Abstract
Scientific Data Analysis Resources (SDARs) such as bioinformatics programs, web servers and databases are integral to modern science, but previous studies have shown that the Uniform Resource Locators (URLs) linking to them decay in a time-dependent manner, with ∼27% decayed to date. Because SDARs are overrepresented among science's most cited papers over the past 20 years, loss of widely used SDARs could be particularly disruptive to scientific research. We identified URLs in MEDLINE abstracts and used crowdsourcing to identify which reported the creation of SDARs. We used the Internet Archive's Wayback Machine to approximate ‘death dates’ and calculate citations/year over each SDAR's lifespan. At first glance, decayed SDARs did not significantly differ from available SDARs in their average citations per year over their lifespan or journal impact factor (JIF). But the most cited SDARs were 94% likely to be relocated to another URL versus only 34% of uncited ones. Taking relocation into account, we find that citations are the strongest predictors of current online availability after time since publication, and JIF modestly predictive. This suggests that URL decay is a general, persistent phenomenon affecting all URLs, but the most useful/recognized SDARs are more likely to persist.
INTRODUCTION
The World Wide Web has accelerated scientific capacity for data analysis, modeling and interpretation of data by providing online access to bioinformatics programs, web servers and databases, hereafter referred to more broadly as Scientific Data Analysis Resources (SDARs). In MEDLINE, the fraction of all scientific publications that reference the use of a program or database within their abstract has been steadily increasing over the past 40 years (1). The use of bioinformatics SDARs such as these in scientific research is ubiquitous, and over the past 20 years, bioinformatics programs have been ∼31-fold over-represented among the top 20 most cited papers each year, relative to the number of bioinformatics papers published in MEDLINE (2), showing that such resources are an integral part of modern science.
Published SDARs are usually linked to via a Uniform Resource Locator (URL), so that readers have an immediate means to locate them online and use them. Unfortunately, a number of studies have found that published URLs decay in a time-dependent manner across all fields studied thus far (3–12), with some even decaying prior to publication (13). A limitation of these prior studies is that they were of URLs in general and not focused specifically on papers reporting the development of new SDARs. A prior study of URL decay in dermatology journals found that most authors did not have direct control over the URLs they published (14). Which brings up an important point that URLs fall in roughly two categories: those that point to external resources not created by the authors (e.g. informational webpages, organizations or even SDARs created by other groups), and those created by the authors. It makes sense to treat these categories separately, when possible. And when authors create an SDAR, they must have had control over the URL, at least during that time. Not all published URLs in MEDLINE link to SDARs, but the majority do. The loss of URLs is problematic because it degrades the integrity and reproducibility of prior research and, with SDARs, it means a method of data analysis is no longer available and anyone who used the method cannot likely have their results reproduced. As a response to URL decay, efforts arose to archive published URLs (5,15,16), although they are limited to archiving static content (i.e. website HTML but not the underlying programs that drive analysis) and their use inconsistent across journals. In terms of SDARs, archiving attempts are not likely to be effective since they are only able to preserve static content (e.g. images and text), but not programmatic interfaces that enable data to be submitted for analysis or querying.
SDARs decay in a time-dependent manner, but the scientific value of the lost SDARs is not clear. That is, we do not know how useful they were, on average, to the scientific community. It has been suggested that bioinformatics, as a field, is somewhat ‘hit or miss’. That is, bioinformatics programs tend to have more of a divide between the highly cited and the poorly cited, with fewer of intermediate success relative to citation patterns in other journals (2). Thus, the average decayed SDAR may not be highly cited, but it could be potentially very alarming and disruptive to the scientific community if highly cited SDARs (e.g. those among the top 20 most cited papers in their year of publication) were to suddenly become unavailable. Prior studies (3–12) have firmly established that time elapsed since publication is highly predictive of current URL availability so, if no other factors mitigate SDAR loss, then loss of the most cited will merely be a function of time.
Aside of the strong correlation with time elapsed since publication, it is still not clear what factors differentiate decayed versus available URLs. In particular, it is of interest to identify why authors that do have control over the availability of their published URL would let availability lapse. We previously thought corresponding author experience and possibly lab or institutional infrastructure might be a factor, but this turned out to not be the case (11). Here, we surveyed several factors that could potentially affect the stability of an SDAR. If an SDAR is published in a higher impact journal, it is possible that the authors may consider continued availability a higher priority, perhaps perceiving that expectations are higher. Or, feedback from the community might encourage them to keep their SDAR available, either by peer-pressure or simply by making them aware that a problem with accessibility has arisen. It is also possible that the source of the feedback may be more influential than the amount of feedback—that others using an SDAR for large, important studies provide more incentive to the developer than, for example someone citing their paper because they are developing a similar SDAR or reviewing the field.
A prior study of URLs in dermatology journals showed that less than half of all authors visited their URL again after publication (14). This suggests that lapses in availability may be more likely to be noticed by visitors/users than authors, and that notifying the authors of availability lapses is likely a function of how many visitors there are and how useful they perceive the site to be. Another study also found that only 5% of URLs mentioned in more than one abstract had decayed versus 20% of those mentioned only once (10), although it is reasonable to presume that decayed URLs are less likely to be mentioned. To account for a citation drop due to decay, we would have to know when the URL decayed. Citations are a reasonable proxy to quantify general interest from the scientific community, as they will likely correlate with the number of people who have used the software and possibly even inquired about features/bugs. Citations might underestimate usage, however, as a couple of studies have reported finding publications that mention bioinformatics programs within the text, but do not officially cite them (17,18). Influential feedback could be approximated with the PageRank algorithm, which weights citations from highly cited studies more heavily. Finally, Journal Impact Factor (JIF) is a well-established (although not uncontroversial) metric to quantify a journal's ‘prestige’.
This survey adds to prior work by classifying which URLs report the development of a new SDAR. This not only restricts analysis to, arguably, the most important class of URLs, but it also overcomes a limitation identified in prior studies that the availability of these URLs is most likely under the direct control of authors. To our knowledge, it is also the first study to estimate URL lifespans, as prior studies were all essentially ‘snapshots’ of URL decay at that moment, and this enables us to estimate citations over the duration of online availability.
URL identification and processing
An expanded version of the methods used can be found in the online supplementary data, and we will attempt to summarize the more pertinent aspects of the survey here. We downloaded MEDLINE records (which includes titles at a minimum and abstracts for many, but not all papers) from the National Center for Biotechnology Information (NCBI), which encompassed over 22 million unique papers as of June 2015 to identify 27 349 unique URLs associated with 25 224 unique PubMed Identifiers (PMIDs). Using methods previously described (6,9–11), URLs were queried for their online availability three times a day over 10 days starting on 8 June 2015. The first chronological mention of each SDAR in MEDLINE was assumed to be the original paper describing its creation and subsequent mentions of the URL associated with the SDAR were assumed to be references to it.
Crowdsourcing classification of SDARs
To identify which of the URLs were links to SDARs, we turned to crowdsourcing (19–21). Crowdsourcing relies upon humans to make a classification decision, and can be effective when a task can be reduced to ‘click and decide’. There are several online services dedicated to matching job providers with workers, and evaluating the competency of the workers. Job providers begin by creating a set of instructions to briefly orient workers to the nature of the task to be performed (our instructions given for the task can be found in the expanded methods section). Usually, the output of the task is in the form of a multiple choice answer. Then, if a worker wishes to proceed after reading the instructions, they will be presented with a test set. The test set is created by the job provider to be representative of the actual work and they have already selected the correct answers. If a worker performs poorly on the test set, they are not allowed to proceed further under the assumption they do not understand the task sufficiently well. For every 10 questions asked of a worker, one is from the test set and is used to judge their continued performance. The job provider is able to judge the efficiency of the test questions by cumulative performance on each question. For example, if one question has a high error rate relative to the rest, it may be too unclear or difficult.
Crowdsourcing is relatively inexpensive (total cost for crowdsourcing 25 274 abstracts was US$960) but it could be argued that automated methods of classifying URLs would be more cost-effective and faster, but two issues led us to believe crowdsourcing was preferable. First, since citations are associated with papers rather than URLs, and the abstracts of these papers may have multiple URLs (e.g. one to a program, one to supplementary information), we needed a way to link citations to SDARs specifically. For example, URLs to both a program and supplementary file, would be associated with the same number of citations since they are part of the same paper, but presumably most (if not all) of the citations are to the program. Second, it was important to determine whether or not the authors were reporting the development of their own SDAR based on the abstract, since URLs mentioned in the abstract can frequently refer to a program or database someone else created and the authors used for analysis. This second issue is a much more challenging problem for an algorithm to recognize accurately. Databases and programs are both SDARs, but we asked they be classified separately so we might be able to detect trends unique to either category.
We used CrowdFlower.com to crowdsource the classification of each abstract to determine whether or not the development of an SDAR was being reported in the abstract. We created an example training set to test performance of the crowd classifiers against our own gold-standard classifications. We required that two independent crowdsourcers both agreed on the classification of the URL before accepting the classification. In the event of disagreement, we had as many as five people submit their classification of the URL content. Fifty abstracts without URLs were added and four options were provided for classification: (i) program, (ii) database, (iii) other, (4) no URL. Random guessing would be expected to yield 25% accuracy. In the event of multiple URLs, crowdsourcers were instructed to choose the ‘highest level’ URL in the abstract (in the order shown above).
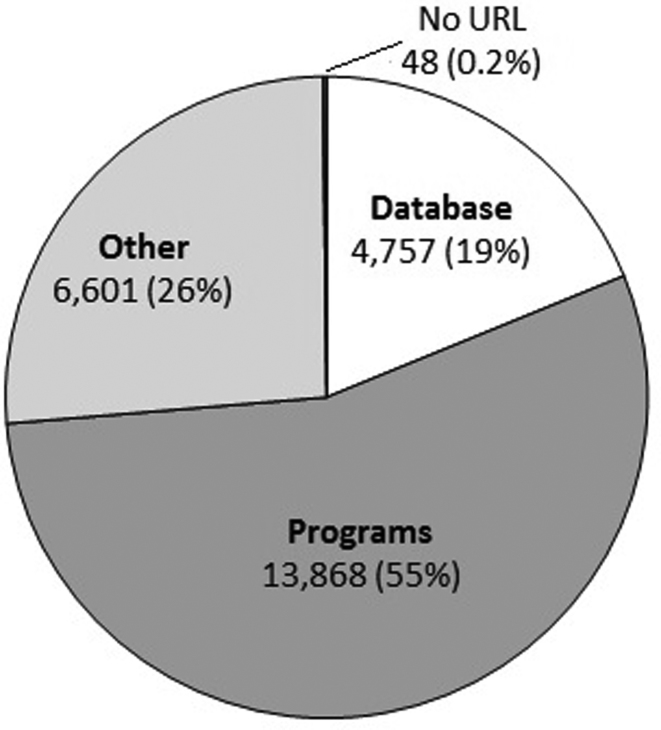
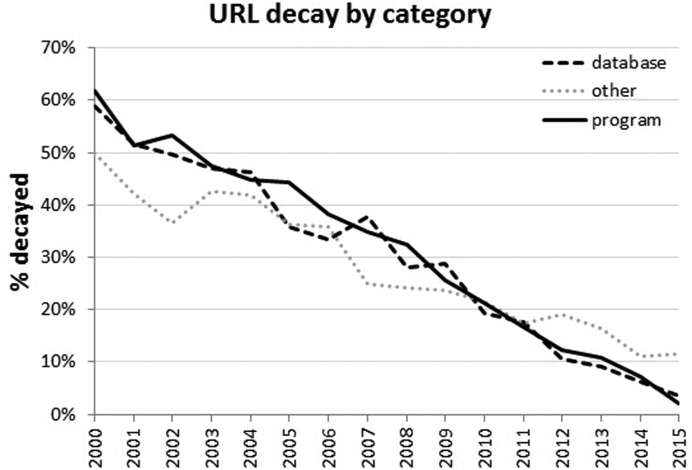
Crowdsourcing took approximately two weeks to complete, although this was non-continuous, as we submitted several increasingly large subsets and then evaluated feedback from the crowdsourcers regarding how fair they thought the test questions were and whether or not certain types of test questions tended to yield lower accuracy than the rest. Figure 1 summarizes the classification of the URLs. Table 1 summarizes how well the crowdsourcers did versus an expert-annotated standard of 600 abstracts (200 for each of the three categories) evaluated by one of the authors (JDW). URL decay by category is shown in Figure 2, with the overall rate of decay consistent with our prior studies. The crowdsourcing results are provided as Supplementary Table S2.
Figure 1.
Crowdsourcing classification of the highest level unique URLs within the abstracts analyzed. Fifty papers with no URL were included to add a fourth category, and 48 of them were correctly classified.
Table 1. Performance of the crowdsourcing by confidence score versus the expert-annotated standard.
| Confidence score | % TP | # of URLs in this range | Est. FP on entire dataset |
|---|---|---|---|
| <0.5 | 50% | 916 | 458 |
| 0.5–0.75 | 61% | 8591 | 3309 |
| 1.0 | 79% | 15 767 | 3335 |
| Total | 72% | 25 274 | 7102 |
The confidence score is given by Crowdflower and is a function of the agreement between evaluators as well as the ‘trust’ score of a particular evaluator (which is based on prior experience). ‘% TP’ refers to the fraction of True Positives (TP) within the sample taken for that confidence score rage. The number of URLs with confidence scores within that range is also shown, and the estimated total False Positives (FP) for the entire dataset based on sample error rates.
Figure 2.
Yearly URL decay by category from 2000–2015. The general rate of URL decay is similar, regardless of the nature of what the URL links to.
Identifying the lifespan of SDARs
The Internet Archive (IA) is the world's largest and oldest public archive of the WWW via its Wayback Machine, available at https://archive.org/web. It periodically accesses URLs across the Internet and takes a ‘snapshot’ of the site, while recording the HyperText Transfer Protocol (HTTP) code (https://tools.ietf.org/html/rfc2616) returned upon each attempt to access a URL (e.g. 404 means the website was not accessible, 200 means success). Querying the access history thus allows one to approximate when a URL was active. To estimate a URL's death date, we used IA's CDX Application Programming Interface (API) (https://archive.org/help/wayback_api.php) to query the history of each URL extracted.
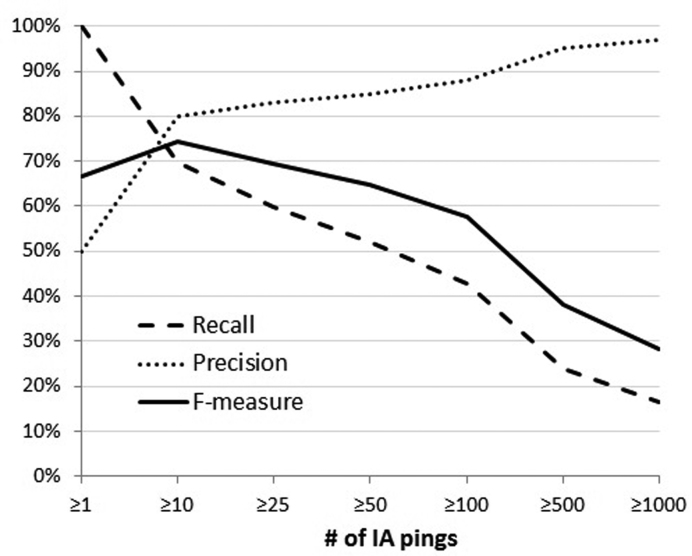
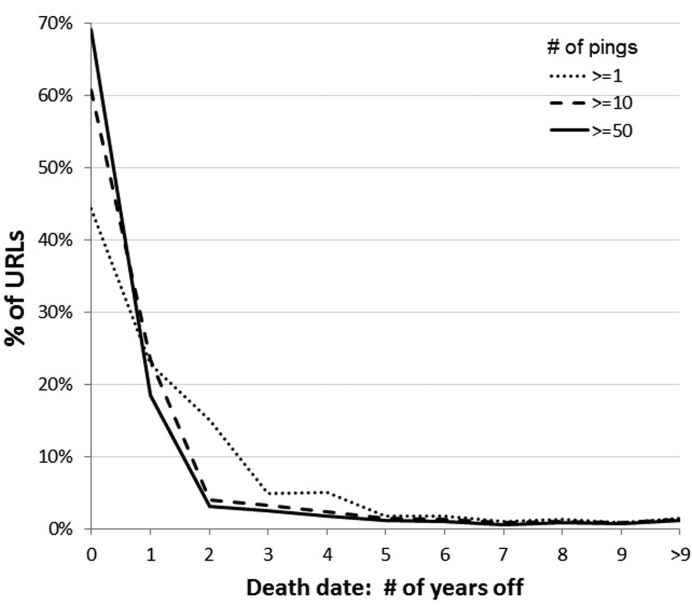
Lifespan was rounded to whole years and URLs dying within the same year as their publication (their ‘birth’ date) were presumed to be up the entire year for the purposes of citation calculation. Whereas birth dates are fairly precise, ‘death’ dates are more granular because URLs are not pinged (checked for accessibility) by IA with equal regularity. We wanted to estimate the number of death dates accurate to within at least a year. However, since the URL survey was conducted in June 2015 (0.5 years into 2015) and birth/death dates are expressed in integer values, our assessment is actually of death dates that are accurate within 1.5 years.. To do this, we took ‘alive’ URLs (i.e. known to be accessible at the time of the study) and calculated what fraction of them had been pinged in the last 1.5 years. Figure 3 shows the trade-off between recall (total URLs queried) and precision (death date correct within 1.5 years). Figure 4 shows a distribution of how far off death dates are by the number of IA access attempts (‘pings’). Based on this and because we consider precision more important than recall for this task, we only conducted analyses on URLs with at least 55 pings (55 was the median). This means that ∼15% of decayed URLs will have their death dates overestimated by at least 1.5 years. Thus, we expect URLs with fewer pings will tend to have slightly overestimated citations per year over their lifespan due to estimated death dates that may be earlier than their actual death date. If a dead URLs last successful IA access date was prior to 1 January 2014 (1.5 years prior to the URL accessibility survey in June 2015), that was considered the official ‘death date’. Otherwise, the date of the survey was used as the death date. The results of the URL survey and the IA pings are provided as supplementary information (Supplementary Table S1).
Figure 3.
Estimating the accuracy of when a URL decayed within 1.5 years. For our URLs within the Internet Archive (IA), recall refers to the percent returned when the number of archival access attempts (# of IA pings) is thresholded. Precision is the number of those URLs whose death date is accurate within 1.5 years. The F-measure shows the trade-off between precision and recall.
Figure 4.
Magnitude of error in estimated death dates by # of IA access attempts (‘pings)’. The Internet Archive (IA) was queried using URLs that were accessible in June 2015, then the date of their last archive attempt was subtracted. The chart shows a histogram of how far off the estimated death dates are when restricting pings to ≥ 1, ≥10 and ≥50.
Relocation of URLs
URLs with automatic redirect pages are counted as ‘accessible’ by the methods we used, as would be any pages containing statements of redirection. For decayed SDARs, we used Google to search for them, using the most unique identifying terms. When the program name was sufficiently unique, that was used alone. When the program name alone was ambiguous (e.g. ‘ArrayDB’, ‘GALAXY’), we combined it with unique terms to further refine the search. Only the first page of Google results was examined.
Examining the decay of ‘important’ SDARs
One way a reader might judge the potential ‘importance’ of an SDAR at the time of publication would be by the prestige of the publishing journal. Over time, however, citations should be a more objective metric. One might also argue that a concept such as PageRank, which weights citations from highly cited papers more heavily, might be an even better metric. So we examined all these factors.
Authors may or not pay attention to the number of citations their published programs receive, but presumably the number of citations to a paper reporting an SDAR strongly correlates with its usage, number of user inquiries, suggestions for updates, and bug reports the authors receive from users. Using citations per year (rather than total citations) controls for the time passed since publication, but to estimate the amount of attention an SDAR received prior to its decay, we need to know when it decayed.
We can approximate death dates using the Internet Archive (IA) Wayback Machine. Because the granularity of the death estimates will affect the results and IA does not cover all URLs equally, we restricted analysis to the 11 523 URLs with at least 55 IA pings (see methods) to calculate average citations/yr over an SDARs lifespan. Comparing decayed versus available SDARs, there was a difference at P < 0.05 that more cited databases were available, but not for programs or SDARs in general (Table 2).
Table 2. Difference between decayed (down) and accessible (up) SDARs by category.
| Mean cites/year | Mean JIF | |||||
|---|---|---|---|---|---|---|
| Using original URL | Down | Up | P-value | Down | Up | P-value |
| Database | 3.4 | 4.5 | 0.04 | 6.5 | 6.5 | 0.89 |
| Program | 5.3 | 5.4 | 0.81 | 6.0 | 6.0 | 0.92 |
| combined | 4.6 | 5.1 | 0.29 | 6.2 | 6.2 | 0.84 |
| Other | 1.4 | 1.0 | 0.0003 | 5.2 | 4.6 | 0.02 |
P-values for significance of the up vs down differences were determined with two tailed t-tests with unequal variance. In the category ‘other’, decayed URLs had significantly more citations and higher JIF. This result may be because available URLs in the ‘other’ category are biased towards links to institutions/organizations (which tend to be more stable) from editorials.
Evaluating the significance of SDARs given the possibility of relocation
Examining the full list of 25 274 SDARs, we took the top 100 most cited, and a random sample of 100 that had not been cited (50 databases and 50 programs for each sample). We found that 94% of the most cited SDARs had been relocated to a different URL versus only 34% of those without citations (P-value for significance < 2.2e–16). Once relocation was taken into account, we found that available SDARs had significantly higher citations per year over their estimated lifespan versus those that had decayed, but no difference in the JIFs (Table 3). However, because we only sampled the extreme ends of the distribution, this alone is insufficient to make a statement regarding how citations or JIF influence current online availability.
Table 3. After considering relocation information on a sample of decayed URLs (no relocation search was attempted for the category ‘other’), the number of citations was significantly different for available SDARs, combined and separate.
| Mean cites/year | Mean JIF | |||||
|---|---|---|---|---|---|---|
| Using relocation info | Down | Up/avail | P-value | Down | Up/avail | P-value |
| Database | 2.2 | 5.5 | 3.9e−16 | 6.5 | 6.5 | 0.88 |
| Program | 3.2 | 6.6 | 3.2e−16 | 6.0 | 6.0 | 0.69 |
| combined | 2.9 | 6.3 | 1.4e−27 | 6.2 | 6.2 | 0.79 |
| Other | n/a | n/a | n/a | n/a | ||
No significant difference in JIF was observed. Similar results were obtained when the analysis was done using the median (Supplementary Table S3).
To model the effect of citations and JIF on SDAR availability, we had to take the possibility of relocation into account. But unfortunately, conducting manual relocation searches on the full list was not feasible. So, to assess potential SDAR relocation to another URL, we manually searched and checked for relocation of a sample of 185 unavailable SDARs from the list of 11 523 URLs with at least 55 IA pings. The sample was selected according to a PPS (proportional to size) design, with size defined by the average number of citations to a category. Thus, SDARs with more citations are more likely to be sampled, but the statistical inference accounts for this by weighting each one in proportion to the inverse probability it was sampled. We did not try to locate URLs in the category ‘other’ because they frequently lacked unique keywords sufficient enough to conduct a Google search.
Citations correlate with SDAR availability
Using logistic regression (more specifically the ‘svyglm’ function in the R package ‘survey’, able to account for the sampled nature of the relocation information), we then modeled the overall availability of SDARs either accessible via their original published URL or relocated, as a function of year of publication, citations/year and JIF. Since IA contains data on the first time a URL was archived, we also included length of URL availability prior to publication, to see if perhaps a prior history of URL availability might influence future availability. We estimated the impact of each of these factors on the probability an SDAR is still available online, and the results are summarized in Table 4.
Table 4. Logistic regression coefficients from modeling SDAR decay as a function of year of publication (PubYear), availability of the URL prior to publication (PriorAvail), citations/year (cites/year) and journal impact factor (JIF).
| Variable | Estimate | Std. error | z value | Pr(>|z|) |
|---|---|---|---|---|
| PubYear | 0.25 | 0.01 | 19 | 9.1e−81 |
| Cites/yr | 0.06 | 0.01 | 5.1 | 3.9e−07 |
| JIF | 0.07 | 0.02 | 3.1 | 0.002 |
| PriorAvail | 0.09 | 0.01 | 1.7 | 0.09 |
Positive values in the ‘Estimate’ column indicate positive correlations between the variable and current SDAR availability. The last column gives the P-value for effect significance. Only terms with significant impact (P-value < 0.05) were included in the model.
As expected, time since publication was the strongest predictor of current availability. Average citations per year was the second strongest predictor, followed by JIF. The availability of a URL prior to the publication of the SDAR did not appear to be predictive of future availability. We also examined PageRank, which was highly predictive of availability (P < 2.1e−67), but decided not to include it in the final analysis because the magnitude of PageRank is highly correlated with and dependent upon PubYear, the strongest predictor.
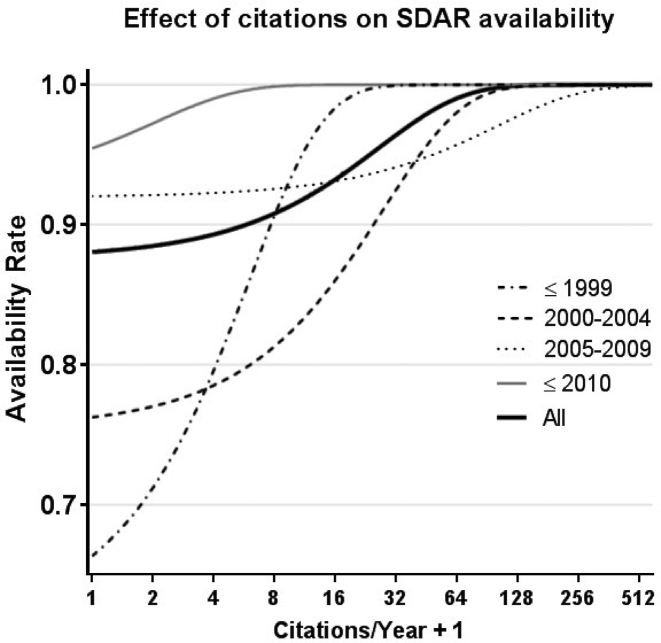
Using a restricted logistic model, including only the two most significant factors (year of publication and number of citations), to predict SDAR availability, the effect is evident—very highly cited SDARs are almost uniformly available. The trend can be seen for SDARs as a group (black line), and by time frame (Figure 5). The impact of the citation number on availability, captured by the slope of the curves, increases progressively from a mild effect on new papers, published after 2009, to a very strong effect on old papers, published before 2000. Impressively, even for the oldest group of SDARs (published in the year 2000 or before), the most cited ones tend to still be available online.
Figure 5.
Mean citations over an SDAR's lifespan is a strong predictor of current availability. SDAR availability curves, one for each of four groups of publication year, depicts the probability the SDAR is still available, predicted with logistic regression, as a function of the number of citations. The thick black curve shows the overall predicted availability rate, when year of publication is removed from the logistic model. Because the x-axis is in log scale and log(0) is undefined, one is added to each number.
CONCLUSION
URL decay in general seems to be a relatively consistent phenomenon, not substantially affected by citations to the paper or JIF of the publication. However, we find that citations to SDARs are highly predictive of whether or not they are still available online at a different URL than the one originally published. This suggests that circumstances necessitating a change in URL arise at a fairly constant rate but the probability of an author expending effort to relocate their SDAR correlates with the amount of attention they receive from the scientific community. Presumably, the citation effect is not direct (i.e. most authors are not closely monitoring citations), but correlates strongly with the number of inquiries, suggestions, bug reports and notifications of lapses in URL availability. Thus, authors likely have a good idea of how much in demand their work is and, although we cannot say what portion of their motivation is altruistic (i.e. to contribute to the greater good) versus selfish (e.g., reduce the number of complaints), both likely play a role.
The initial reaction to biomedical URL decay was one of general alarm (7,15,22). And although URL decay is certainly an undesirable phenomenon, this study suggests that the highest impact (most cited) published SDARs tend to persist. Figure 5 illustrates this nicely - the earliest SDARs (≤1999) have the highest aggregate decay rate, yet those that are highly cited are almost all still available online today. When an SDAR meets a need for scientists, they adopt and use it, and our data suggests this motivates the developers (or possibly other groups to take over the project) to maintain it and perhaps even further its development. If an SDAR is not used or rarely used, then as time goes by, its initial decay may not even be noticed. For the developers, who have likely moved on to other projects, there may be little incentive to re-establish availability, particularly if nobody is requesting it. We also found that the use of source code repositories (e.g. code.google.com, github.com, Bitbucket.org, cran.r-project.org, sourceforge.net, Bioconductor.org) has been on the rise in recent years, going from being <1% of published URLs in 2004 to over 8% by 2015. This is an exciting trend that could result in not only increased availability for SDARs but also a historical record of program versions, which will likely benefit reproducible research (23). Unfortunately, this is not a panacea as repositories of this nature will not obviate the need for active web servers which might have intense data storage, computational or complex configuration requirements.
Our survey has several limitations. First, since only 36% of the most cited SDARs had an URL in their abstract, this means many of the URLs may appear in the full text instead, and would be missed by our approach. Second, we could not measure the relative importance of an SDAR within its research niche. For example, a program may be highly cited within a certain field, but if the field is small, then the potential number of citers is also small. Third, because of the granularity of IA coverage, we could only approximate when an SDAR decayed.
Both the ‘hit or miss’ phenomenon (2) and the trend of SDAR decay due to lack of use/interest suggests that time and effort are being spent developing bioinformatics solutions that are not significantly used, and it may be worth further examining what the source of disconnect may be between the developers of unused SDARs and their intended user audience. That is, if the authors believed it would be useful enough to spend time developing and publishing it, why wasn’t it? By studying which SDARs have been successful and which have not, we may be able to understand more about what makes the difference between a widely adopted bioinformatics approach and one that is not. There are many possibilities, including the existence of other programs within the competitive niche of the unused SDAR, differences in the ease of use and/or the utility of the output, and perhaps a lack of awareness of the existence of new SDARs among the intended user audience.
Supplementary Material
ACKNOWLEDGEMENTS
The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Science Foundation.
Author contributions: J.D.W. conceived of the project, supervised, gathered data and wrote most of the paper. C.B.G. compiled the citation and PageRank statistics, C.G. conducted the statistical analyses and J.H. determined URL accessibility and compiled IA statistics. All authors helped write the final paper.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Foundation [NSF grant #ACI-1345426]. Funding for open access charge: Institutional funding (OMRF).
Conflict of interest statement. None declared.
REFERENCES
- 1. Perez-Iratxeta C., Andrade-Navarro M.A., Wren J.D.. Evolving research trends in bioinformatics. Brief. Bioinformatics. 2007; 8:88–95. [DOI] [PubMed] [Google Scholar]
- 2. Wren J.D. Bioinformatics programs are 31-fold over-represented among the highest impact scientific papers of the past two decades. Bioinformatics. 2016; 32:2686–2691. [DOI] [PubMed] [Google Scholar]
- 3. Carnevale R.J., Aronsky D.. The life and death of URLs in five biomedical informatics journals. Int. J. Med. Informatics. 2007; 76:269–273. [DOI] [PubMed] [Google Scholar]
- 4. Dellavalle R.P., Hester E.J., Heilig L.F., Drake A.L., Kuntzman J.W., Graber M., Schilling L.M.. Information science. Going, going, gone: lost Internet references. Science. 2003; 302:787–788. [DOI] [PubMed] [Google Scholar]
- 5. Ducut E., Liu F., Fontelo P.. An update on Uniform Resource Locator (URL) decay in MEDLINE abstracts and measures for its mitigation. BMC Med. Informatics Decision Making. 2008; 8:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hennessey J., Ge S.. A cross disciplinary study of link decay and the effectiveness of mitigation techniques. BMC Bioinformatics. 2013; 14(Suppl. 14):S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kelly D.P., Hester E.J., Johnson K.R., Heilig L.F., Drake A.L., Schilling L.M., Dellavalle R.P.. Avoiding URL reference degradation in scientific publications. PLoS Biol. 2004; 2:E99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wagner C., Gebremichael M.D., Taylor M.K., Soltys M.J.. Disappearing act: decay of uniform resource locators in health care management journals. J. Med. Library Assoc.: JMLA. 2009; 97:122–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wren J.D. 404 not found: the stability and persistence of URLs published in MEDLINE. Bioinformatics. 2004; 20:668–672. [DOI] [PubMed] [Google Scholar]
- 10. Wren J.D. URL decay in MEDLINE–a 4-year follow-up study. Bioinformatics. 2008; 24:1381–1385. [DOI] [PubMed] [Google Scholar]
- 11. Hennessey J., Georgescu C., Wren J.D.. Trends in the production of scientific data analysis resources. BMC Bioinformatics. 2014; 15(Suppl. 11):S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Habibzadeh P. Decay of references to Web sites in articles published in general medical journals: mainstream vs small journals. Appl. Clin. Informatics. 2013; 4:455–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Thorp A.W., Schriger D.L.. Citations to Web pages in scientific articles: the permanence of archived references. Ann. Emerg. Med. 2011; 57:165–168. [DOI] [PubMed] [Google Scholar]
- 14. Wren J.D., Johnson K.R., Crockett D.M., Heilig L.F., Schilling L.M., Dellavalle R.P.. Uniform resource locator decay in dermatology journals: author attitudes and preservation practices. Arch. Dermatol. 2006; 142:1147–1152. [DOI] [PubMed] [Google Scholar]
- 15. Eysenbach G., Trudel M.. Going, going, still there: using the WebCite service to permanently archive cited web pages. J. Med. Internet Res. 2005; 7:e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Eysenbach G. Going, going, still there: using the WebCite service to permanently archive cited Web pages. Annu. Symp. Proc./AMIA Symp. 2006; 919. [PMC free article] [PubMed] [Google Scholar]
- 17. Howison J., Bullard J.. Software in the scientific literature: problems with seeing, finding, and using software mentioned in the biology literature. JASIST. 2015; doi:10.1002/asi.23538. [Google Scholar]
- 18. Rung J., Brazma A.. Reuse of public genome-wide gene expression data. Nat. Rev. Genet. 2013; 14:89–99. [DOI] [PubMed] [Google Scholar]
- 19. Good B.M., Su A.I.. Crowdsourcing for bioinformatics. Bioinformatics. 2013; 29:1925–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Parvanta C., Roth Y., Keller H.. Crowdsourcing 101: a few basics to make you the leader of the pack. Health Promot. Pract. 2013; 14:163–167. [DOI] [PubMed] [Google Scholar]
- 21. Ranard B.L., Ha Y.P., Meisel Z.F., Asch D.A., Hill S.S., Becker L.B., Seymour A.K., Merchant R.M.. Crowdsourcing–harnessing the masses to advance health and medicine, a systematic review. J. Gen. Intern. Med. 2014; 29:187–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Whitfield J. Web links leave abstracts going nowhere. Nature. 2004; 428:592. [DOI] [PubMed] [Google Scholar]
- 23. Peng R.D. Reproducible research in computational science. Science. 2011; 334:1226–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.