were normalized for sequencing depth differences using the normalize function from the scran package version 1.0.4.
Down-sampling of reads
After mapping, cell index and UMI extraction, a BAM file with reads for 47 single cells (mean = 1.43 × 106 reads/cell) was down-sampled to 0.05, 0.1, 0.2 0.4, 0.5, 0.75, 1.0, 1.25 million mean reads per cell. mRNA molecule counting was done with the dropseq.jar java pipeline (16).
Analysis of public single cell RNAseq data
Data were downloaded from Gene Expression Omnibus, reads were matched to the reference genomes and UMIs were counted as described above.
HEK293 Dropseq data are from GEO accession GSE63473 (16). 259 cells were selected for further analysis using the same quality filtering as described above.
CEL-seq2 data of 96 cells processed in the Fluidigm C1 are from GEO accession GSE 78779 (samples GSM 2076519–GSM2076614) (14). The UMIs in this dataset have just five nucleotides and not six nucleotides as stated in Hashimshony et al. (14).
Accession codes
Data were deposited in Gene Expression Omnibus (GSE79136).
RESULTS
Library preparation strategy
We sought to design a 5΄ selective library preparation strategy that uses UMIs and fulfills the following criteria: (i) unbiased introduction of cell indices before the costly and labor intensive fragmentation step; (ii) compatibility with the Fluidigm C1 microfluidic device design; (iii) essentially sequencing platform independent; (iv) cost and labor effective.
A simple option for pre-fragmentation barcoding is to introduce cell indices during reverse transcription. All current 3΄ selective single cell profiling approaches use this strategy and barcode via indexed reverse transcription primers. A similar strategy was also used in an early version of a 5΄ selective protocol by Islam et al. (12) who used barcoded TSOs to introduce the cell index at the 5΄ end of the cDNA during reverse transcription. However, introducing the cell index via barcoded TSOs has essentially two major disadvantages. First, an increase in TSO length has a negative impact on capture efficiency (10,24), a critical parameter in single cell transcriptome profiling. Secondly, use of different TSOs during reverse transcription was shown to cause differential capture of transcripts (24). Thus, the use of barcoded TSOs during reverse transcription would likely introduce bias that cannot be corrected with UMIs since UMIs are introduced during this step.
To avoid any barcode induced bias we opted for barcoding during PCR amplification of the cDNA, as eventual barcode dependent amplification bias can easily be detected and corrected by counting Unique Molecule Identifiers (UMIs) introduced during reverse transcription.
Performing reverse transcription in tiny volumes in Fluidigm microfluidic devices was shown to yield superior mRNA capture than carrying out the same protocol in tubes or microtiter plates (18). Although the cost of the disposable microfluidic device is substantial, it is compensated by the >100-fold lower amount of required reagents and enzymes.
We designed a 5΄ single cell transcriptome sequencing workflow that is compatible with the Fluidigm C1 microfluidic device (Figure 1). We initially performed pilot experiments in tubes that mimicked the reaction conditions in the microfluidic device to select the optimal UMI and TSO design. The proposed protocol is therefore highly versatile and can easily be adapted to other instruments.
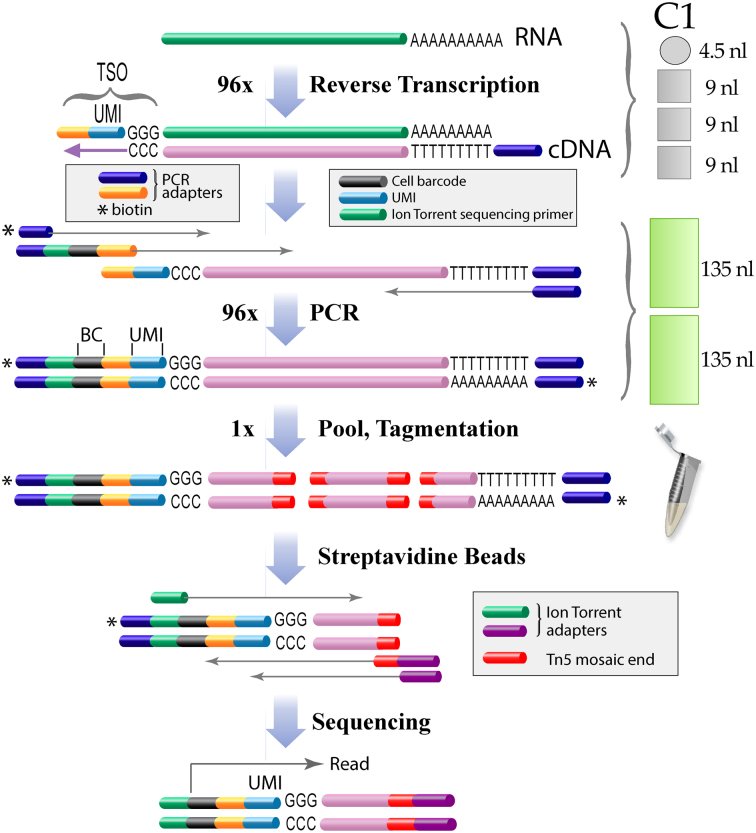
Figure 1.
On chip barcoding workflow. After cell lysis in 4.5 nl poly-adenylated RNA is reverse-transcribed in 31.5 nl with an anchored oligodT primer. A PCR primer sequence and unique molecular identifiers (UMIs) are added to the 3΄ end of the cDNA via reverse transcriptase template switching. The cDNA is subsequently amplified and cell index sequences (barcode) as well as terminal biotins are introduced by PCR in the microfluidic device. The barcoded cDNAs are pooled, fragmented by tagmentation with Tn5 transposase and the biotinylated terminal fragments are isolated on streptavidin beads. 5΄ terminal fragments are selectively amplified and additional sequences required for Ion Torrent sequencers are introduced by PCR. For a detailed protocol see Supplementary Data and for Illumina sequencers see Supplementary Data.
TSO and UMI design
Rather short UMIs with five degenerate nucleotides were previously used by others for 5΄ selective single cell mRNA sequencing (7,10). However, the 1024 distinct sequences of a N5 UMI are clearly insufficient to uniquely tag each copy of an abundant transcript with one and only one UMI. Several strategies were used by others to count abundant transcripts with short UMIs. One approach considers two reads with identical UMI of a given transcript as distinct molecules if both reads start at different positions on the transcript (10). Theoretical considerations of UMI usage saturation or UMI collision were also used to extrapolate the number of molecules for abundant transcripts (25). The used equations contain logarithms that tend to infinity and exaggeratedly overestimate the number of molecules when the number of detected UMIs approaches the maximal complexity of the UMI: they are thus not reliable for highly abundant transcripts.
To overcome the limitations of short UMIs, we rather increased the length of the UMI to 7 nucleotides. The complexity of a N7 UMI (n = 16 384) should be sufficient to tag each copy of even abundant transcripts in a single cell with a unique UMI.
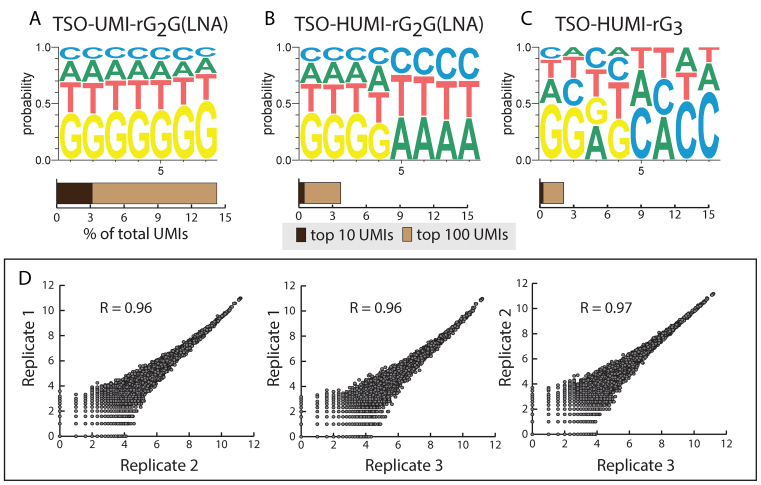
Ideally, UMIs should be introduced randomly, without bias for particular UMI sequences. However, we noticed that UMIs that are G-rich, particularly at the 3΄ end of the UMI, were highly enriched. Fifteen % of the UMI::transcript combinations were associated with a limited subset of just 100 G-rich UMIs (Figure 2a). This G-bias is likely due to the variable number of template independent nucleotides that are added by the reverse transcriptase. Previous studies showed that mainly three to four but sometimes up to six nucleotides, mainly cytosines, are added to the end of a cDNA (24) by the intrinsic terminal transferase activity of Superscript II. In the TSO, the seven Ns of the UMI are followed by three guanosines to allow annealing to the 3΄ terminal cytosines of the cDNA. When more than three cytosines are added by the reverse transcriptase, a TSO with a longer stretch of 3΄ terminal Gs and thus UMIs that have Gs at their 3΄ terminus are likely selected, leading to a G bias at the 3΄ of the UMI.
Figure 2.
UMI optimization and reproducibility of the protocol. (A–C) Impact of the TSO design on UMI usage bias. We examined TSOs with either a N7N6 UMI (A) and N4H4 UMI (HUMI) (B, C). The 3΄ terminal nucleotide of the TSO was either a LNA-guanosine (A, B) or a ribo guanosine (C). The weblogos represent the frequency at which we found each nucleotide at the given positions of the UMI in our genome matched sequencing reads. The bar graphs below show the percentage of the total transcript molecules associated with the top 10 and top 100 most frequently found UMI sequences. Data are from 100 pooled HEK293 cells processed in tubes. (D) Pairwise correlations of transcript (UMI) counts for three biological replicates of 100 HEK293 cells with the TSO-HUMI-rG3 (C). Data shown are log2(counts+1), R: Pearson correlation coefficient.
To overcome such a G bias, we tested a N4H4 UMI where the last four nucleotides are constrained to either A, T or C. The new UMI design resulted in a far better balanced usage of UMI sequences than the initial N7 UMI (Figure 2B and C).
In single cell transcriptome profiling efficient transformation of a limited number of mRNA molecules into amplified cDNA is crucial. Most currently used highly efficient single cell transcriptome sequencing approaches exploit the template switching activity of reverse transcriptases to add a priming site required for subsequent PCR amplification to the 3΄ end of the cDNA (10,11,16). Different TSO designs were recently proposed for efficient template switching. Islam et al. (12) used a TSO with three 3΄ terminal riboguanosines (TSO_rG3) while Picelli et al. (11) claimed superior template switching efficiency when the 3΄ terminal nucleotide of the TSO is a LNA base (TSO_LNA). Conversely, another study reported superior efficiency of TSOs with three terminal riboguanosines over TSOs with LNA bases (26). In our experimental conditions both TSO designs performed rather similarly (cDNA yield TSO_rG3/ TSO_LNA = 1.03 ± 0.26 S.E.M., n = 3 means of triplicates). Since the UMI usage was slightly better balanced with the TSO_rG3 (Figure 2B and C), we used this TSO for all further experiments. The final protocol is highly reproducible with pools of HEK293 (correlation coefficients > 0.96, Figure 2D).
We next examined how our on chip barcoding protocol performs with single HEK293 cells in the Fluidigm C1 96 cell integrated fluidic circuit (IFC). The 96 amplified cDNAs were pooled without normalization, libraries were prepared and sequenced on an Ion Proton sequencer (Figure 1, Supplementary Data, Supplementary Data).
Introducing UMIs during cDNA synthesis theoretically allows correction of all bias induced by steps downstream of cDNA synthesis (e.g. PCR). However UMI counting and error correction strategies need to be critically considered to avoid bias.
UMI counting
Different UMI counting strategies were previously used. Islam et al. (10) used a N5 UMI with a maximal complexity of 1024. To count abundant transcripts they considered two identical UMIs as distinct molecules when the read start positions on the transcript were distinct. Conversely, Macosko et al. (16), who used a high complexity N8 UMI (n = 65 536), simply eliminated all duplicate UMIs for a given gene. We frequently noted identical UMIs at different start positions even for low expressed transcripts. For example, for TCEB2 we found 50 distinct UMIs with one UMI at three distinct start positions and seven UMIs at two start positions (Supplementary Data). This is statistically highly unlikely with our N4H4 UMI (complexity = 20 736) and we rather suspect this start position heterogeneity results from soft clipping of the 5΄end of some lower quality reads by the read mapper. To avoid any counting of fake UMIs we counted UMIs only once for a given gene.
UMI error correction
PCR amplification errors and sequencing errors can generate novel UMI sequences, which would be falsely counted as distinct molecules.
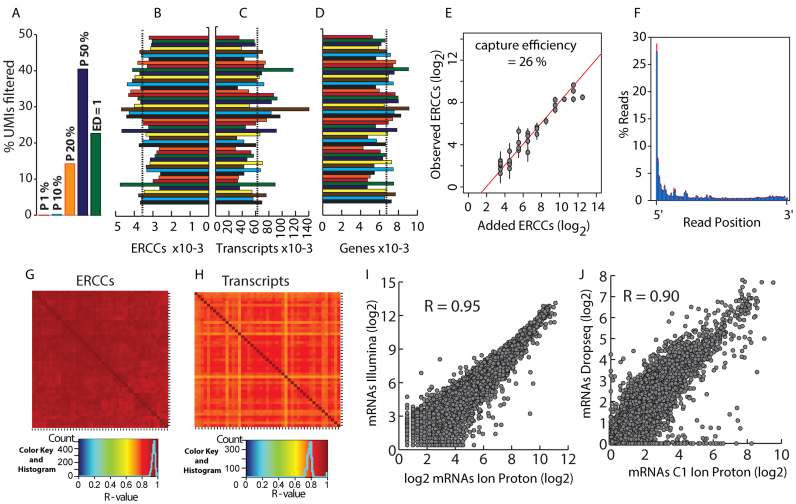
The impact of PCR errors is probably small with high fidelity polymerases such as the Kappa HiFi polymerase we used. PCR error rates are far below 10−6 and PCR amplification of an 8 nucleotide UMI for 30 cycles will introduce PCR errors in <0.024% of the amplified UMIs. Sequencing errors are a more serious issue, since benchtop sequencers have substitution rates >0.1%, meaning that >0.8% of the sequenced 8 nucleotide UMIs have at least one substitution (>8000 erroneous UMIs per million reads). In consequence, efficient identification and elimination of such false UMIs is critical for reliable molecule counting. We examined various options to correct those errors and deduce molecule counts from UMI counts. Since sequencing errors affect a minority of reads, real UMIs should, on average, be covered by far more reads than UMIs generated by sequencing errors. Islam et al. (10) exploited the anticipated low read coverage of fake UMIs to correct for UMI sequencing errors, using a ‘percentile filtering’ approach. For each gene, they discarded UMIs with a read coverage <1% of the median coverage for all UMIs of the respective gene. However, this approach requires a quite high UMI sequencing depth to reliably identify 100-fold differences in read coverage. In our dataset, with a mean UMI sequencing depth of 12.6 (before filtering), only 0.01% of the transcript molecules were eliminated (Figure 3A), a rate far below the expected number of UMI sequencing errors. A more stringent filtering where UMIs with <10% of the mean UMI read coverage were discarded only increased the number of filtered UMIs to 0.2% (Figure 3A). A further increase of the cutoff to 20% had a pronounced impact on the number of retained UMIs. However, such stringent cutoffs (≥20%) capped the number of detected molecules, which barely increased when more reads were generated (Supplementary Data).
Figure 3.
Single cell sequencing. (A) Impact of UMI error filtering strategies. Percentage of filtered UMIs for different UMI error correction strategies. Filtering strategies were: Percentile, UMIs with a read coverage of less than the indicated fraction (P 1%, P 10%, P 20%, P 50%) of the average UMI read coverage of the corresponding gene were discarded; Edit distance (ED) = 1, UMIs that differ in just one nucleotide were merged into a single UMI. The percentages of eliminated UMIs were: P1%, 0.01%; P10%, 0.20%; P20%, 14.25%; P50%, 40.43%; ED = 1, 22.61%. Data are from one cell. (B, C) Number of ERCC (B) and transcript molecules (C) detected for each cell (means (dashed lines)/c.v.: ERCCs, 3558/15.7%; transcripts, 62 841/36.7%). (D) Number of genes detected for each cell (mean = 6679 (dashed line); c.v. = 16.9%). (E) Scatter plot showing the number of input ERCCs vs. the number of detected ERCCs (means ± SD). The capture efficiency (26%) was calculated from the intercept of the regression line and the y-axis. (F) Distribution of read starts on annotated transcripts in one % bins between the 5΄ (0%) and the 3΄ end (100%). Data are means ± SD (red bars) for 47 cells. (G) Heatmap of the pairwise correlation of ERCC molecules for 47 cells. (H) As (G) but for mRNAs. (I) Correlation between transcript (UMI) counts (log2(counts + 1)) for pools of 100 HEK293 cells sequenced on an Ion Proton or Illumina Nextseq 500, respectively. Data are means from two pools of 100 HEK293 cells processed in tubes. (J) Correlation of HEK293 single cell transcript (UMI) counts (log2(counts+1)) between our Fluidigm C1 data and previously published Dropseq data (16). Transcript counts are means from 47 cells (Fluidigm) or 259 cells (Dropseq). Average numbers of transcript molecules detected per cell were: Fluidigm, 62,841; Dropseq, 36,746. R: Pearson correlation coefficient.
An alternative approach that does not rely on high sequencing depths was recently introduced (16). This approach, called ‘edit distance filtering’, merges UMIs of a given transcript when they differ by just one base and eliminates UMIs generated by substitution errors during PCR or sequencing. With our dataset, this filtering method eliminates 22.6% of the UMIs (Figure 3A) and is already effective at low sequencing depths. Edit distance filtering eliminated preferentially UMIs with low read coverage and increased the average UMI sequencing depth from 12.6 to 16.3 (Supplementary Data).
Despite an average UMI sequencing depth of 12.6 before filtering, 18% of the UMI::transcript combinations were read just once (Supplementary Data). A similar heterogeneity in UMI sequencing depth was reported by others (7). Yet, 66% of the UMIs covered by just one read are retained by the ‘edit distance’ filtering, an approach that eliminates UMIs with single substitution errors. In consequence, the majority of those single read UMIs are likely not erroneous UMIs but rather correspond to real mRNA molecules. Conversely, those single read coverage UMIs are preferentially (10) or completely (7) discarded with UMI error filtering strategies that are simply based on UMI sequencing depth. Both the higher sensitivity at reasonable sequencing depths and the higher selectivity for erroneous UMIs led us to select the ‘edit distance’ filtering for UMI error correction. The Ion Proton sequencer adds a particular challenge since it generates a pretty high number of indel errors (up to 0.5%) which are hardly detected by the ‘edit distance’ UMI error filtering. To eliminate UMIs erroneously generated by indels, we took advantage of our UMI design where no guanosine is present in the last four bases of the UMI (Supplementary Data). Any insertion or deletion in the UMI sequence thus results in a right or left shift of the first G following the UMI, respectively and can thus be filtered out.
Performance and reproducibility
Analysis of the 47 cells that passed our quality filters (see methods section, Supplementary Data) showed that reads were preferentially located close to the 5΄ end of transcripts (27.3% of the reads started within the first percent of the mRNA, Figure 3F) what is consistent with our 5΄ selective library preparation strategy (Figure 1).
Individual ERCCs were highly correlated between cells (average R = 0.94, Figure 3G), gross ERCC molecule counts (Figure 3B) and UMI counts for individual ERCC spike in RNA (Figure 3E, Supplementary Data) were similar for all cells indicating that mRNA was captured with comparable efficiency. The average ERCC cloning efficiency was 26% (Figure 3E), close to the efficiencies recently reported after stringent UMI error correction (7,16).
Conversely, gross counts for mRNA molecules (Figure 3C) and reads (Supplementary Data) were more heterogeneous. The cell-to-cell differences in UMI and read counts likely represent real differences in the number of mRNA molecules rather than experimental variability: (i) ERCC cloning efficiencies were similar among cells (Figure 3B); (ii) while the number of transcript reads varied by as much as a factor of 6.8 between cells, the average transcript sequencing depth varied <2-fold (Supplementary Data). In consequence, library normalization and generation of the same number of reads for each cell would likely lead to a higher heterogeneity in sequencing depth for the individual cells than simple library pooling without normalization.
Despite the rather high cell-to-cell heterogeneity of mRNA molecule (UMI) counts we observed a good correlation of mRNA expression between all cells (average R = 0.77, Figure 3H). The correlation between single cells was, as expected, somewhat lower than between pools of 100 HEK293 cells (Figure 2D) where cell-to-cell variations (e.g. cell cycle, etc.) are averaged out.
We performed two additional HEK293 cell single cell sequencing experiments to test the reproducibility of our protocol. Mean mRNA expressions from the three experiments correlated well (Supplementary Data), despite the fact that the experiments were performed over a period of six months with HEK293 cells at different passage numbers.
To test our protocol in a biologically relevant context, we profiled human airway epithelial cells cultured at an air-liquid interface, a model that contains several distinct cell populations. The data from two independent primary cultures, which slightly differed in their cell culture conditions correlated well (R = 0.95, Supplementary Data). We anticipated the detection of at least two cell populations, namely multiciliated and basal cells. Hierarchical clustering of RNA sequencing data from two independent IFC runs (one for each cell culture) identified three main clusters that were further characterized based on the expression of specific markers (Supplementary Data). One cluster clearly corresponded to multiciliated cells, as evidenced by the expression of ciliated cells markers such as TPPP3, FOXJ1 and ROPN1. A second cluster was reminiscent of basal cells, as evidenced by a robust expression of basal cell markers such as KRT5, KRT6A, KRT17 or S100A2. A third cluster is characterized by high levels of BPIFA1 and BPIFB1, which are associated to the innate immune response. Further experiments will be necessary to understand the cell types in this cluster. A secondary sub-clustering by cell culture / donor within those clusters is likely due to the different differentiation state of both cultures (culture 1, 52 days; culture 2, 33 days).
Taken together, the high reproducibility shown for pools of 100 HEK293 cells, for single HEK 293 cells and for primary epithelial cultures illustrates well the robustness of our SmartSeq based single cell library preparation technique.
While all other current single cell transcriptome profiling approaches were specifically designed for Illumina sequencers, our approach is essentially platform independent. After replacement of some oligonucleotides, the protocol designed for Ion Torrent sequencers was adapted for sequencing on Illumina sequencers (Supplementary Data, Supplementary Data). Interestingly, the correlation between two distinct biological replicates sequenced on two different sequencers (R = 0.95; Figure 3i) is close to what we obtained when replicates were sequenced on the same platform (R = 0.96-0.97; Figure 2D). The precision of UMI based molecule counting is further illustrated by the high correlation (R = 0.90) between our data and Dropseq single cell transcriptome data previously published for HEK293 cells (Figure 3J). This is particularly noteworthy, considering the use of two distinct single cell isolation approaches (Dropseq vs. microfluidic device), two different sequencing strategies (3΄ versus 5΄ end sequencing) and two different sequencer specific library preparations (Illumina versus Ion torrent).
Our on chip barcoding strategy reduces library preparation cost for the Fluidigm 96 cell IFC to essentially the cost of the microfluidic chip. With decreasing library preparation cost, sequencing of the libraries becomes the major cost factor in single cell transcriptome profiling. To estimate how many sequencing reads are required for profiling, we examined the impact of the number of sequencing reads on the number of detected transcript molecules and genes (Supplementary Data). With an average of 1.43 million reads per cell, transcript and gene discovery rates approached a maximum with 62841 transcript molecules and 6679 expressed genes per HEK293 cell. The transcript detection rate is principally capped by the mRNA cloning efficiency, which is slightly above 26% with our protocol. (Figure 3e). Increasing further the sequencing depth would bring the transcript discovery rate somewhat closer to this limit but would also increase sequencing cost drastically. The number of required sequencing reads depends on the question to be addressed. Shallow sequencing with just 50 000 reads per cell was shown to be sufficient for cell type classification and biomarker identification (5). With 50 000 reads per cell we detect 54% and 28% of the maximally detected genes and transcript molecules, respectively. Reliable identification of expression changes for weakly expressed transcripts will require more reads. With 0.5–1 million reads we detect 80–93% of the transcript molecules and 90–97% of the expressed genes that we find at our maximal, almost saturating sequencing depth (Supplementary Data). This should be sufficient for most routine single cell transcriptome profiling studies.
DISCUSSION
We present a robust cost and labor effective 5΄ selective single cell transcriptome profiling approach where all the barcoding is done prior to fragmentation. This is to our knowledge the first 5΄ selective single cell mRNA sequencing protocol that allows pooling of the amplified, barcoded cDNA before fragmentation and does not require labor intensive and costly fragmentation of the individual libraries. We adapted the workflow for the Fluidigm integrated fluidic circuit (C1) and we detail workflows for Ion Torrent Proton and for Illumina sequencers. Yet, the method could likely be adapted for any other sequencing platform including long read sequencers. Adapters and barcodes that are used during cDNA amplification have just to be replaced by ad hoc sequences required for respective specific sequencer.
Most current single cell transcriptome profiling approaches use UMIs, which are introduced during reverse transcription. In consequence, any cell barcode bias introduced during reverse transcription remains uncorrected and should be avoided. Unlike all other currently popular single cell transcriptome profiling approaches that use UMIs (10,16,17), our method does not use barcoded primers during reverse transcription to exclude any barcode induced bias during this first highly critical step of mRNA capture and UMI tagging.
Another source of bias comes from false UMIs generated by sequencing errors. Such false UMIs can even outnumber real UMIs in heavily over-sequenced samples, for instance in experiments with low capture efficiency or for leaky cells with few mRNA molecules when the read number is boosted by library normalization. Since current UMI error correction strategies (16) hardly eliminate indel errors, we developed a novel UMI design that allows reliable identification and elimination of erroneous UMIs with indels (Supplementary Data). This improved UMI design will be of interest not just for transcriptome profiling but also for the increasing number of NGS applications that rely on UMI based molecule counting or identification (27,28).
ERCC spike-in RNAs combined with UMIs were used to probe mRNA capture efficiencies in several studies. However, UMI lengths and UMI error correction strategies differ widely. We used an error correction strategy for our N4H4 UMI that merges UMIs that differ in just one nucleotide (edit distance 1) and obtain 26% ERCC capture efficiency. Jaitin et al. (13) used the same UMI error filtering for a N4 UMI in a 3΄ selective isothermal amplification based approach in microplates (Mars-seq) and claimed just 1–2% capture efficiency. This is likely highly underestimated since a N4 UMI has a complexity of just 256 and edit distance filtering reduces the effective complexity even further, resulting in elimination of UMIs that correspond to real RNA molecules. Conversely, 22% capture efficiency was recently reported with a isothermal amplification approach (CEL-seq2) which is similar to the Mars-seq approach in a microfluidic device (14). However the authors did not correct for UMI errors and used UMI collision extrapolations to correct the UMI counts for abundant transcripts upwards. We reanalyzed the CEL-seq2 data (see methods section) with ‘edit distance 1’ UMI error filtering, the error correction used in our and previous studies (16) and obtained 13.8% capture efficiency. The highest capture efficiency reported for single cell transcriptome sequencing was 48%. (10). However this value was obtained with the low stringency percentile filtering that barely filters any UMIs in our data. A more recent study by the same group used the same approach with more stringent UMI error filtering and reported 22% capture efficiency (7). Those differences in UMI design and error correction make any direct comparison of capture efficiencies reported in different studies difficult.
Our protocol and other published high efficiency single cell transcriptome profiling techniques (10,11,16) use a Smartseq based mRNA cloning strategy that relies on the template switching activity of reverse transcriptase, a process which is thought to favor capped RNAs, since reverse transcriptase preferentially adds non template dependent nucleotides to the cDNA when the RNA is capped (29). Thus, capture of mRNAs is likely cap selective but definitively not cap specific since the uncapped ERCC spike-in RNAs are also cloned with high efficiency by us (Figure 3) and others (10). ERCC RNAs (NIST #2374, https://www-s.nist.gov/srmors/view_cert.cfm?srm=2374) are in vitro transcribed RNAs that all start with the same pT7T318 plasmid sequence including three 5΄ terminal Gs. The resulting cDNAs have 3΄ terminal Cs which are complementary to the 3΄ terminal Gs of the TSO. This might explain why template switching on uncapped ERCCs is efficient.
In our opinion, the ERCC capture efficiency should not be used to extrapolate absolute mRNA molecule counts from cDNA (UMI) counts since: (i) it is currently unknown whether those uncapped ERCCs are captured with the same efficiency as capped cellular mRNAs. (ii) Although capture of individual ERCCs (Figure 3E) and mRNAs (Figures 2D, and 3H–J) was highly reproducible in replicated experiments, the capture efficiency of two distinct but equally abundant ERCC molecules can vary almost by a factor of four in one sample (Figure 3E). Yet, the use of spike-in RNAs is crucial for the comparison of different protocols and the identification of badly performing samples or channels in a microfluidic device.
The transcript discovery rate did not completely saturate with an average of 1.43 million reads per cell (Supplementary Data). This is essentially due to the high UMI sequencing depth heterogeneity. After UMI error filtering, 16% of the UMI::transcript combinations were sequenced just once despite a mean UMI sequencing depth of 16.3 (Supplementary Data). One likely reason for this broad heterogeneity is PCR amplification bias (30). Isothermal cRNA amplification, which is typically less biased than PCR, was recently proposed as an alternative to PCR in a 3΄ selective single cell sequencing approach (CEL-Seq, (15); Mars-Seq, (13), CEL-seq2 (14)). However, comparison of our data (PCR based approach) with recently published CEL-seq2 data (Isothermal amplification) reveals a similar UMI sequencing depth heterogeneity and thus amplification bias for both approaches (Supplementary Data). Although bias downstream of reverse transcription is efficiently corrected by UMIs, reducing amplification and library preparation bias remains an important future challenge since this would profoundly reduce the required sequencing depth and sequencing cost. This will be of particular importance for high throughput droplet based approaches where thousands of cells are analyzed.
Recent developments in single cell transcriptome profiling focused on an increased throughput mainly with droplet based techniques. However, ERCC capture efficiencies are apparently lower for droplet based approaches (12.8% (16), 7.1% (17) than for the Fluidigm microfluidic device (26% (this study), 22% (7)). The lower capture efficiency is not restricted to ERCC spike-in RNAs and was also observed for mRNAs. Macosko et al. (16) reported a mean Dropseq mRNA capture efficiency of 10.7%. Our observations are consistent with this. With our microfluidic approach we detected an average of 62 841 transcript molecules in a single HEK293 cell (Figure 3). In a published HEK293 Dropseq dataset (16) we identified 36 746 mRNA molecules per cell (Figure 3j).
While droplet based techniques are currently clearly the method of choice when thousand of cells are analyzed, microfluidic devices are in our opinion better suited for small to medium size projects. With the on chip barcoding strategy we present, the Fluidigm C1 96 cell IFC allows a robust and highly efficient capture of the single cell transcriptome with little hands on time and negligible reagent cost. For routine single cell transcriptome profiling one Proton P1 chip (108 reads) should be sufficient for a 96 cell microfluidic device. This reduces the overall cost of a 96 single cell transcriptome profiling study to about 1400 USD (Supplementary Data) and thus into a cost range where single cell transcriptome profiling becomes highly accessible and competitive with qPCR assays.
Supplementary Material
ACKNOWLEDGEMENTS
We thank all members from PB's group for discussions and Virginie Magnone for valuable advice on high throughput sequencing. This work was developed together with the functional genomics platform of Nice Sophia Antipolis, a partner of the National Infrastructure France Génomique.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Cancéropôle PACA and the Commissariat aux Grands Investissements [ANR-10-INBS-09-03 and ANR-10-INBS-09-02]; Fondation pour la Recherche Médicale [DEQ20130326464 to P.B.]; Vaincre la Mucoviscidose [RF20140501158/1/1/70]; Agence Nationale pour la Recherche [ANR-12-BSVI-0023-02]; labex Signalife [ANR-11-LABX-0028-01]; Conseil Départemental 06. Funding for open access charge: ANR.
Conflict of interest statement. None declared.
REFERENCES
- 1. Ramskold D., Luo S., Wang Y.-C., Li R., Deng Q., Faridani O.R., Daniels G.A., Khrebtukova I., Loring J.F., Laurent L.C. et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat. Biotechnol. 2012; 30:777–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., Lennon N.J., Livak K.J., Mikkelsen T.S., Rinn J.L.. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014; 32:381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Treutlein B., Brownfield D.G., Wu A.R., Neff N.F., Mantalas G.L., Espinoza F.H., Desai T.J., Krasnow M.A., Quake S.R.. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature. 2014; 509:371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shalek A.K., Satija R., Adiconis X., Gertner R.S., Gaublomme J.T., Raychowdhury R., Schwartz S., Yosef N., Malboeuf C., Lu D. et al. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature. 2013; 498:236–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pollen A.A., Nowakowski T.J., Shuga J., Wang X., Leyrat A.A., Lui J.H., Li N., Szpankowski L., Fowler B., Chen P. et al. Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nat. Biotechnol. 2014; 32:1053–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Usoskin D., Furlan A., Islam S., Abdo H., Lonnerberg P., Lou D., Hjerling-Leffler J., Haeggstrom J., Kharchenko O., Kharchenko P.V. et al. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat. Neurosci. 2015; 18:145–153. [DOI] [PubMed] [Google Scholar]
- 7. Zeisel A., Munoz-Manchado A.B., Codeluppi S., Lonnerberg P., La Manno G., Jureus A., Marques S., Munguba H., He L., Betsholtz C. et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015; 347:1138–1142. [DOI] [PubMed] [Google Scholar]
- 8. Tang F., Barbacioru C., Wang Y., Nordman E., Lee C., Xu N., Wang X., Bodeau J., Tuch B.B., Siddiqui A. et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods. 2009; 6:377–382. [DOI] [PubMed] [Google Scholar]
- 9. Kolodziejczyk A.A., Kim J.K., Svensson V., Marioni J.C., Teichmann S.A.. The technology and biology of single-cell RNA sequencing. Mol. Cell. 2015; 58:610–620. [DOI] [PubMed] [Google Scholar]
- 10. Islam S., Zeisel A., Joost S., La Manno G., Zajac P., Kasper M., Lonnerberg P., Linnarsson S.. Quantitative single-cell RNA-seq with unique molecular identifiers. Nat. Methods. 2014; 11:163–166. [DOI] [PubMed] [Google Scholar]
- 11. Picelli S., Björklund Å.K., Faridani O.R., Sagasser S., Winberg G., Sandberg R.. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods. 2013; 10:1096–1098. [DOI] [PubMed] [Google Scholar]
- 12. Islam S., Kjallquist U., Moliner A., Zajac P., Fan J.B., Lonnerberg P., Linnarsson S.. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Res. 2011; 21:1160–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jaitin D.A., Kenigsberg E., Keren-Shaul H., Elefant N., Paul F., Zaretsky I., Mildner A., Cohen N., Jung S., Tanay A. et al. Massively parallel single-cell RNA-Seq for marker-free decomposition of tissues into cell types. Science. 2014; 343:776–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hashimshony T., Senderovich N., Avital G., Klochendler A., de Leeuw Y., Anavy L., Gennert D., Li S., Livak K.J., Rozenblatt-Rosen O. et al. CEL-Seq2: sensitive highly-multiplexed single-cell RNA-Seq. Genome Biol. 2016; 17:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hashimshony T., Wagner F., Sher N., Yanai I.. CEL-Seq: single-cell RNA-Seq by multiplexed linear amplification. Cell Rep. 2012; 2:666–673. [DOI] [PubMed] [Google Scholar]
- 16. Macosko E.Z., Basu A., Satija R., Nemesh J., Shekhar K., Goldman M., Tirosh I., Bialas A.R., Kamitaki N., Martersteck E.M. et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015; 161:1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Klein A.M., Mazutis L., Akartuna I., Tallapragada N., Veres A., Li V., Peshkin L., Weitz D.A., Kirschner M.W.. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 161:1187–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wu A.R., Neff N.F., Kalisky T., Dalerba P., Treutlein B., Rothenberg M.E., Mburu F.M., Mantalas G.L., Sim S., Clarke M.F. et al. Quantitative assessment of single-cell RNA-sequencing methods. Nat. Methods. 2014; 11:41–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Marcet B., Chevalier B., Luxardi G., Coraux C., Zaragosi L.-E., Cibois M., Robbe-Sermesant K., Jolly T., Cardinaud B., Moreilhon C. et al. Control of vertebrate multiciliogenesis by miR-449 through direct repression of the Delta/Notch pathway. Nat. Cell Biol. 2011; 13:693–699. [DOI] [PubMed] [Google Scholar]
- 20. Picelli S., Faridani O.R., Björklund Å.K., Winberg G., Sagasser S., Sandberg R.. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 2014; 9:171–181. [DOI] [PubMed] [Google Scholar]
- 21. Picelli S., Bjorklund A.K., Reinius B., Sagasser S., Winberg G., Sandberg R.. Tn5 transposase and tagmentation procedures for massively scaled sequencing projects. Genome Res. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang Q., Gu L., Adey A., Radlwimmer B., Wang W., Hovestadt V., Bähr M., Wolf S., Shendure J., Eils R. et al. Tagmentation-based whole-genome bisulfite sequencing. Nat. Protoc. 2013; 8:2022–2032. [DOI] [PubMed] [Google Scholar]
- 23. Ilicic T., Kim J.K., Kolodziejczyk A.A., Bagger F.O., McCarthy D.J., Marioni J.C., Teichmann S.A.. Classification of low quality cells from single-cell RNA-seq data. Genome Biol. 2016; 17:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zajac P., Islam S., Hochgerner H., Lönnerberg P., Linnarsson S.. Base Preferences in Non-Templated Nucleotide Incorporation by MMLV-Derived Reverse Transcriptases. PLoS One. 2013; 8:e85270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fu G.K., Hu J., Wang P.-H., Fodor S.P.A.. Counting individual DNA molecules by the stochastic attachment of diverse labels. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:9026–9031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Harbers M., Kato S., de Hoon M., Hayashizaki Y., Carninci P., Plessy C.. Comparison of RNA- or LNA-hybrid oligonucleotides in template-switching reactions for high-speed sequencing library preparation. BMC Genomics. 2013; 14:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Borgstrom E., Redin D., Lundin S., Berglund E., Andersson A.F., Ahmadian A.. Phasing of single DNA molecules by massively parallel barcoding. Nat. Commun. 2015; 6:7173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kinde I., Wu J., Papadopoulos N., Kinzler K.W., Vogelstein B.. Detection and quantification of rare mutations with massively parallel sequencing. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:9530–9535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schmidt W.M., Mueller M.W.. CapSelect: A highly sensitive method for 5΄ CAP-dependent enrichment of full-length cDNA in PCR-mediated analysis of mRNAs. Nucleic Acids Res. 1999; 27:e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kebschull J.M., Zador A.M.. Sources of PCR-induced distortions in high-throughput sequencing data sets. Nucleic Acids Res. 2015; 43:e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.