Abstract
The detection of mRNAs undergoing transcription or decay is challenging, because both processes are fast. However, the relative proportion of an mRNA in synthesis or decay increases with mRNA size and decreases with mRNA half-life. Based on this rationale, I have exploited a 22 200 nucleotide-long, short-lived endogenous mRNA as a reporter for mRNA metabolism in trypanosomes. The extreme 5΄ and 3΄ ends were labeled with red- and green-fluorescent Affymetrix® single mRNA FISH probes, respectively. In the resulting fluorescence images, yellow spots represent intact mRNAs; red spots are mRNAs in transcription or 3΄-5΄ decay, and green spots are mRNAs in 5΄-3΄ degradation. Most red spots were nuclear and insensitive to transcriptional inhibition and thus likely transcription intermediates. Most green spots were cytoplasmic, confirming that the majority of cytoplasmic decay in trypanosomes is 5΄-3΄. The system showed the expected changes at inhibition of transcription or translation and RNAi depletion of the trypanosome homologue to the 5΄-3΄ exoribonuclease Xrn1. The method allows to monitor changes in mRNA metabolism both on cellular and on population/tissue wide levels, but also to study the subcellular localization of mRNA transcription and decay pathways. I show that the system is applicable to mammalian cells.
INTRODUCTION
The life of a eukaryotic mRNA starts with transcription and processing in the nucleus, followed by nuclear export to the cytoplasm where it can act as a template for protein translation. It ends with degradation, mainly by one of two decay routes. Both pathways start with the removal of the mRNA's poly(A) tail by a deadenylase complex, with the catalytic subunits Ccr4 and Caf1. Next, the mRNA can either be degraded 3΄-5΄ by the cytoplasmic exosome or, alternatively, be decapped followed by 5΄-3΄ degradation by the exoribonuclease Xrn1. mRNA translation and decay appear to be inversely linked, with the process of translation acting as an mRNA stabilizer (1,2), although there is also increasing evidence for co-translational degradation (3,4). For each individual mRNA, the expression level is thus mainly determined by its synthesis and decay rate, which can be regulated on multiple levels. Gene expression is also regulated globally: external triggers, such as stresses or differentiation signals for example can cause global changes in transcriptional, translational or decay activities, either individually or in any combination. Specific genes necessary for stress response or differentiation may be excluded. For example, in Trypanosoma brucei, a pathogenic protist responsible for sleeping sickness (human African trypanosomiasis) and related cattle diseases, heat shock stress causes both a translational arrest and an increase in mRNA decay, with the exception of some mRNAs encoding heat shock proteins (5).
The study of mRNA metabolism is challenging. One reason is that the detection of mRNA decay or transcription intermediates is difficult, because both processes occupy only a small fraction of an mRNA's lifetime. In mammals for example, the average gene is transcribed in about 20 min (6,7) but mRNAs have half-lives between 7 and 10 h (8,9). There are less data available about the kinetics of mRNA decay, because decay rates are highly variable between individual mRNAs dependent on secondary structures (10–14). However, the time of mRNA decay is also much shorter than the mRNA's lifetime.
There are several methods available to specifically detect mRNA metabolism intermediates. For example, 5΄-3΄ decay intermediates can be biochemically enriched for by specifically selecting for uncapped (5΄ phosphorylated) mRNAs (3,15). Another example is the fluorescent detection of transcription intermediates in vivo - binding sites for the MS2 phage coat protein (MS2) can be introduced into a reporter mRNA and elongation can be followed using a combination of photobleaching and photoactivation of fluorescent MS2 protein (6). However, none of the currently available methods allows the simultaneous detection of global changes in mRNA transcription and decay pathways in single cells or even subcellular level. Questions such us: ‘Which mRNA pathways are affected in which way after an experimentally induced or naturally occurring interference with mRNA metabolism?’ or ‘Where does mRNA decay takes place?’ remain difficult to address.
The aim of this work was to seek for a new, simple tool to monitor global changes in mRNA metabolism with cellular and subcellular resolution, based on the following reasoning: The time an mRNA spends in synthesis and decay increases proportionally with mRNA size and with decreasing mRNA half-life. Thus, very long, short-lived transcripts should be enriched for mRNA synthesis and decay intermediates and could be employed as native reporters for transcription and decay intermediates. If the extreme 5΄ end of such a reporter mRNA is labelled in red, and the far 3΄ end is labelled in green, this would result in various colour combinations representing different metabolic states when visualized using fluorescence microscopy. Yellow spots would represent complete mRNA molecules with both 5΄ and 3΄ ends intact; green spots would represent mRNA molecules with no 5΄ end – in other words, 5΄-3΄ decay intermediates; red spots would represent mRNA molecules with no 3΄ end – either 3΄-5΄ decay intermediates or mRNAs in transcription. Labelling can be done very efficiently by single molecule RNA fluorescence in situ hybridization (FISH) using the Affymetrix® system. Here, up to 20 pairs of adjacent antisense oligonucleotides hybridize to the target mRNA. This is followed by signal amplification using branched DNA technology (16,17). In this way, single mRNA molecules can be detected using a standard fluorescence microscope with very low background staining.
The establishment of the assay was done in Trypanosoma brucei. The general characteristics of mRNA metabolism in trypanosomes are very similar to those in budding yeast: the median mRNA half-life is about 20 min (18), the average length of an open reading frame is about 1500 nucleotides (19) and with only two exceptions genes have no introns (19–21). The major cytoplasmic mRNA decay pathway is most likely 5΄-3΄ degradation by XRNA, the trypanosome orthologue to the 5΄-3΄ exoribonuclease Xrn1, with the exosome playing no or only a minor role (22–24).
Here, I show by dual colour single molecule mRNA FISH that a very long, short-lived trypanosome mRNA (Tb427.01.1740) is indeed highly enriched for mRNA metabolism intermediates. The average cell has 0.9, 2.1 and 1.3 yellow, green and red mRNA molecules, respectively, thus more than three quarters in synthesis or decay. The suitability of the system to report global changes in mRNA metabolism was tested by adding transcriptional and translational inhibitors and by RNAi depletion of XRNA. Finally, I show that the system is also applicable to mammalian cells.
MATERIALS AND METHODS
Cells and plasmid
Trypanosoma brucei Lister 427 procyclic cells were used for most experiments. The XRNA RNAi experiment was done in Lister 427 pSPR2.1 cells (25) as previously described (24). Cells were cultured in SDM-79 (26) at 27°C and 5% CO2. Transgenic trypanosomes were generated using standard procedures (27). All experiments used logarithmically growing trypanosomes. NIH3T3 cells were grown at 37°C and 5% CO2 in DMEM (Invitrogen) supplemented with 10% FCS and 1% penicillin/streptomycin.
Affymetrix single mRNA FISH
A total of 10 ml trypanosomes at ≈5 × 106 cells/ml were harvested by centrifugation (5 min, 1400 g), resuspended in 1 ml SDM79 without serum and haemin and fixed by the addition of 1 ml 8% PFA (10 min, RT, orbital mixer). A total of 13 ml of PBS were added and cells were harvested by centrifugation (5 min, 1400 g). The cell pellet was resuspended in 1 ml PBS and spread on glass microscopy slides (previously incubated at 180°C for 2 h for RNAse removal) within circles of hydrophobic barriers (PAP pen, Sigma). The cells were allowed to settle (≈20 min, RT), and the slides were then washed once in PBS and used for Affymetrix FISH experiments, essentially as described in the manual of the QuantiGene® ViewRNA ISH Cell Assay (Affymetrix), protocol for glass slide format. The protease of the kit was usually diluted between 1:5000 and 1:10 000, although sometimes higher concentrations were necessary. I found that protease activity decreased with time stored at 4°C (within a few weeks) and increased with temperature (protease digest was therefore performed at 25°C rather than at a variable room temperature). In some experiments a self-made washing buffer (0.1xSSC (saline sodium citrate); 0.1% SDS) was used instead of the washing buffer from the kit. NIH3T3 cells were cultured on poly-L-lysine coated cover slips, fixed in freshly-made 4% formaldehyde solution for 30 min and processed following the standard protocol for 24 well plates. The protease was used 1:10 000. All Affymetrix probe sets used in this work are described in Supplementary Table S1.
Microscopy and image analysis
Z-stacks (100 slices, 100-nm step size) were recorded using a custom-built TILL Photonics iMIC microscope equipped with a 100x, 1.4 numerical aperture objective (Olympus, Tokyo, Japan) and a sensicam qe CCD camera (PCO, Kehlheim, Germany). Images were deconvolved using Huygens Essential software (SVI, Hilversum, The Netherlands). Unless otherwise stated, images are presented as Z-projections (summed slices) produced by and quantified with ImageJ. The numbers of red, green and yellow FISH spots were counted manually.
RESULTS
Detection of mRNA transcription and degradation intermediates by dual colour single molecule mRNA FISH
The sequences of the four T. brucei genes with the longest open reading frames were examined for their suitability to report mRNA metabolism by dual colour single mRNA FISH; in particular, they were searched for the presence of unique sequence stretches at both ends that could be used for probe design.
The mRNA with the longest open reading frame (Tb427tmp.160.1200) encodes GB4, a protein that localizes to the posterior end of the cortical cytoskeleton (28). The open reading frame of GB4 has a 2256 nucleotides-long unique sequence at its 5΄ end, followed by a 22 388 nucleotides-long sequence at the 3΄ end that consists of 37 repeats of 600 nucleotides each. The 3΄ and 5΄ untranslated regions (UTRs) are about 272 and 1250 nucleotides, respectively (21,29). GB4 has no unique sequence at the 3΄ end (the 3΄ UTR is too short for probe design) and is therefore not suitable to report 3΄-5΄ decay or transcription intermediates. GB4 is, however, a very valuable tool to estimate the length extension of such very long mRNAs, because the repetitive sequence at the 3΄ end uniquely allows to probe the entire length of the mRNA molecule. In this work it was therefore used for control experiments.
The mRNA with the second-longest open reading frame (Tb427tmp.57.0008) has inconsistent genome annotations between different trypanosome strains and was therefore not considered. The third-longest mRNA, Tb427.01.1740, is a T. brucei gene encoding a hypothetical protein with a repetitive internal sequence, but unique sequences at both ends. Its open reading frame is 21 465 nucleotides long (Figure 1A). Its 5΄ and 3΄ UTRs are 288 and 444 nucleotides long, respectively (29). The fourth-longest mRNA, Tb427.04.310, encodes a putative ubiquitin-transferase, has an open reading frame of 19 841 nts and a non-repetitive, unique sequence throughout. The 3΄ and 5΄ UTRs are 499 and 187 nts, respectively (29). Both the third and the fourth longest mRNA are therefore candidates for an mRNA metabolism reporter.
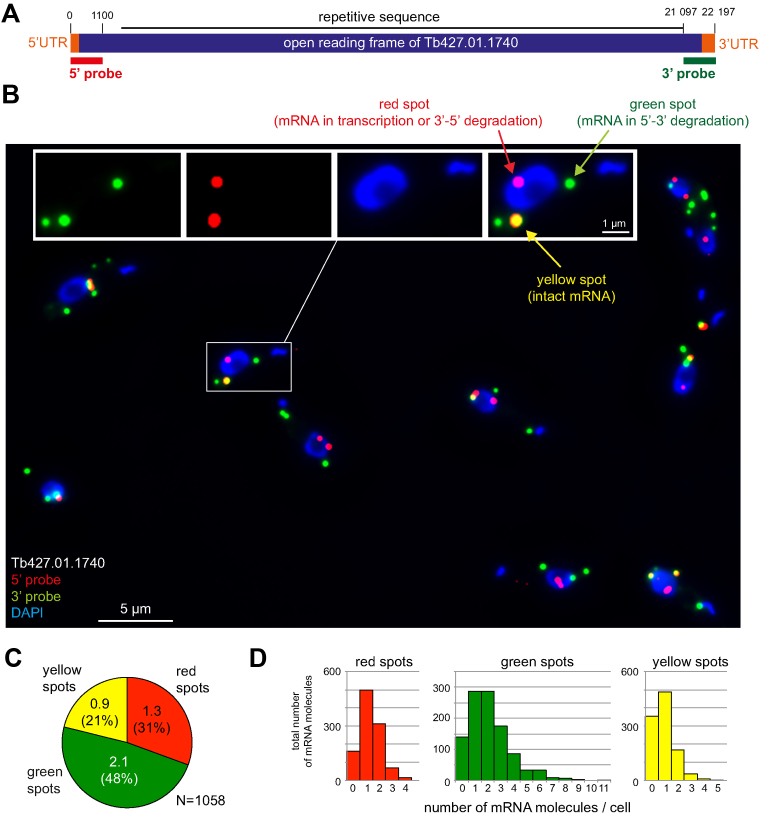
Figure 1.
A long mRNA as a reporter for mRNA metabolism. (A) Schematic of the mRNA metabolism reporter mRNA Tb427.01.1740. The 5΄ and 3΄ untranslated regions (UTRs) (orange) and the open reading frame (purple) are shown above the probes used for the detection of the 5΄ end (5΄ probe, red) or 3΄ end (3΄ probe, green). All elements are shown to scale. (B) Sample Z-stack projection image of trypanosome cells probed with both the 5΄ and the 3΄ probes and stained with DAPI. A merged image of all three fluorescence channels is shown; one cell is shown enlarged in separate channels in the inset. In trypanosomes, DAPI stains both the nucleus (large blue oval) and the kinetoplast (small blue spot or line), the mitochondrial genome. (C) Quantification of the different mRNA states. The number of red, green and yellow spots in each cell was counted. The values shown are average values of 1058 trypanosome cells from six independent experiments. Standard deviations are indicated. (D) Histogram displaying the distribution of the differently-coloured spots between individual cells. Data used are the same as in panel C.
Affymetrix® single molecule mRNA FISH probe sets were designed antisense to the 1100 most 5΄ nucleotides (red fluorescence) and the 1100 most 3΄ nucleotides (green fluorescence) of Tb427.01.1740 and Tb427.04.310. Trypanosomes were fixed, probed simultaneously with both the 3΄ and the 5΄ probes and analysed microscopically. When probed for the Tb427.01.1740 mRNA, the average cell had 1.3 ± 0.9 red, 2.1 ± 1.7 green and 0.9 ± 0.8 yellow mRNA molecules (Figure 1B–D). When probed for Tb427.04.310, there were 0.8 ± 0.9 red, 3.0 ± 1.8 green and 2.3 ± 1.2 yellow mRNA molecules per cell (Supplementary Figure S1). Thus, putative transcription and decay intermediates (red and green spots) were detectable for both mRNAs accounting for more than half of the total mRNA molecules. Moreover, both mRNAs had suitable expression levels, there were enough molecules to perform analyses, but not too many to prevent the detection of individual molecules. Both mRNAs appear equally suited to report mRNA metabolism and I will focus on Tb427.01.1740 for all trypanosome experiments described below.
Technical controls and the definition of a yellow spot
One obvious concern was whether the observed single-coloured spots were true mRNA synthesis and degradation intermediates or technical artefacts. Theoretically, single-coloured spots could also arise from differences in accessibility to the target mRNA between the two differently coloured fluorophores. Such differences could be caused by innate properties of the fluorophores themselves. Alternatively, one end of the target mRNA could be less accessible to the rather bulky Affymetrix probes than the other end, perhaps because it is bound by more proteins and/or has a stronger secondary structure. Moreover, background labelling could also result in non-specific red and green spots. To address these points, two control experiments were performed. First, a red and a green probe were designed antisense to the same part of an average-sized mRNA molecule (Tb427.10.14550, open reading frame of 1980 nucleotides) (Figure 2A, further examples in Supplementary Figure S2). Almost every spot was yellow, indicating that there is no major difference in access to mRNAs between probes labelled with red or green fluorescent dyes and that background fluorescence can be neglected under the FISH conditions used here. As a second control, a red and a green probe were designed to the adjacent 5΄ and 3΄ ends of this mRNA (Figure 2B, further examples in Supplementary Figure S3). Again, the majority of the spots were yellow, indicating that, at least for this mRNA, there was no major difference in probe accessibility between the mRNAs 3΄ and 5΄ end. Moreover, the very small fraction of red and green spots confirms the assumption that a large mRNA size is essential for the detection of significant amounts of decay and transcription intermediates.
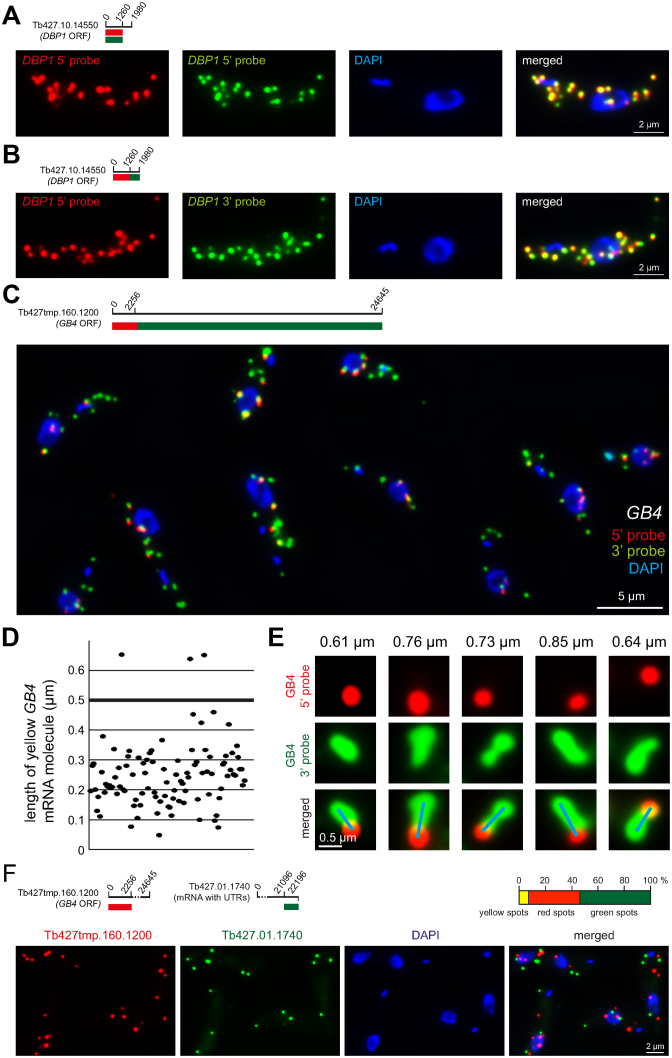
Figure 2.
Technical controls and the definition of a yellow spot. (A) Red and green fluorescence in situ hybridization (FISH) probes were designed to anneal to the same mRNA sequence of an average-sized mRNA (Tb427.10.14550). The signals from the two probes showed a strong overlap. One representative cell is shown; more cells are shown in Supplementary Figure S2. (B) Red and green FISH probes were designed to anneal to adjacent mRNA sequences of the same, average-sized mRNA. The signals from the two probes again showed a strong overlap. One representative cell is shown; more cells are shown in Supplementary Figure S3. (C–E) Trypanosome cells were simultaneously probed with the 5΄ probe (red) and the 3΄ probe (green) specific for the GB4 mRNA (Tb427tmp.160.1200) (details on top left of C). Together, these probes cover the entire 24 645 nucleotides of the GB4 open reading frame: the 5΄ probe binds to the 2256 nucleotides at the 5΄ end and the 3΄ probe to the remaining 22 389 nucleotides. (C) One representative Z-stack projection image with merged fluorescence channels is shown. (D) The length of 100 yellow GB4 mRNA molecules was measured from Z-stack projection images. In total, 97% are less than 0.5 μm in length (D). (E) Example images of very extended GB4 mRNA molecules; the length is indicated on top and the blue line shows how the length measurement was performed (from the middle of the ‘red circle’ to the middle of the most distant ‘green circle’ at the end of the green string). These very long mRNA molecules are rare and never longer than 1.0 μm. (F) Trans-probing: dual colour mRNA FISH using probes antisense to two different mRNAs (details on top left). The percentage of red, green and yellow spots was quantified from 152 cells (top right). The average cell had 7.3 ± 2.8 mRNA molecules. A representative image is shown (bottom).
The enormous size of the Tb427.01.1740 reporter mRNA also raises the question of how to define a yellow spot. The maximal theoretical distance between a red and a green spot on the same mRNA molecule is 7.5 μm, if the mRNA would exist in its fully extended form. This is almost half the length of the trypanosome cell: red and green spots would be clearly separated over such a long distance and therefore impossible to recognize as belonging to the same mRNA molecule. However, mRNA molecules are unlikely to exist in such fully extended forms. To estimate the true extension of very long mRNA molecules, I took advantage of the, above-mentioned, unique features of the trypanosome mRNA with the longest open reading frame (Tb427tmp.160.1200, encoding GB4). The highly repetitive sequence at the 3΄ end of this mRNA allows to probe over the entire length of the mRNA's 3΄ end with one single green Affymetrix probe set. The short unique sequence at the 5΄ end of Tb427tmp.160.1200 was simultaneously probed with a red Affymetrix probe set. Elongated mRNA molecules are thus visible as green strings with a red spot at their 5΄ ends (Figure 2C–E). The vast majority of Z-stack projected mRNA strings (97%, N = 100) was shorter than 0.5 μm (Figure 2D). Even the most extended molecules I found by analysing many images were shorter than 1 μm (Figure 2E). Thus, most red and green spots that are more than 0.5 μm apart are very unlikely to belong to the same mRNA molecule.
If 0.5 μm maximal distance between the centre of a red and a green spot is used to define a yellow spot, how many yellow spots are false positives, e.g. in fact two different mRNA molecules? To get an estimation, the number of red, green and yellow spots were quantified from 152 cells probed by dual colour FISH for two different mRNA molecules (trans-probing) (Figure 2F). The average cell had 7.3 ± 2.8 mRNA molecules, of these were 3.9 ± 1.6 red, 2.9 ± 1.9 green and 0.5 ± 0.6 yellow. Thus, by trans-probing, 7.1% of all spots were classified as yellow and are therefore false positives, in contrast to 23% of yellow spots that were detected for the Tb427.01.1740 reporter mRNA by the dual colour FISH (compare Figure 1C). These data suggest that about one third of all yellow spots could be false positives. However, this is an overestimation and the true fraction of false positives will be lower for two reasons. Firstly, the number of total mRNA molecules per cell for the probing in trans was 7.3 ± 2.8 – almost double the number of total mRNA molecules found for the Tb427.01.1740 reporter mRNA (4.3 ± 3, compare Figure 1C). Therefore, a higher fraction of false positive spots is expected for the trans-probing, simply because individual red and green spots will be on average closer together. Secondly, the red and green spots of the Tb427.01.1740 reporter mRNA were on average closer together than for the trans-probed mRNAs: the majority of the yellow spots of the reporter mRNA Tb427.01.1740 (54%) were completely overlapping and only 12% were lying adjacent (>0.4 μm apart) to each other. In contrast, for the mRNAs probed in trans, the majority of yellow spots (53%) were lying adjacent to each other (distance of >0.4 μm) and only 16% were completely overlapping.
In all experiments in this work, I have defined yellow spots as red and green single spots that have their centres <0.5 μm apart. The distance chosen in this definition is a compromise that tries to minimize both the number of red and green spots wrongly classified as yellow (false positive yellow spots), as well as the number of yellow spots wrongly classified as one red and one green spot (missed yellow spots).
Subcellular localisation and decay rates
I next examined the subcellular distribution of the different-coloured mRNA spots of the reporter mRNA Tb427.01.1740 in more detail. The mRNA molecules were classified as nuclear, partially nuclear, cytoplasmic close to the nucleus or cytoplasmic distant from the nucleus, based on their relative intracellular localization to the DAPI-stained nucleus on Z-stack projections (Figure 3A).
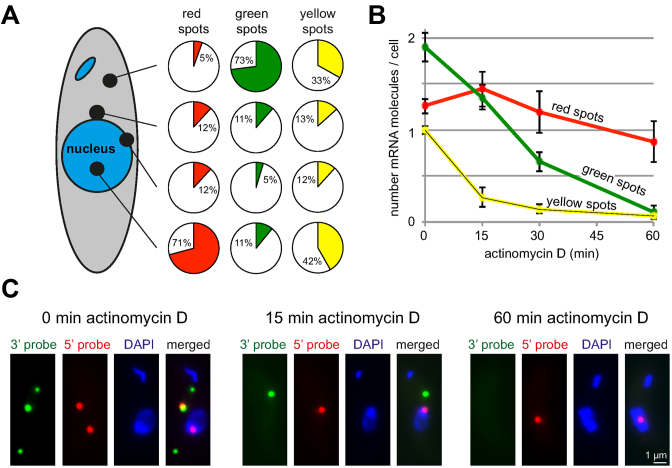
Figure 3.
Subcellular localization and mRNA decay rates. (A) The red, green and yellow spots of 371 cells were classified according to their subcellular localization relative to the DAPI stained nucleus. For each colour, the fraction at each position is indicated in a pie diagram. (B) Cells were treated with actinomycin D (10 μg/ml) over a time-course of 60 min and probed with the 5΄ and 3΄ probes for Tb427.01.1740. Red, green and yellow spots were counted at the times indicated. Averages of the average values from three independent experiments are shown; in each experiment between 160 and 200 cells were analysed per time-point. Error bars represent the standard deviation between the three different experiments. (C) Z-stack projection image of a single representative cell probed with the 5΄ and 3΄ probes of Tb427.01.1740 after 0, 15 and 60 min actinomycin D treatment. More cells are shown in Supplementary Figure S4.
A total of 83% (71 + 12) of the red spots (mRNAs with only the 5΄ end detectable) overlapped either completely or partially with the nucleus. A further 12% of the red spots were found close to the nucleus. Only 5% of red spots were in the cytoplasm and distant from the nucleus. Therefore, the 12% of red spots close to the nucleus account for more than two-thirds of all cytoplasmic red spots. This is much higher than expected from a random cytoplasmic distribution: only 13% of the green cytoplasmic spots were close to the nucleus. A large fraction of the red spots close to the nucleus may therefore not be true cytoplasmic mRNAs but mRNAs being transcribed as they are being exported from the nucleus (assuming the existence of simultaneous export and transcription). These data indicate that the majority of red spots are nuclear or nucleus-associated and must therefore be mRNAs in transcription or nuclear 3΄-5΄ exosomal decay.
Green spots, in contrast, were overwhelmingly localized to the cytoplasm (84%, 73+11) and are therefore likely 5΄-3΄ degradation intermediates. Only 16% (11+5) of the green spots partially or completely overlapped with the nucleus on a Z-stack projection. These could correspond either to nuclear 5΄-3΄ decay intermediates or to cytoplasmic 5΄-3΄ decay intermediates in top or bottom of the nucleus. Yellow spots were found in the nucleus or cytoplasm at approximately equal amounts.
To determine the decay rates of the different mRNA metabolites, cells were treated with actinomycin D, a transcriptional inhibitor that intercalates with DNA and stalls the RNA polymerase (7) (Figure 3B and C and Supplementary Figure S4). Yellow spots decreased to 27% within 15 min, indicating that the mRNA has a half-life of less than 15 min – shorter than the mean half-life of 20 min measured for trypanosome mRNAs (18). The number of green spots gradually decreased to 34% within 30 min and to almost 0 within 60 min. From these data, the total degradation time for the reporter mRNA can be roughly estimated to be about 60 min, which corresponds to a degradation speed of ∼6 nucleotides per second. Red spots, in contrast, first slightly increased in number and then only decreased very slowly, with 69% of the spots still left after 60 min of actinomycin D treatment. The minor increase in red spots is difficult to explain but consistent with the observation in trypanosomes that actinomycin D initially causes a delay in mRNA degradation or even a minor stabilization of some transcripts (30). The slow decrease in red spots suggests that the majority of red spots are mRNA molecules in transcription rather than 3΄-5΄ decay intermediates – 3΄-5΄ decay would be expected to continue in the presence of actinomycin D and should cause a larger reduction in red spots. A block in transcription, however, would probably not allow 3΄-5΄ decay to happen for the transcripts stalled in transcription, because the RNA polymerase II would still be at the mRNA's 3΄ end. Within the short time-scale of the experiment, a block in transcription should therefore not cause a significant reduction in red spots.
In summary, the subcellular localization of mRNA spots in combination with transcriptional inhibition supports the interpretation that red spots are mainly transcription intermediates, and green spots are mainly 5΄-3΄ decay intermediates. This is consistent with the assumption that the cytoplasmic 5΄-3΄ decay pathway is the main mRNA degradation pathway in trypanosomes (22–24).
Detection of experimentally induced changes in mRNA decay
One major aim of the dual colour mRNA FISH system is the detection of global changes in mRNA metabolism. This was tested by adding inhibitors of translation with known effects on mRNA stability (Figure 4) and by inducible RNAi depletion of XRNA (Figure 5).
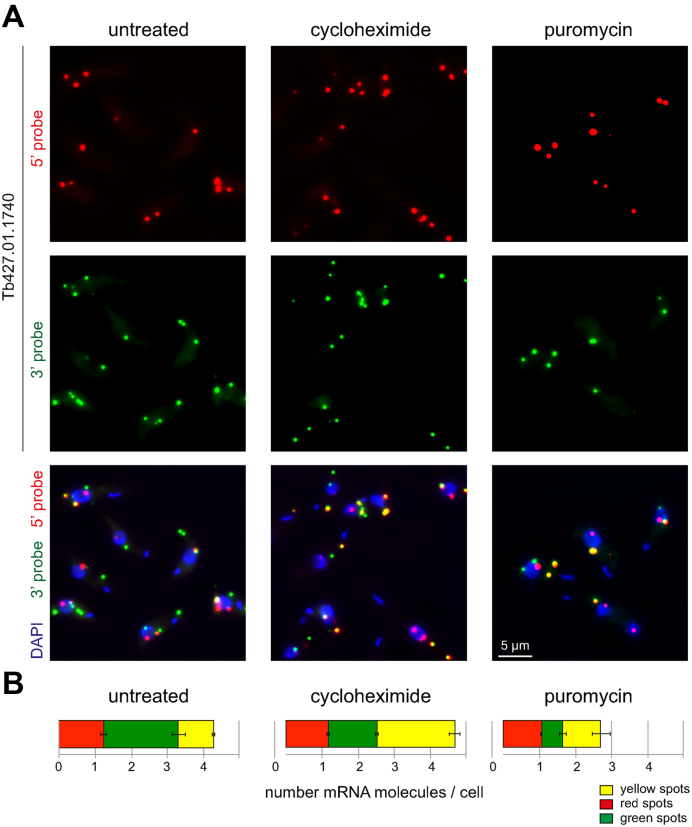
Figure 4.
Translational inhibition by cycloheximide and puromycin. Untreated cells or cells treated with cycloheximide (50 μg/ml) or puromycin (50 μg/ml) for 60 min were probed with the 5΄ and 3΄ probes for Tb427.01.1740. (A) Representative Z-stack projection images from each of the three conditions. (B) Red, green and yellow spots were counted. Averages of the average values from three independent experiments are shown; in each experiment between 127 and 200 cells were analysed for each treatment. Error bars represent the standard deviation between the three different experiments.
Figure 5.
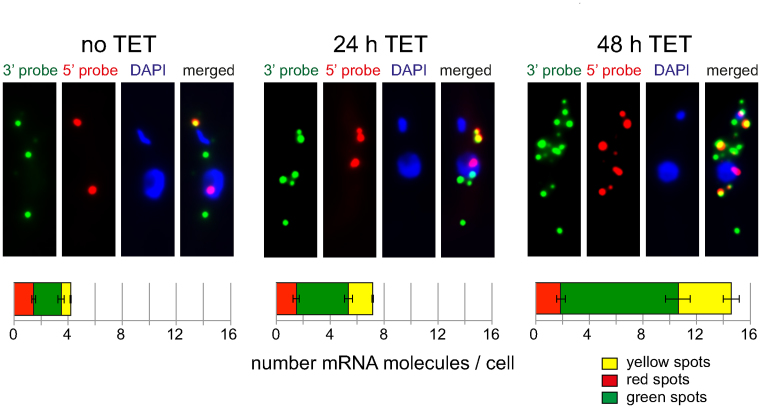
Inducible RNAi depletion of XRNA. Tetracycline inducible RNAi depletion of XRNA was done as previously described (24). Cells induced for RNAi depletion of XRNA for 0, 24 or 48 h were probed with the 5΄ and 3΄ probes specific for Tb427.01.1740. Z-stack projections of representative cells are shown for each time-point; more cells are shown in Supplementary Figure S6. Red, green and yellow spots were counted. Averages of the average values of three independent clonal cell lines are shown; for each clone, between 100 and 200 cells were analysed for each time-point. Error bars represent the standard deviation between the three different clones.
Cycloheximide is an inhibitor of translation that acts by binding to the ribosomal exit site, thereby stalling ribosome translocation (31,32). mRNAs become trapped in translation and, in general, this causes a decrease in mRNA degradation and consequently mRNA stabilization, because translation and mRNA degradation are inversely related (1). These effects of cycloheximide—mRNA stabilization and polysome increase—are conserved in trypanosomes (5,33,34). Addition of cycloheximide to trypanosomes for 60 min caused a 2.2-fold increase in yellow spots, almost entirely on expense of the green spots (Figure 4A and B). This result is consistent with the expected stabilization of the mRNA (increase in yellow spots = increase in intact mRNA molecules) and the decrease in mRNA degradation (decrease in green spots = decrease in 5΄-3΄ degradation intermediates).
A second translation inhibitor is puromycin, a tRNA mimic that causes premature chain termination (35). In contrast to cycloheximide, it causes polysome decrease and does not stabilize mRNAs. Consistent with this, there was no significant change in the number of yellow spots (intact mRNA molecules), when cells were treated with puromycin for 1 h (Figure 4A and B). There was also a 3.5-fold decrease in the number of green spots, likely because non-translated mRNAs outside polysomes become unstable. Neither translational inhibitor had any effect on the number of red spots; this is expected, as red spots are mainly transcription intermediates.
Most mRNA degradation in trypanosomes is carried out by the 5΄-3΄ exoribonuclease XRNA, the orthologue to yeast Xrn1 (18,36). To inhibit mRNA degradation, tetracycline inducible RNAi depletion of XRNA was done as previously described (24). Both the levels of XRNA mRNA and cell growth is greatly reduced within 24 h of RNAi induction (24) and there is an increase in total mRNAs due to mRNA stabilization (18,36) (Supplementary Figure S5). Imaging of the Tb427.01.1740 reporter mRNA by dual colour FISH showed a clear increase in the total number of spots per cell (Figure 5, further examples in Supplementary Figure S6). Quantification of the mRNA spots revealed that while the number of red spots remained essentially unchanged, there was a 1.9-/4.3-fold increase in green and a 2.5-/5.6-fold increase in yellow spots after 24/48 h of RNAi induction (Figure 5). The increase in yellow spots is consistent with the expected stabilization of mRNAs following XRNA depletion. Why is there an increase in green spots? If XRNA would be 100% processive (e.g. never leaving its mRNA substrate before it has been fully degraded), a reduction in XRNA molecules would only cause a reduction in decay initiation and thus an increase in intact mRNA molecules (yellow spots). Once started, decay would be as fast as in the absence of RNAi and therefore not cause an increase in 5΄-3΄ degradation intermediates (green spots). The easiest explanation for the increase in green spots is therefore to assume that XRNA is not 100% processive over the extreme length of this mRNA substrate. Any release of the enzyme from its substrate (even it this just happens in average once per mRNA substrate) would increase the number of green spots, because the time of decay-re-initiation is longer with a reduced number of XRNA molecules.
I have experimentally manipulated mRNA metabolism in three different ways. Each experiment changed the numbers of red, green and yellow spots in a significant and characteristic way, consistent or at least not contradictory to the expectations.
Application to mammalian cells
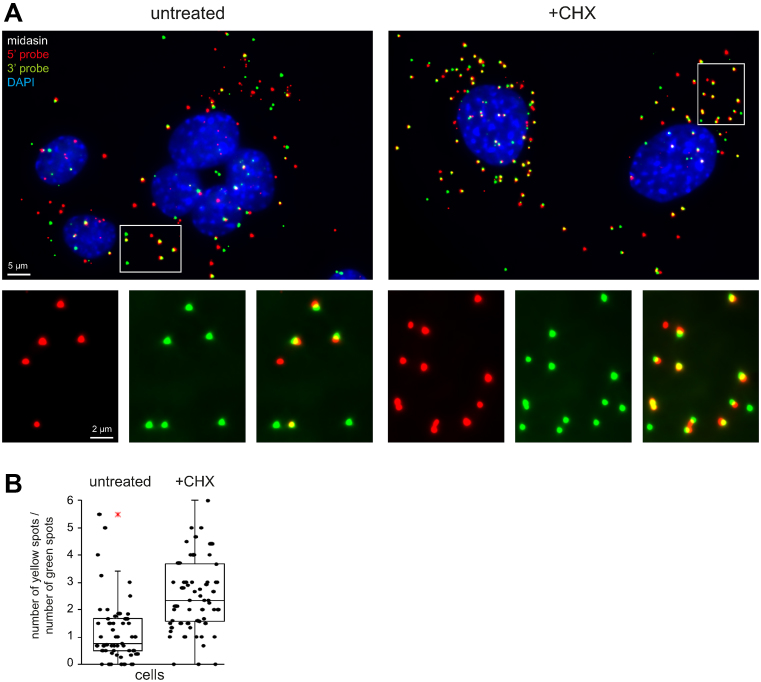
To test the system in mammalian cells, I searched the data on mRNA stability and mRNA copy number of (37) for a suitable mammalian reporter. Midasin (Mdn1, NM_001081392) is with 17 959 nucleotides one of the longest mRNAs of NIH3T3 cells with a copy number of 8.32 molecules per cell and a half-life of 9.23 h. NIH3T3 cells were probed simultaneously with a probe antisense to the 1100 most 5΄ nucleotides (5΄ probe, red) and to the 1100 most 3΄ nucleotides (3΄ probe, green) of midasin mRNA (Figure 6A). On average, there were 2.3, 2.5 and 2.7 red, green and yellow spot in the cytoplasm of each cell, respectively (N = 73), suggesting that decay/transcription intermediates are visible in the mammalian system. Importantly, when cells were treated with cycloheximide, the average numbers of red, green and yellow spots changed to 3.0, 2.9 and 7.4, respectively (N = 88). Thus, as seen in trypanosomes, cycloheximide resulted in a significant increase in the ratio between intact mRNAs and 5΄-3΄ decay intermediates from 1.2 ± 1.3 to 2.6 ± 1.3 (unpaired, two-tailed t-test < 7.5 × 10−10) (Figure 6B). The data show that the system is not restricted to trypanosomes but easily applicable to mammalian cells.
Figure 6.
Detection of mRNA decay intermediates in mammalian cells. NIH3T3 cells were treated with 5 μM cycloheximide for 60 min and simultaneously probed with probes specific to the 5΄ end (red) or 3΄ end (green) of midasin mRNA. The DNA was stained with DAPI. (A) Z-stack projection images of untreated cells and cells treated with cycloheximide. A merged image of all three fluorescence channels is shown; a section is shown enlarged in separate channels in the inset. (B) The number of green and yellow spots in the cytoplasm of each cell was quantified from Z-stack projections and the ratio (yellow/green) is blotted for each cell. The cytoplasm was defined as the cellular space outside the nucleus, ignoring spots that lay on top or below the nucleus. A cell was defined as having one nucleus; in dividing cells, or when individual cells could not be clearly distinguished, the spots were counted together and then divided through the number of nuclei. The data are presented as box plots (waist is median; box is interquartile range (IQR); whiskers are ±1.5 IQR; only the smallest and largest outliers are shown; n ≥ 73). These are representative data from one of two independent experiments.
DISCUSSION
To my knowledge, a system to monitor changes in mRNA decay and transcription on a cellular level is not yet available. Here, I have tested the suitability of a very long endogenous trypanosome mRNA as reporter for transcription and mRNA decay. The mRNA was labelled at its far 5΄ and 3΄ ends with two different colours and allowed the simultaneous detection of both transcription and decay intermediates by dual colour single molecule RNA FISH. The determination of the mRNA's subcellular localization (nuclear versus cytoplasmic) in combination with transcriptional inhibitors allowed transcription intermediates to be distinguished from 3΄-5΄ decay intermediates. The mRNA correctly reported the expected changes in mRNA decay when cells were challenged with different translational inhibitors or RNAi depletion of XRNA.
What kind of questions can be addressed? One important application is to test the function of an unknown protein. Is there a change in transcription and/or mRNA decay following any experimental manipulation with a protein? A combination of the FISH assay with RNAi, as shown here with XRNA as an example, is easy. Another application is to determine which mRNA decay pathways (5΄-3΄ versus 3΄-5΄) dominate in an organism of interest, under certain conditions or in a certain tissue. Are there variations in transcription / mRNA decay within a population of cells, within a tissue or between different tissues and are there changes in response to external triggers? Finally, one very important question in the mRNA decay field has been whether there are specific loci of mRNA decay. Cytoplasmic P-bodies for example are enriched in mRNA decay proteins (38). It still remains unclear however whether they actually function as decay centres (3,4,39). Affymetrix FISH can be easily combined with antibody staining (40–42). For example, the absence of hepatitis C virus mRNA from P-bodies was shown by combining Affymetrix FISH staining with P-body staining via an Xrn1 antibody in mammalian cells (41).
Where are the limitations of the method? With only one single reporter mRNA, the question arises as to how representative this mRNA is for reporting mRNA metabolism. Different types of mRNAs for example are degraded via different decay pathways. This problem can be partially overcome by using more than one reporter mRNA. More important is the inclusion of control experiments. The exact design of these will depend on what is known about mRNA decay and transcription in the respective organism/tissue, and on the exact question that the assay aims to address. For example, in an organism that is known to utilise both 5΄-3΄ and 3΄-5΄ cytoplasmic mRNA decay, the sensitivity of the reporter to both pathways could be tested by Xrn1 and exosome depletion, if the aim is to distinguish between the pathways.
Another problem is that 3΄-5΄ decay intermediates cannot be easily distinguished from transcription intermediates, as both will result in red mRNA molecules. One way to distinguish is to determine the subcellular localization of the red spots: cytoplasmic red spots are likely 3΄-5΄ decay intermediates, while nuclear ones can be either transcription intermediates or decay intermediates of the nuclear exosome. The later could be distinguished by selective depletion of components the nuclear exosome (if known) or, as done here, by testing the sensitivity to transcriptional inhibitor actinomycin D.
The method is also not suitable to determine global transcription and decay parameters. In this work, I have estimated the average 5΄-3΄ mRNA decay rate for the reporter Tb427.01.1740 to be about 6 nucleotides per second, based on the disappearance of the green spots after transcriptional inhibition by actinomycin D. In addition, the finding that there are less red than green spots allows the conclusion that transcription is faster than mRNA decay. However, RNA polymerase II elongation is not processive but appears to be a mixture of very high speed (4.3 kb/min) and very long pausing (6). Similarly, decay rates are highly dependent on the individual mRNAs; strong secondary structures for example decrease decay rates (10–14). Thus, the parameters determined with one single reporter mRNA are not representative for the organism.
The assay was established in trypanosomes, protists with mostly very short-lived mRNAs. How easily could it be transferred to other eukaryotes? The key is the identification of a suitable reporter mRNA. Such a reporter mRNA would ideally have (i) a very large size to increase the total time spent in transcription and degradation, (ii) a short half-life to increase mRNA turn-over time, (iii) a unique nucleotide sequence at both the 5΄ and 3΄ ends for probe design. Also important is a suitable steady-state expression level - if the expression level is too low, a statistical analysis will require too many cells; if the expression level is too high, mRNA spots may overlap and be difficult to count and to assign to the different colours. In trypanosomes, the finding of a reporter was simply done by looking at the four mRNAs with the longest open reading frames. Of these, one mRNA was excluded because genome annotation was inconsistent between different trypanosome strains, another (GB4) had no unique sequence at its 3΄ end and also a slightly too high expression level. The remaining two mRNAs instantly proved suitable reporters.
Yeast and many protists have mRNA half-lives in the same short range as trypanosomes: S. cerevisiae has mean mRNA half lives of between 11 and 32 min (43–47) and Plasmodium falciparum mRNAs have average half-lives of between 9.5 and 65 min depending on the life-cycle stage (48). The adaptation of the assay should therefore be straightforward. Transcripts of higher eukaryotes tend to be more stable: mammalian mRNAs have half-lives between 7 and 10 h (8,9). However, with RNA sequencing technology advancing, global data on mRNA decay have/will become available for many organisms and can be explored to identify short-lived transcripts of large size. The fact that in most organisms the length of the transcript negatively correlates with its stability (18,43,49) increases the likelihood that a long mRNA will be short-lived. As a proof of principle, I have tested the system in mouse fibroblast cells. I had chosen a candidate for testing based on size and half-life using data of (37), and this mRNA instantly proved a suitable reporter. It actually had a half-life of 9.23 h, indicating that a short half-life of the reporter may be less important than transcript size and a suitable copy-number.
The novel assay presented here is easy to apply, as it requires nothing else but the identification of an endogenous short lived mRNA of large size. It can be performed rapidly, and does not need any special technology other than a standard fluorescence microscope. It is cheaper than global approaches that rely on RNA sequencing. It adds another tool to the kit that is currently available to study mRNA metabolism.
Supplementary Material
ACKNOWLEDGEMENTS
The author would like to thank Mark Carrington (University of Cambridge, UK) for valuable discussions and together with Sophie Piper (then University of Cambridge, UK) for providing the XRNA plasmid. Keith Gull (University of Oxford, UK) is acknowledged for providing the Trypanosoma brucei Lister 427 procyclic cells. The author is very grateful to Markus Engstler for providing lab space, access to infrastructure and support. The comments of Brooke Morriswood and Nicolai Siegel (both University of Würzburg) significantly improved the manuscript. Silke Braune is acknowledged for essential help with culturing NIH3T3 cells.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft [Kr4017/1-2]. Funding for open access charge: Deutsche Forschungsgemeinschaft [Kr4017/1-2].
Conflict of interest statement. None declared.
REFERENCES
- 1. Beelman C.A., Parker R.. Differential effects of translational inhibition in cis and in trans on the decay of the unstable yeast MFA2 mRNA. J. Biol. Chem. 1994; 269:9687–9692. [PubMed] [Google Scholar]
- 2. Parker R. RNA degradation in Saccharomyces cerevisae. Genetics. 2012; 191:671–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pelechano V., Wei W., Steinmetz L.M.. Widespread Co-translational RNA decay reveals ribosome dynamics. Cell. 2015; 161:1400–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hu W., Sweet T.J., Chamnongpol S., Baker K.E., Coller J.. Co-translational mRNA decay in saccharomyces cerevisiae. Nature. 2009; 461:225–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kramer S., Queiroz R., Ellis L., Webb H., Hoheisel J.D., Clayton C.E., Carrington M.. Heat shock causes a decrease in polysomes and the appearance of stress granules in trypanosomes independently of eIF2(alpha) phosphorylation at Thr169. J. Cell Sci. 2008; 121:3002–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Darzacq X., Shav-Tal Y., de Turris V., Brody Y., Shenoy S.M., Phair R.D., Singer R.H.. In vivo dynamics of RNA polymerase II transcription. Nat. Struct. Mol. Biol. 2007; 14:796–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kimura H., Sugaya K., Cook P.R.. The transcription cycle of RNA polymerase II in living cells. J. Cell Biol. 2002; 159:777–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sharova L.V., Sharov A.A., Nedorezov T., Piao Y., Shaik N., Ko M.S.H.. Database for mRNA Half-Life of 19 977 genes obtained by DNA microarray Analysis of pluripotent and differentiating mouse embryonic stem Cells. DNA Res. 2009; 16:45–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang E., van Nimwegen E., Zavolan M., Rajewsky N., Schroeder M., Magnasco M., Darnell J.E.. Decay rates of human mRNAs: correlation with functional characteristics and sequence attributes. Genome Res. 2003; 13:1863–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vreken P., Raué H.A.. The rate-limiting step in yeast PGK1 mRNA degradation is an endonucleolytic cleavage in the 3΄-terminal part of the coding region. Mol. Cell. Biol. 1992; 12:2986–2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stevens A. 5΄-exoribonuclease 1: Xrn1. Methods Enzymol. 2001; 342:251–259. [DOI] [PubMed] [Google Scholar]
- 12. Muhlrad D., Decker C.J., Parker R.. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5΄–>3΄ digestion of the transcript. Genes Dev. 1994; 8:855–866. [DOI] [PubMed] [Google Scholar]
- 13. Decker C.J., Parker R.. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 1993; 7:1632–1643. [DOI] [PubMed] [Google Scholar]
- 14. Poole T.L., Stevens A.. Structural modifications of RNA influence the 5΄ exoribonucleolytic hydrolysis by XRN1 and HKE1 of Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 1997; 235:799–805. [DOI] [PubMed] [Google Scholar]
- 15. Harigaya Y., Parker R.. Global analysis of mRNA decay intermediates in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:11764–11769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kern D., Collins M., Fultz T., Detmer J., Hamren S., Peterkin J.J., Sheridan P., Urdea M., White R., Yeghiazarian T. et al. An enhanced-sensitivity branched-DNA assay for quantification of human immunodeficiency virus type 1 RNA in plasma. J. Clin. Microbiol. 1996; 34:3196–3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Collins M.L., Irvine B., Tyner D., Fine E., Zayati C., Chang C., Horn T., Ahle D., Detmer J., Shen L.P. et al. A branched DNA signal amplification assay for quantification of nucleic acid targets below 100 molecules/ml. Nucleic Acids Res. 1997; 25:2979–2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fadda A., Ryten M., Droll D., Rojas F., Färber V., Haanstra J.R., Merce C., Bakker B.M., Matthews K., Clayton C.E.. Transcriptome-wide analysis of trypanosome mRNA decay reveals complex degradation kinetics and suggests a role for co-transcriptional degradation in determining mRNA levels. Mol. Microbiol. 2014; 94:307–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Berriman M., Ghedin E., Hertz-Fowler C., Blandin G., Renauld H., Bartholomeu D.C., Lennard N.J., Caler E., Hamlin N.E., Haas B. et al. The genome of the African trypanosome Trypanosoma brucei. Science. 2005; 309:416–422. [DOI] [PubMed] [Google Scholar]
- 20. Mair G., Shi H., Li H., Djikeng A., Aviles H.O., Bishop J.R., Falcone F.H., Gavrilescu C., Montgomery J.L., Santori M.I. et al. A new twist in trypanosome RNA metabolism: cis-splicing of pre-mRNA. RNA. 2000; 6:163–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Siegel T.N., Hekstra D.R., Wang X., Dewell S., Cross G.A.M.. Genome-wide analysis of mRNA abundance in two life-cycle stages of Trypanosoma brucei and identification of splicing and polyadenylation sites. Nucleic Acids Res. 2010; 38:4946–4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fadda A., Färber V., Droll D., Clayton C.E.. The roles of 3΄-exoribonucleases and the exosome in trypanosome mRNA degradation. RNA. 2013; 19:937–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Manful T., Fadda A., Clayton C.E.. The role of the 5΄-3΄ exoribonuclease XRNA in transcriptome-wide mRNA degradation. RNA. 2011; 17:2039–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kramer S., Piper S., Estevez A.M., Carrington M.. Polycistronic trypanosome mRNAs are a target for the exosome. Mol. Biochem. Parasitol. 2016; 205:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sunter J., Wickstead B., Gull K., Carrington M.. A new generation of T7 RNA polymerase-independent inducible expression plasmids for Trypanosoma brucei. PLoS One. 2012; 7:e35167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Brun R., Schönenberger M.. Cultivation and in vitro cloning or procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Short communication. Acta Trop. 1979; 36:289–292. [PubMed] [Google Scholar]
- 27. McCulloch R., Vassella E., Burton P., Boshart M., Barry J.D.. Transformation of monomorphic and pleomorphic Trypanosoma brucei. Methods Mol. Biol. 2004; 262:53–86. [DOI] [PubMed] [Google Scholar]
- 28. Rindisbacher L., Hemphill A., Seebeck T.. A repetitive protein from Trypanosoma brucei which caps the microtubules at the posterior end of the cytoskeleton. Mol. Biochem. Parasitol. 1993; 58:83–96. [DOI] [PubMed] [Google Scholar]
- 29. Kolev N.G., Franklin J.B., Carmi S., Shi H., Michaeli S., Tschudi C.. The transcriptome of the human pathogen Trypanosoma brucei at single-nucleotide resolution. PLoS Pathog. 2010; 6:e1001090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Clayton C.E., Shapira M.. Post-transcriptional regulation of gene expression in trypanosomes and leishmanias. Mol. Biochem. Parasitol. 2007; 156:93–101. [DOI] [PubMed] [Google Scholar]
- 31. Obrig T.G., Culp W.J., McKeehan W.L., Hardesty B.. The mechanism by which cycloheximide and related glutarimide antibiotics inhibit peptide synthesis on reticulocyte ribosomes. J. Biol. Chem. 1971; 246:174–181. [PubMed] [Google Scholar]
- 32. Schneider-Poetsch T., Ju J., Eyler D.E., Dang Y., Bhat S., Merrick W.C., Green R., Shen B., Liu J.O.. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat. Chem. Biol. 2010; 6:209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Webb H., Burns R., Ellis L., Kimblin N., Carrington M.. Developmentally regulated instability of the GPI-PLC mRNA is dependent on a short-lived protein factor. Nucleic Acids Res. 2005; 33:1503–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Holetz F.B., Correa A., Avila A.R., Nakamura C.V., Krieger M.A., Goldenberg S.. Evidence of P-body-like structures in Trypanosoma cruzi. Biochem. Biophys. Res. Commun. 2007; 356:1062–1067. [DOI] [PubMed] [Google Scholar]
- 35. Pestka S. Inhibitors of ribosome functions. Annu. Rev. Microbiol. 1971; 25:487–562. [DOI] [PubMed] [Google Scholar]
- 36. Li C.-H., Irmer H., Gudjonsdottir-Planck D., Freese S., Salm H., Haile S., Estevez A.M., Clayton C.E.. Roles of a Trypanosoma brucei 5΄->3΄ exoribonuclease homolog in mRNA degradation. RNA. 2006; 12:2171–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M.. Global quantification of mammalian gene expression control. Nature. 2011; 473:337–342. [DOI] [PubMed] [Google Scholar]
- 38. Sheth U., Parker R.. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003; 300:805–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Eulalio A., Behm-Ansmant I., Schweizer D., Izaurralde E.. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol. Cell. Biol. 2007; 27:3970–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Garaigorta U., Heim M.H., Boyd B., Wieland S., Chisari F.V.. Hepatitis C Virus (HCV) induces formation of stress granules whose proteins Regulate HCV RNA replication and virus assembly and egress. J. Virol. 2012; 86:11043–11056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li Y., Masaki T., Yamane D., McGivern D.R., Lemon S.M.. Competing and noncompeting activities of miR-122 and the 5΄ exonuclease Xrn1 in regulation of hepatitis C virus replication. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:1881–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Solga A.C., Gianino S.M., Gutmann D.H.. NG2-cells are not the cell of origin for murine neurofibromatosis-1 (Nf1) optic glioma. Oncogene. 2013; 33:289–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Geisberg J.V., Moqtaderi Z., Fan X., Ozsolak F., Struhl K.. Global analysis of mRNA isoform half-lives reveals stabilizing and destabilizing elements in yeast. Cell. 2014; 156:812–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Miller C., Schwalb B., Maier K., Schulz D., Dümcke S., Zacher B., Mayer A., Sydow J., Marcinowski L., Dölken L. et al. Dynamic transcriptome analysis measures rates of mRNA synthesis and decay in yeast. Mol. Syst. Biol. 2011; 7:458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Holstege F.C., Jennings E.G., Wyrick J.J., Lee T.I., Hengartner C.J., Green M.R., Golub T.R., Lander E.S., Young R.A.. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998; 95:717–728. [DOI] [PubMed] [Google Scholar]
- 46. Wang Y., Liu C.L., Storey J.D., Tibshirani R.J., Herschlag D., Brown P.O.. Precision and functional specificity in mRNA decay. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:5860–5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Grigull J., Mnaimneh S., Pootoolal J., Robinson M.D., Hughes T.R.. Genome-wide analysis of mRNA stability using transcription inhibitors and microarrays reveals posttranscriptional control of ribosome biogenesis factors. Mol. Cell. Biol. 2004; 24:5534–5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shock J.L., Fischer K.F., Derisi J.L.. Whole-genome analysis of mRNA decay in Plasmodium falciparum reveals a global lengthening of mRNA half-life during the intra-erythrocytic development cycle. Genome Biol. 2007; 8:R134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Feng L., Niu D.-K.. Relationship between mRNA stability and length: an old question with a new twist. Biochem. Genet. 2007; 45:131–137. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.