Abstract
Recent evidence indicates that shRNAs with a relatively short basepaired stem do not require Dicer processing, but instead are processed by the Argonaute 2 protein (Ago2). We named these molecules AgoshRNAs as both their processing and silencing function are mediated by Ago2. This alternative processing yields only a single RNA guide strand, which can avoid off-target effects induced by the passenger strand of regular shRNAs. It is important to understand this alternative processing route in mechanistic detail such that one can design improved RNA reagents. We verified that AgoshRNAs trigger site-specific cleavage of a complementary mRNA. Second, we document the importance of the identity of the 5΄-terminal nucleotide and its basepairing status for AgoshRNA activity. AgoshRNA activity is significantly reduced or even abrogated with C or U at the 5΄-terminal and is enhanced by introduction of a bottom mismatch and 5΄-terminal nucleotide A or G. The 5΄-terminal RNA nucleotide also represents the +1 position of the transcriptional promoter in the DNA, thus further complicating the analysis. Indeed, we report that +1 modification affects the transcriptional efficiency and accuracy of start site selection, with A or G as optimal nucleotide. These combined results allow us to propose general rules for the design and expression of potent AgoshRNA molecules.
INTRODUCTION
RNA interference (RNAi) is a post-transcriptional gene silencing mechanism that is widely conserved among vertebrates and invertebrates and that uses microRNAs (miRNAs) to control gene expression (1,2). Cellular miRNAs are typically processed by the Drosha endonuclease in the nucleus and subsequently by Dicer in the cytoplasm. This canonical route yields the mature miRNA duplex of 20–24 base pairs (bp), of which one strand is preferentially loaded into an Ago protein-containing complex to form the RNA-induced silencing complex (RISC). The miRNA-loaded RISC complex targets partially complementary messenger RNA (mRNA) transcripts for degradation and/or translational repression. Thermodynamic properties determine which strand of the miRNA duplex will be selected as guide (3,4). The passenger strand is cleaved and degraded. The RNAi pathway can also be triggered by artificial short hairpin RNAs (shRNAs) that are processed by Dicer into small interfering RNAs (siRNAs) that subsequently program RISC (5–7). Although Dicer is essential for the processing of the majority of miRNAs, some exceptions have recently been reported. Notably, miR-451 with a short 17 bp stem and 4 nucleotide (nt) loop does not require Dicer for processing (8). It was suggested that the pre-miR-451 duplex is processed instead by Ago2 with cleavage between bp 10 and 11 at the 3΄ side of the stem generating an extended ∼30 nt miRNA (9–11). Subsequent 3΄ processing by poly(A)-specific ribonuclease (PARN) creates the ∼22–26 nt mature miR-451, but this modification appears redundant for miR-451 activity (12).
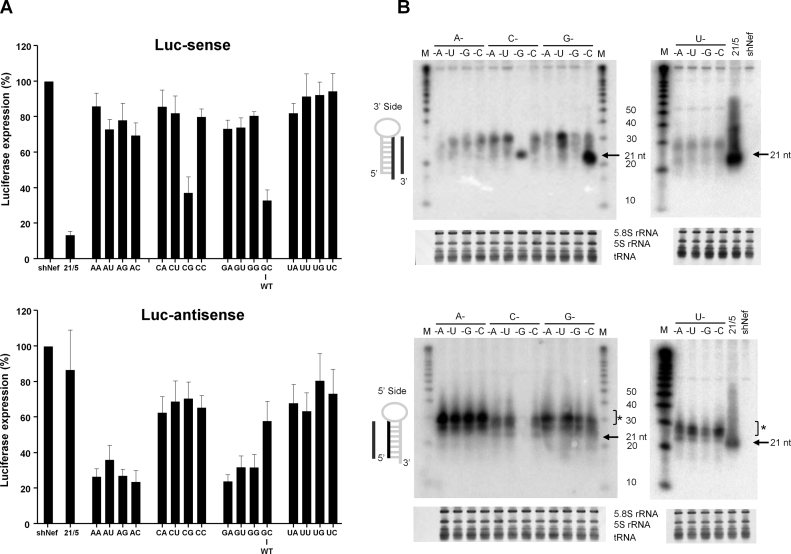
More recently, Dicer-independent shRNAs have also been described (13–15). We proposed a hairpin design with a short stem and small loop that triggered this alternative pathway. We named these molecules AgoshRNAs because Ago2 is involved both in the processing and subsequent silencing reaction (15). Figure 1A depicts the two possible processing routes for a shRNA substrate. Regular shRNAs are processed by Dicer, but hairpins of 18 bp or shorter are too small for Dicer recognition and shift to Ago2-processing. Regular Dicer-mediated cleavage removes the loop and generates a duplex siRNA consisting of two candidate active strands of ∼21 nt, marked as black and white arrows for the 3΄ and 5΄ strand, respectively (Figure 1A, top). Asymmetry in the thermodynamic stability of the ends of the siRNA duplex determines which strand is selected as guide (3,4). The activity of the 3΄ and 5΄ strand can be scored by silencing of the Luc-sense and Luc-antisense reporter, respectively (Figure 1B), but it thus far remains unknown whether the mRNA reporter is inactivated by Ago2-mediated cleavage when a fully complementary AgoshRNA inhibitor is used.
Figure 1.
Schematic of canonical (Dicer dependent) and non-canonical (Ago2 dependent) shRNA processing routes. (A) Secondary structure of short hairpin RNA (shRNA). In the canonical pathway the stem of the shRNA is cleaved by Dicer into an siRNA duplex of ∼21 bp with 3΄ overhangs that is loaded into RISC. The passenger strand (the 5΄ strand, white arrow) is cleaved and degraded, the 3΄ strand acts as guide (black arrow) in RNAi silencing. In the alternative AgoshRNA route, the shRNA is recognized directly by Ago2 such that it is cleaved on the 3΄ side between bp 10 and 11 into an RNA molecule of ∼33 nt (gray arrow), which subsequently instructs Ago2 for RNAi-silencing. The predicted Dicer and Ago2 cleavage sites are marked with black and grey arrows, respectively. (B) Luc reporter constructs with sense and antisense sequences. The Luc-sense reporter scores canonical shRNA 3΄ strand activity, the Luc-antisense reporter scores both shRNA 5΄ strand and AgoshRNA activity. (C) mRNA cleavage by AgoshRNA-Gag4 and shRNA-Gag4 was analyzed by 3΄ RACE. The expected cleavage site (position 11/10) is marked as black triangle in the mRNA. The obtained fragments were aligned with the mRNA; we only show the 3΄ end nt, and the post-cleavage addition of nt (mostly U, occasionally A or C) is marked by underlining. The 3΄ RACE reads were grouped per actual cleavage site, and ranked according to size. (D) Design of AgoshRNAs with variation in the bottom bp. AgoshRT5 and AgoshPol47 with 19 bp stem and 5 nt loop were used as wild-type backbones. The bottom bp (encircled) was mutated into all 16 dinucleotide combinations (insert below).
Alternative Dicer-independent processing is mediated by the Ago2 nuclease that cleaves halfway the 3΄ side of the hairpin as determined by deep sequencing (16). An extended RNA molecule of ∼33 nt (gray arrow) is created (Figure 1A, bottom) that can anneal exclusively to the Luc-antisense reporter (Figure 1B). Ago2 co-immunoprecipitation revealed that these alternatively processed AgoshRNA molecules are indeed incorporated into RISC (15). A detailed mutational analysis indicated that the length of the basepaired stem is a major determinant for shRNA activity via the regular Dicer route versus AgoshRNA activity via the non-canonical Ago2 route (15,17). However, other sequence or structural features may also influence the activity of AgoshRNA molecules.
There is accumulating evidence that the AgoshRNA pathway mimics miR-451 biogenesis (18). Interestingly, miR-451 is special among the miRNAs with an A-nucleotide at the 5΄-terminal instead of the more common U. In contrast to regular shRNAs, miR-451 has a mismatch (AC) at the bottom of the hairpin stem. A recent study indicated that the creation of a bp at the bottom of the pre-mir-451 stem substantially impaired miR-451 processing (19). Inspired by these miR-451 findings, we recently tested AgoshRNA molecules with a mismatch AC or UC at the bottom of the stem (20). Much to our surprise, the AC variants showed a substantially enhanced silencing activity, whereas the UC variants showed minimal silencing activity. In this study, we performed a systematic analysis of the AgoshRNA bottom bp to identify the optimal 5΄-terminal nt and the opposing 3΄ nt (Figure 1D). In addition, we attempted to separate RNA 5΄-end effects from DNA effects at the corresponding +1 position of the transcriptional promoter. We evaluated the generality of the optimized AgoshRNA design for a second unrelated AgoshRNA molecule and in different cell types.
MATERIALS AND METHODS
DNA constructs and synthetic AgoshRNA molecules
DNA constructs used in this study were made by annealing complementary oligonucleotides (containing BamHI and HindIII sites) and inserting them into the BglII and HindIII sites of the pSUPER vector, as previously described (5,21,22). The RNA secondary structure of the shRNA transcript was predicted by the Mfold web server (23). Firefly luciferase reporter plasmids were constructed by insertion of a 50–70 nt HIV-1 sequence, with the target region in the center, into the EcoRI and PstI sites of the pGL3 plasmid (24). The luciferase reporters with the sense and antisense target sequences were previously described (15). H1-Luc constructs with the H1 promoter and +1 nt variation (N: A, G, C and U) were constructed in two cloning steps. First, annealed complementary oligonucleotides followed by an extra NcoI site and target sequence for the northern blot probe (5΄-GATCCCCNCCATGGAAGTGAAGGGGCAGTAGTAATATACC–GGTGATATCTTTTTGGAAA-3΄) were inserted in between the BamHI and HindIII sites of the pSuper vector, resulting in four H1-N44 variants (Figure 5A). Second, the H1-N44 variants were digested with SacI and NcoI and the H1 promoter cassette was inserted in between the SacI and NcoI sites of the pGL3 control plasmid (Promega, Madison, WI, USA), resulting in H1-Luc variants (Figure 4A). The U6 promoter was PCR amplified from pSilencer 2.0-U6 (Ambion, Austin, TX. USA). Mutations around the +1 position were introduced by specific reverse primers. The N44 constructs were created by inserting the annealing complementary oligonucleotides into the corresponding enzyme sites of the vector as previously described.
Figure 5.
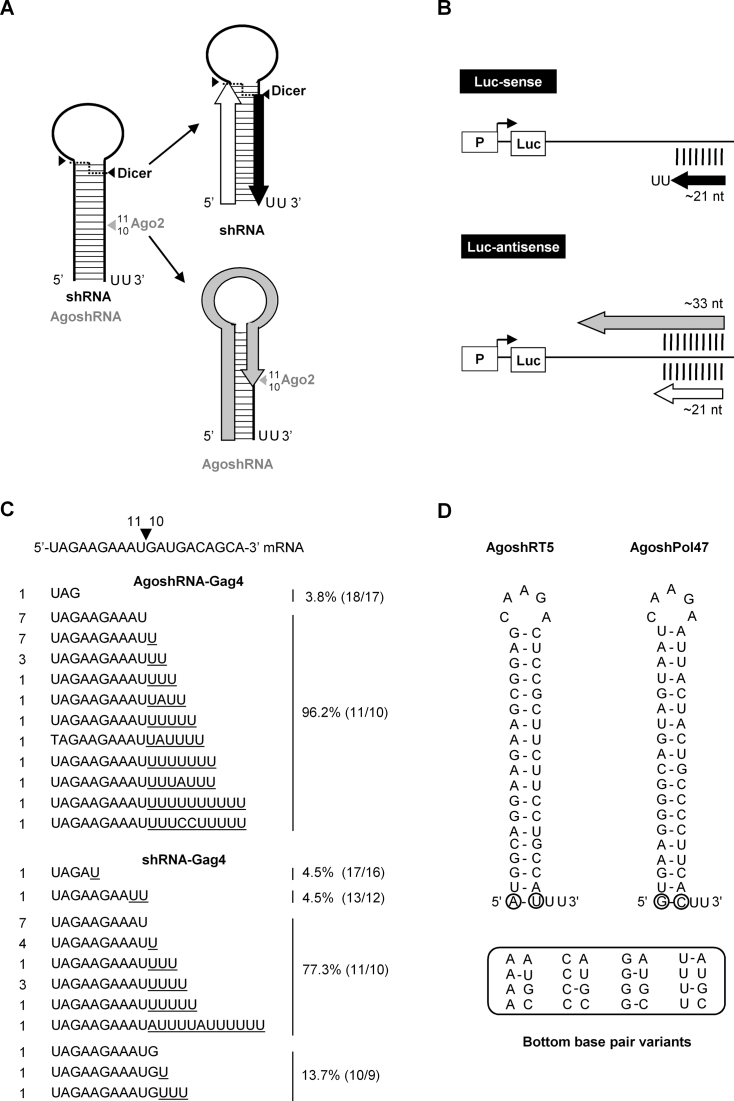
H1 and U6 transcription efficiency of short transcripts with +1 variation. (A and D) Design of short constructs with variation at the +1 position (Human H1-N44 constructs and human U6-N44 constructs, respectively). (B and E) Total RNA was analyzed by northern blot. Size markers are indicated. AgoshPol47 and shNef were included as controls. Ethidium bromide staining of small rRNAs and tRNAs are shown as loading controls below the blot. (C and F) The RNA products were quantitated using ImageQuant. The mean values are based on three independent northern blots.
Figure 4.
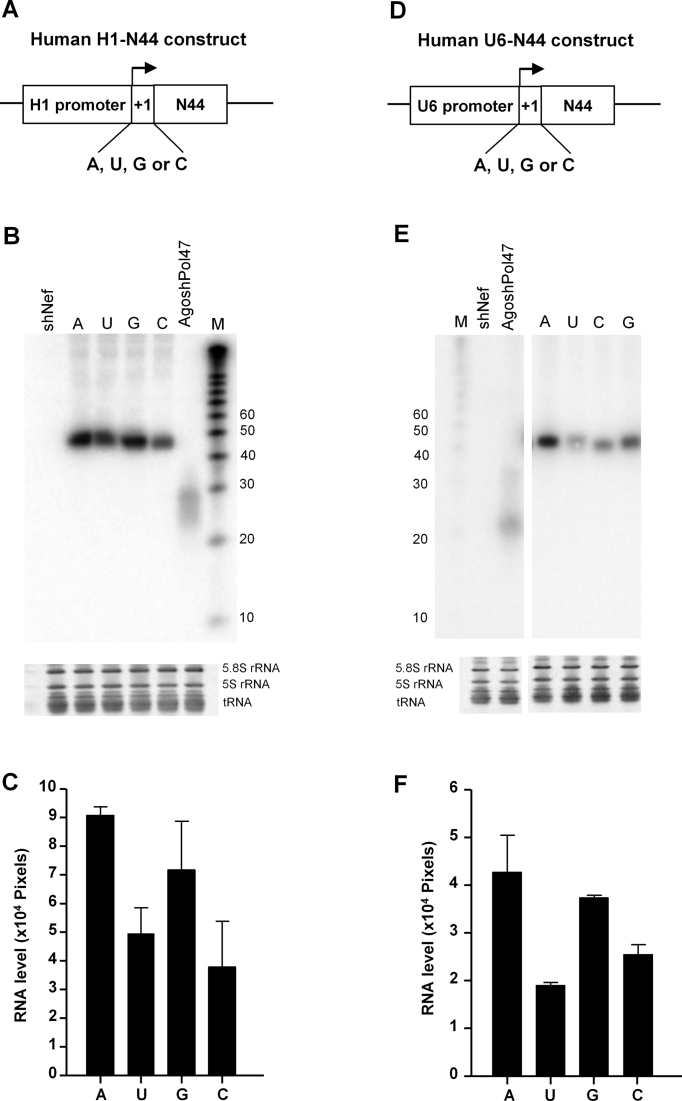
Gene expression by H1-Luc +1 variants. (A) Design of human H1-Luc constructs with variation at the +1 position. (B) Luciferase activity of the different H1-Luc variants was measured. HEK293T cells were co-transfected with 1, 5 and 25 ng of H1-Luc construct and 1 ng of pRL. pBluescript SK- (pBS) was used as negative control. The ratio between firefly and renilla luciferase activity was used for normalization of experimental variations such as differences in transfection efficiencies. We performed three independent transfections, each in duplicate, and standard deviations were calculated and shown as error bar.
Cell culture and transfection
Human embryonic kidney (HEK) 293T, Vero, HCT116 and C33A cells were grown as monolayer in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 10% fetal calf serum (FCS) (Hybond), penicillin (100 U/ml), streptomycin (100 μg/ml) and minimal essential medium non-essential amino acids (DMEM/10% FCS) at 37°C and 5% CO2. For luciferase assays, all cell lines were plated one day before transfection in 24-well plates at a density of 1.4×105 cells per well in 0.5 ml DMEM/10%FCS without antibiotics. Cells were transfected with 100 ng of the firefly luciferase expression plasmid, 1 ng of Renilla luciferase expression plasmid (pRL) and 1, 5 or 25 ng of RNA vector or differing amount of synthetic AgoshRNA (1 μl of 64 pM, 0.32 nM, 8 nM, 40 nM, 200 nM or 1 μM) using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. pBluescript SK- (pBS) (Promega) was added to the transfection mixtures to obtain equal DNA concentration. Cells were lysed 48 h post transfection to measure firefly and renilla luciferase activities using the Dual Luciferase Reporter Assay System (Promega). An unrelated shRNA (shNef) or synthetic AgoshRNA (synthetic AgoshGag4) served as negative control, which was set at 100% luciferase expression. We performed three independent transfections, each in duplicate. The ratio between firefly and renilla luciferase activity was used for normalization of experimental variations such as differences in transfection efficiencies. The luciferase data were subsequently corrected for between session variation as described previously (25). The resulting six values were used to calculate the standard deviation. To estimate the inhibitory concentration (IC50) of synthetic AgoshRNAs we used nonlinear regression models, assuming a symmetrical sigmoidal four-parameters curve for the relationship (GraphPad Prism software). The response was used in the log form (log10(dose)) (26).
Probing the mRNA cleavage site by 3΄ RACE
3΄ RACE was performed using the Life Technologies RACE system according to the manufacturer's instructions. Briefly, 5 μg of Luc reporter construct and 1 μg of AgoshRNA or shRNA construct were co-transfected into HEK293T cells. Total cellular RNA was isolated 48 h post-transfection. Total RNA (500 ng) was ligated to an adenylated 3΄ adapter [5΄-(rApp)-GGAACCATCAATATCTCGTATGCCGTCTTCTGCTTG-(3ddC)-3΄] by a truncated T4 RNA Ligase 2 (New England Biolabs) according to the manufacturer's protocol. The RNA-3΄ adapter product was reversed-transcribed using ThermoScript™ RT-PCR System (Invitrogen) with the specific primer 5΄-CAAGCAGAAGACGGCATACG-3΄. PCR amplification was performed using specific primers (5΄ primer 5΄-AGGTCTTACCGGAAAACTCG-3΄, 3΄ primer 5΄-CAAGCAGAAGACGGCATACG-3΄). The PCR products were run on a 1.5% agarose gel and products of the expected size were extracted from the gel. Purified DNA was cloned into the pCR2.1 TOPO vector (Invitrogen) by TA cloning. After transformation, colony PCR was performed with T7 and M13R primers. The inserts were subjected to sequencing using the BigDye Terminator v1.1 Cycle Sequencing Kit (ABI). The mRNA cleavage sites were determined by aligning the 3΄ RACE reads with the mRNA sequence.
Ago2 immunoprecipitation, RNA isolation and library preparation
At 48 h post-transfection, the cells were washed several times with cold phosphate-buffered saline and Ago2-FLAG was immunoprecipitated as previously described (27). In short, the cells were incubated with IsoB-NP40 (10 mmol/l Tris–HCl pH 7.9, 150 mmol/l NaCl, 1.5 mmol/l MgCl2, 1% NP40) for 20 min on ice. The cell-lysates were centrifuged at 12 000 × g for 10 min at 4°C to clear cell debris. The supernatant was incubated with 75 μl anti-FLAG M2 Agarose bead-suspension (Sigma, St Louis, MO, USA) with constant rotation for 16 h at 4°C. The supernatant (depleted fraction) was separated from the beads (enriched fraction). The beads were washed three times with NET-1 buffer (50 mmol/l Tris–HCl pH 7.5, 150 mmol/l NaCl, 2.5% Tween-20) and resuspended in IsoB-NP40. RNA was isolated by phenol–chloroform extraction followed by DNAse treatment using the TURBO DNA-free kit (Life technologies). The isolated RNA was size separated on a 15% denaturing polyacrylamide gel electrophoresis gel next to a size marker (gene ruler ultra low range DNA ladder; Thermo Scientific, Waltham, MA, USA) for size estimation. The 15–55 nt RNA fragments were purified from gel using a spin column (Ambion, Carlsbad, CA, USA). The quality and percentage of miRNA was assayed on a Bioanalyzer 2100 (Agilent, Santa Clara, CA, USA) using a small RNA chip. The SOLiD Small RNA Library Preparation protocol (Applied Biosystems, Carlsbad, CA, USA) was used to prepare an RNA library that was subsequently analyzed on the SOLiD Wildfire system (Applied Biosystems). Analysis of the SOLiD color space reads was performed with LifeScope Genomic Analysis Software version 2.5 (Applied Biosystems) using the small RNA pipeline.
siRNA detection by northern blotting
Northern blot experiments were performed as previously described (15,28). Briefly, HEK293T cells were transfected with 5 μg shRNA constructs using Lipofectamine 2000. Total cellular RNA was isolated after 48 h with the mirVana miRNA isolation kit (Ambion) according to the manufacturer's protocol. The RNA concentration was measured using the Nanodrop 1000 (Thermo Fisher Scientific). Isolated RNA was analysed by denaturing 15% polyacrylamide gel electrophoresis (precast Novex TBU gel, Invitrogen) using a 32P-labeled Decade Marker (Ambion) for size estimation. To check for equal sample loading, the gel was stained with 2 μg/ml ethidium bromide for 20 min. The ribosomal RNA (5.8S and 5S rRNA) and tRNA bands were visualized with UV light. The RNA was electro-transferred to a positively charged nylon membrane (Boehringer Mannheim, GmbH, Mannheim, Germany) and cross-linked to the membrane using UV (254 nm, 0.12 J). LNA oligonucleotide probes were 5΄ end labeled with the kinaseMax kit (Ambion) in the presence of 1 μl of [γ-32P]ATP (0.37 MBq/μl Perkin Elmer). We used the following oligonucleotides probes (LNA positions underlined) to detect the 5΄ and 3΄ strand of the siRNA, respectively 5΄-CTCCGCTTCTTCCTGCCAT-3΄ and 5΄-ATGGCAGGAAGAAGCGGAG-3΄ for AgoshRT5 and 5΄-ATTACTACTGCCCCTTCAC-3΄ and 5΄-GTGAAGGGGCAGTAGTAAT-3΄ for AgoshPol47. The 5΄-ATTACTACTGCCCCTTCAC-3΄ was also used to detect the H1-N44 and the U6-N44 variants (Figure 5A). To remove unincorporated nt, the probes were purified on Sephadex G-25 spin columns (Amersham Biosciences). The blot was incubated in 10 ml ULTRAhyb hybridization buffer (Ambion) at 42°C for 30 min. After addition of the labeled LNA oligonucleotide, hybridization was performed at 42°C for 16 h. The blot was washed twice for 5 min at 42°C in 2× SSC/0.1% SDS and twice for 15 min at 42°C in 0.1× SSC/0.1% SDS and subsequently analysed using a PhosphorImager (Amersham Biosciences) and the ImageQuant (v5.1) software package. For each experiment, at least two independent northern blots were performed.
Detection of the transcript 5΄-end by 5΄ RACE
HEK293T cells were transfected with 5 μg of RNA constructs using Lipofectamine 2000. Total cellular RNA was isolated after 48 h with the mirVana miRNA isolation kit (Ambion) according to the manufacturer΄s protocol. The RNA concentration was measured using the Nanodrop 1000 (Thermo Fisher Scientific). 5΄ RACE analysis was performed on 2 μg of total RNA. The RNA was directly ligated to the 5΄ adaptor (5΄-CGACTGGAGCACGAGGACACTGACATGGACTGAA-GGAGTAGAAA-3΄) by T4 RNA ligase (New England Biolabs) according to the manufacturer's protocol (29). Then the 3΄ adaptor containing a domain (underlined) complementary to the 3΄ end of the RNA transcript (3΄-ATTATATGGCCACTATAGGTACAGCCGGCGGAGCCGGA-GATCTTA-5΄) was used to prime reverse transcription (ThermoScript™ RT-PCR System, Invitrogen). A primer set, one pairs with the 5΄ adaptor and the other with the 3΄ adaptor, was used to amplify the small RNA into DNA (5΄ primer 5΄-GGACACTGACATGGACTGAAGGAGTA-3΄, 3΄ primer 5΄-ATTCTAGAGGCCGAGG-CGGCCGACATG-3΄). The PCR products were gel purified and cloned into the pCR2.1 TOPO vector (Invitrogen) and individual clones were subsequently sequenced with the T7 or M13R primers.
RESULTS
AgoshRNA-mediated mRNA cleavage
We first addressed the important question whether AgoshRNA molecules designed to be perfectly complementary to a specific mRNA target do trigger cleavage of that mRNA or lead to translational suppression. To do this, we co-transfected a Luc-reporter construct with the fully complementary AgoshRNA-Gag4 construct in HEK293T cells. As a positive control, we included the shRNA-Gag4 construct that was designed to attack the same target sequence in the Luc-reporter. We probed the Luc mRNA cleavage event by 3΄ RACE. We observed site-specific mRNA cleavage for the AgoshRNA inhibitor and the shRNA control (Figure 1C). In fact, AgoshRNA-mediated cleavage seems more accurate by cleaving predominantly at the 11/10 position (96.2%), whereas the regular shRNA achieved only 77.3% cleavage at the 11/10 position with a significant 10/9 byproduct (13.7%). This result may indicate imprecise Dicer-processing of the shRNA molecule, a step that is avoided by the AgoshRNA design. Similar to miRNA-mediated mRNA cleavage, we found that the 3΄ end of both the AgoshRNA- and shRNA-cleaved mRNA is prone to U addition (occasionally A or C), which may mark the RNA for degradation (30,31).
Importance of the bottom basepair position in AgoshRT5 molecules
A systematic analysis of the optimal bottom stem composition of RNA hairpins with a 19 bp stem and a 5 nt loop (19/5) was performed. Two unrelated AgoshRNA molecules with a different sequence were selected as substrates for the current mutational analysis (Figure 1D, AgoshRT5 and AgoshPol47). We tested all 16 dinucleotide combinations at the bottom of the stem (Figure 1D, see box), representing strong bp (G-C and C-G), moderate bp (A-U and U-A), weak bp (GU and UG) and mismatches (all others). We named the AgoshRNAs accordingly, e.g. the AA variant or AG.
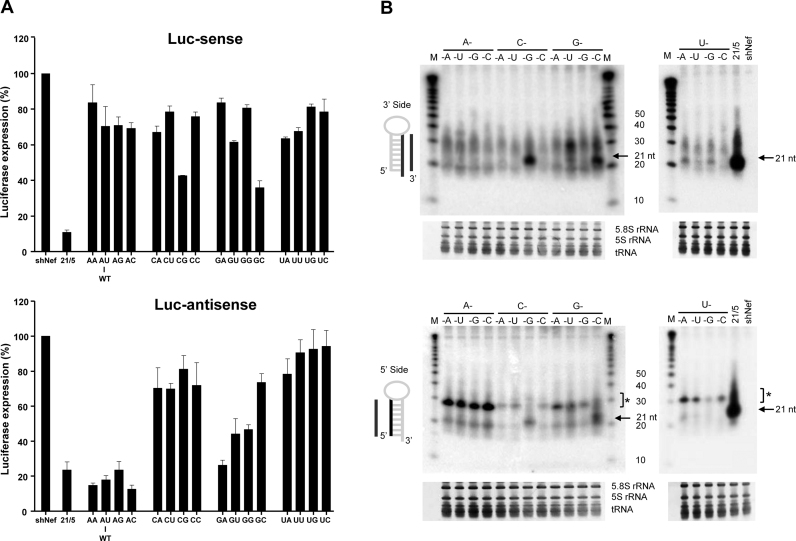
The knockdown activity mediated by the 5΄ and 3΄ strand of the hairpin was tested using two luciferase reporters with complementary target sequences. Luc-sense reporter detects 3΄ strand activity in the regular Dicer-route and Luc-antisense reporter detects 5΄ strand activity of AgoshRNAs, but also potential 5΄ strand activity in the Dicer-route (Figure 1B). To determine the knockdown activity of the AgoshRNA variants, we performed transfection series in HEK293T cells in which the AgoshRNA construct was titrated (1, 5 and 25 ng in Supplementary Figures S1 and S2). A fixed amount of renilla luciferase plasmid was used to control for the transfection efficiency. Firefly and renilla luciferase was measured two days post-transfection and the ratio was used to calculate the relative luciferase activity. An irrelevant shRNA (shNef) served as negative control for which luciferase expression was set at 100%. A clear dose-dependent knockdown activity was measured for all AgoshRNA variants on the Luc-reporters (Supplementary Figure S1 and S2). For the sake of simplicity, we will show the results obtained with an intermediate AgoshRNA concentration (5 ng, see Figure 2). We first describe results for the Dicer-route on the Luc-sense reporter and then the Ago2-route on the Luc-antisense reporter. Regular shRNA molecules with a duplex of 21 bp and a 5 nt loop (21/5) were included as controls and showed good inhibitory activity on the Luc-sense reporter, with luciferase levels dropping to <15%, while most AgoshRNA variants show little or no knockdown activity as expected (Figure 2A, upper panel) (17). The bottom bp apparently did not influence the knockdown activity on the Luc-sense reporter. Only the CG and GC variants exhibited a moderate knockdown activity. This indicates that a strong GC and CG bottom bp will cause a partial shift from Ago2 to Dicer processing, likely by stabilization of the duplex, allowing partial recognition by Dicer. Consistent with this scenario, we observed a 3.5-fold shift in the ratio of typical Ago2 versus Dicer products for the CG mutant compared to AU (WT) by deep sequencing of the Ago2 bound small RNAs (results not shown).
Figure 2.
Knockdown activity and processing of AgoshRT5 variants. (A) The knockdown activity of the different AgoshRNA variants was determined by co-transfection with a luciferase reporter containing either the sense or antisense target sequence (upper and lower panel, respectively). The AgoshRNAs varied in the bottom bp. HEK293T cells were co-transfected with 100 ng of the respective firefly luciferase reporter plasmid, 1 ng renilla luciferase plasmid as internal control and 5 ng of the corresponding AgoshRNA construct. The regular shRT5 (21/5) was used as control. An unrelated shRNA (shNef) served as negative control, this activity was set at 100% luciferase expression. We performed three independent transfections, each in duplicate, and standard deviations were calculated. (B) Processing of the 3΄ strand (upper panel) and 5΄ strand (lower panel) of the AgoshRT5 variants was analyzed by northern blot. HEK293T cells were transfected with 5 μg of the indicated constructs. Size markers were included and length indicated in nt. The regular shRT5 (21/5) was included as control. An unrelated shRNA (shNef) was included as negative control. The regular shRNA ∼21 nt products are marked and * indicates the AgoshRNA ∼30 nt products. Ethidium bromide staining of small rRNAs and tRNAs are shown as loading controls below the blot.
In contrast, the activity scored on the Luc-antisense reporter is profoundly influenced by the identity of the bottom bp/mismatch (Figure 2A, lower panel). The four CN variants, where N is any nt, showed little knockdown activity and the four UN variants were inactive. All four AN variants exhibited good silencing activity with luciferase levels dropping to ≤20%, while striking differences were noticed among the four GN variants. The GC variant exhibited very little activity on the Luc-antisense reporter, which combined with the increased activity on the Luc-sense reporter supports the idea that GC shifts from Ago2 to Dicer, such that the 3΄ strand of the hairpin is produced as guide. The GU and GG variants exhibited moderate silencing activity, while the GA variant showed a silencing activity comparable to that of the 5΄ guide strand of the Dicer-processed control shRNA 21/5. Only the AN variants showed enhanced knockdown activity when compared to the regular shRNA 21/5. AA and AC variants exhibited a subtly improved knockdown activity (85% and 87%, respectively) compared to the original AgoshRNA (AU variant or WT) with 82% knockdown activity). These trends were confirmed in the titration (Supplementary Figure S1). Apparently a bottom mismatch improves AgoshRNA activity when the 5΄ nt is A or G.
We next analyzed the AgoshRNA processing products by northern blotting using probes that detect either the 3΄ or 5΄ strand of the hairpin (Figure 2B, upper and lower panel, respectively). We included the regular shRNA shRT5 (21/5) as control that produces the ∼21 nt Dicer products from both sides of the hairpin. Consistent with the luciferase activity tests, such regular Dicer products of ∼21 nt were detected exclusively for the CG and GC variants, whereas most AgoshRNA constructs produces the predicted Ago2 product of ∼30 nt (marked with * in Figure 2B, lower panel). In fact, this same ∼30 nt product is detected with the 3΄ side probe (Figure 2B, upper panel) that can form nine bp with the processed AgoshRNA, which may explain the reduced signal. Although the expected size of these Ago2 products is approximately 33 nt, a shift in size is apparent that is consistent with these products being trimmed by the PARN enzyme after Ago2 cleavage (27). The ∼30 nt RNA products derived from the 5΄ side were quantitated and revealed striking differences among the AgoshRNAs variants (Supplementary Figure S3A). Consistent with the luciferase data, AN variants exhibited the most abundant ∼30 nt Ago2 products, the GN variants were 2-fold less abundant. Although no knockdown activity was apparent for UN variants on the Luc-antisense reporter, UN variants produced moderate amounts of ∼30 nt Ago2 products. In contrast, unpaired CN variants showed little ∼30 nt RNA fragments that yet are subtly active on the Luc-antisense reporter. Thus, the abundance of RNA variants (5΄ nt: A > U ≈ G> C) is consistent with the measured knockdown activity, except for the UN variant which is less active than expected based on the abundance of the RNA signal.
General AgoshRNA 5΄ improvements
It is important to determine whether the observed trends specify a general AgoshRNA requirement. We therefore used the unrelated AgoshPol47 to construct an identical set of 16 variants (Figure 1D). Silencing of luciferase reporters with the Pol47 target or the complementary sequence revealed similar trends to that observed for AgoshRT5 (Figure 3A). No knockdown activity was apparent for most of the AgoshRNA variants on the Luc-sense reporter, except for the CG and GC variants, which reduced luciferase levels <40%. Little to no knockdown activity was apparent on the Luc-antisense reporter for the CN and UN variants. The unpaired AN and GN variants showed good silencing activity with luciferase levels <30%. Unpaired AN variants (AA, AG and AC) exhibited more than 2-fold the knockdown activity of the WT (GC) variant, which underscores the importance of the bottom mismatch and the 5΄ A identity.
Figure 3.
Knockdown activity and processing of AgoshPol47 variants. (A) The knockdown activity of the 3΄ strand on Luc-sense (upper panel) and 5΄ strand on Luc-antisense (lower panel) of the AgoshPol47 was determined by co-transfection of a luciferase reporter encoding the sense and antisense target sequence, respectively, in HEK293T cells. The regular shPol47 (21/5) was used as control. We performed three independent transfections, each in duplicate, and standard deviations were calculated. (B) Total RNA was analyzed by northern blot for processing products derived from the 3΄ strand (upper panel) and 5΄ strand (lower panel). Size markers are indicated. The regular shRNA ∼21 nt products are marked and * indicates the AgoshRNA ∼30 nt products. See the legend to Figure 2 for more details.
We next analyzed the processing products by northern blot analysis (Figure 3B). Consistent with the luciferase knockdown data, we observed regular Dicer products of ∼21 nt for the control shRNA (21/5) and the CG and GC variants, the latter most prominently from the 3΄ side (Figure 3B, upper panel). The 5΄ side probe detected the predicted Ago2 product of ∼30 nt for most of the AgoshRNA constructs (marked with * in Figure 3B, lower panel), and again a weak signal is also detected with the 3΄ side probe (which has only 9 bp complementarity to the processed AgoshRNA) (Figure 3B, upper panel). There was a loss of the signal for CG and GC variants, consistent with the shift towards the 3΄ strand Dicer product. Again, we observed the most prominent ∼30 nt RNA fragments for the AN variants, followed by GN and UN that are equally active and finally the CN variants (Supplementary Figure S3B). To summarize, differential activity was scored for the AgoshRNA variants that are either due to differential processing (Dicer versus Ago2) or differences in the amount of RNA product that is generated, with AN variants being the most abundant and consequently the most active AgoshRNA species. Three possibilities can be envisaged: the AN variants are transcribed at a higher level, the transcripts are processed more efficiently or the transcripts are more stable upon processing. As we never observed the precursor RNA transcripts, we assume that there is no difference in processing efficiency.
The 5΄-terminal nucleotide determines the transcript abundance
We used the polymerase III H1 promoter to express the AgoshRNAs. The human H1 transcript starts with A, but there is considerable variation in +1 nt usage among mammals. A quick screen among 18 mammalian species in the NCBI database indicates that all nucleotides can be found at the +1 position (9xG, 5xA, 2xC and 2xU). To examine the relative transcription efficiency of +1 variants we generated H1-Luc reporters (Figure 4A). Reporter gene expression was measured upon titration of the transfected H1-Luc constructs (1, 5 and 25 ng). We used a fixed amount of renilla luciferase plasmid to control for the transfection efficiency. The pBluescript plasmid (pBS) mock-transfection served as negative control. Firefly and renilla luciferase was measured two days post-transfection and the ratio was determined to calculate the relative luciferase activity. We should treat these results with caution as polymerase III transcripts are uncapped and not polyadenylated, and thus not optimally suited for translation of the reporter gene. On the other hand, there are reports on polymerase II activity on the U6 and H1 polymerase III promoter (32,33). A clear dose-dependent activity was measured for all four +1 constructs (Figure 4B). High firefly expression was measured for the A construct, which may be considered the WT. Similar firefly expression was apparent for the G construct, but firefly expression was 2-fold reduced for the C and U constructs. Two possibilities can be envisaged: A and G constructs are either transcribed at a higher level or the transcripts are more stable.
To mimic the short AgoshRNA transcript, we next cloned a sequence encoding an unstructured RNA of similar length in the pSuper vector (Figure 5A, H1-N44 constructs). We varied the +1 position (A, U, G and C) and analyzed the RNA by northern blot analysis (Figure 5B). The RNA blot revealed RNA fragments of the expected size (∼44 nt) and we observed small differences in the intensity of the RNA product. We quantitated these bands and plotted the transcript levels (Figure 5C). In particular, A and G transcripts were slightly more abundant and likely more efficiently transcribed than the U and C constructs. To see if these results also apply to another polymerase III promoter, we created a similar set of N44 constructs with the frequently used U6 promoter in the pSilencer plasmid (Figure 5D). The blot shows ∼44 nt transcripts (Figure 5E). The +1 sequence variation yielded the same ranking order as observed for the H1 promoter: A and G were more abundant than the U and C transcripts (Figure 5F). We note that the A and U constructs yield slightly longer transcripts that the C and G constructs.
The 5΄-terminal nucleotide identity influences transcription accuracy
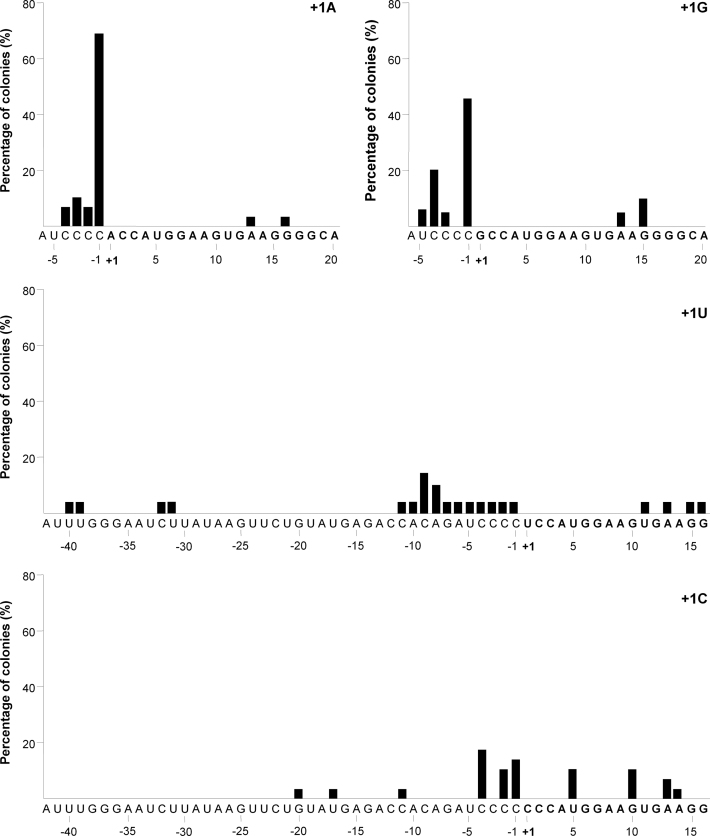
We next tested whether the +1 nt changes in the H1 promoter affect the transcription start site usage. H1-N44 constructs were transfected into HEK293T cells and total cellular RNA was isolated and tested by a designed 5΄ RACE PCR. The PCR products were TOPO-TA cloned and positive clones were sequenced. The incidence of transcription start sites for each human H1-N44 construct (+1A, U, G or C) is plotted in Figure 6 as percentage of the colonies that were screened. We found that the original H1 promoter starts transcription at multiple sites, resulting in considerable 5΄ end variability, consistent with a recent literature report (34). In fact, not the regular +1 start, but -1 was the most frequent start position for the human WT (+1A) and G constructs, consistent with a recent study (27). A minority of transcripts started in the -4/-2 region for the A construct and in the −5/−3 region for the G construct. The C and U promoters loose the prominent −1 signal, showing an array of low intensity start sites in the −11/−1 region (U construct) or the −4/+14 region (C construct). The prominent changes in the transcription start site of the C and U constructs maybe linked to their reduced silencing activity.
Figure 6.
Transcription start usage of +1-mutated H1 promoters. The transcription initiation site for H1-N44 variants (A, G, U and C) was determined by 5΄ RACE. We plotted the frequency of start site usage (depicted as % of the total number of colonies analyzed) along the sequence.
These results were largely confirmed by a deep sequencing analysis of the transcripts generated by the AgoshRNA constructs. The wt A construct starts predominantly at the -1 position and the G construct exhibited a more diffuse start site usage in the −3/+3 area (results not shown). We scored a considerable loss of the −1 start site for the C and U constructs, revealing a much more diffuse start site selection. The finding that transcription for the H1 promoter starts at -1C seems at odd with previous findings for the same H1-AgoshRT5 construct (16). Close inspection of the data generated in that study for all variant constructs indicates that -1C is indeed the major start site, but a noticeable +1A start was described for the WT AgoshRT5 construct, a result we could not reproduce in the current study. Similar H1-shRNA constructs were recently analyzed by deep sequencing and again demonstrated -1C usage (27). We also probed the transcription start site usage of some of the U6 promoter constructs to explain the minor size difference on the northern blot between A/U (longer) versus C/G constructs (shorter). For this, we performed 5΄ RACE PCR on the A and C constructs, which revealed the expected +1 start site for the A construct and striking shift to the -1 position for the C construct, thus explaining the shift in transcript size (results not shown). These results are consistent with the Ma et al. study in that a +1 purine is critical for efficient transcription and accurate +1 usage (35).
Cell type independent effects
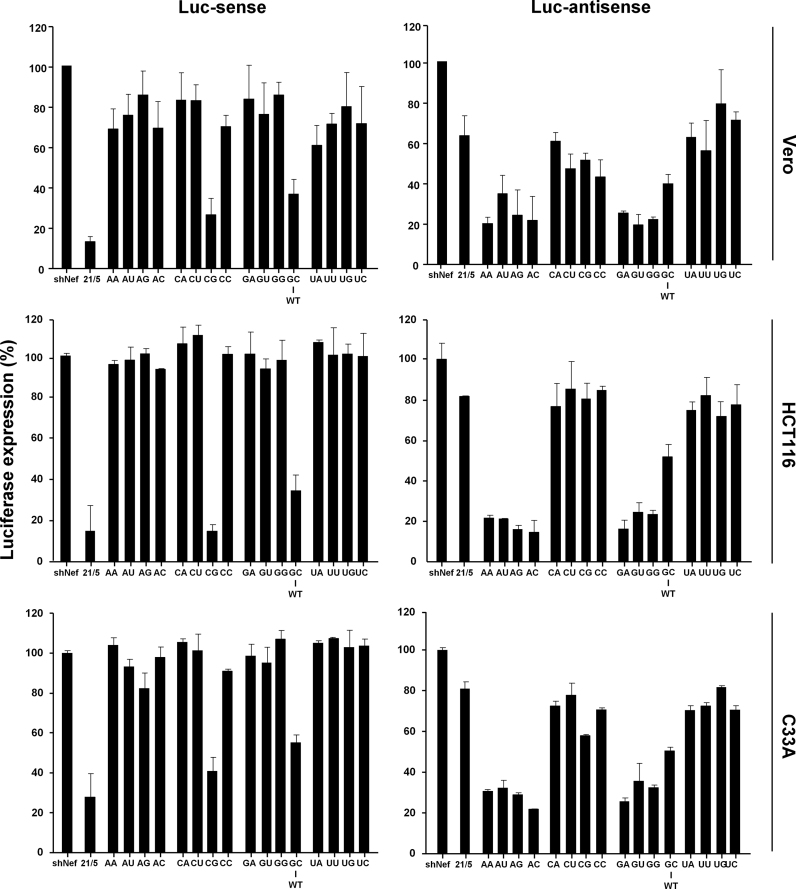
To investigate whether the influence of base pairing on AgoshRNA activity observed in HEK293T cells specify a general AgoshRNA requirement, we tested the set of AgoshPol47 inhibitors in others cell types (Vero, C33A and HCT116 cells). Vero is an African green monkey kidney-derived cell line, C33A is a human cervical cancer cell line and HCT116 is a human colon cancer cell line. Cells were co-transfected with the AgoshRNA constructs and the appropriate reporter plasmid (Figure 7). Silencing of the luciferase reporters revealed similar trends as observed for AgoshRT5 and AgoshPol47 in HEK293T cells. The regular shRNA (21/5) showed good inhibitory activity on the Luc-sense reporter, with luciferase levels dropping to <30%. In contrast, most AgoshRNA variants show little or no knockdown activity. Only the CG and GC variants exhibited a moderate knockdown activity. No to moderate knockdown activity was scored on the Luc-antisense reporter for the CN and UN variants. Unpaired AN and GN variants showed good silencing activity with luciferase levels <30%. These combined results indicate that the 5΄-terminal nt identity has a significant impact on AgoshRNA biogenesis and activity.
Figure 7.
AgoshRNA-mediated is cell line independent. The knockdown activity of AgoshPol47 of the 3΄ strand on Luc-sense (left panel) and 5΄ strand on Luc-antisense (right panel) of the AgoshPol47 was determined in transfected Vero, HCT116 and C33A cells. The mean values are based on three independent transfections that were performed in triplicate. See the legend to Figure 3 for more details.
The 5΄-terminal nucleotide may influence binding to the Ago2 protein
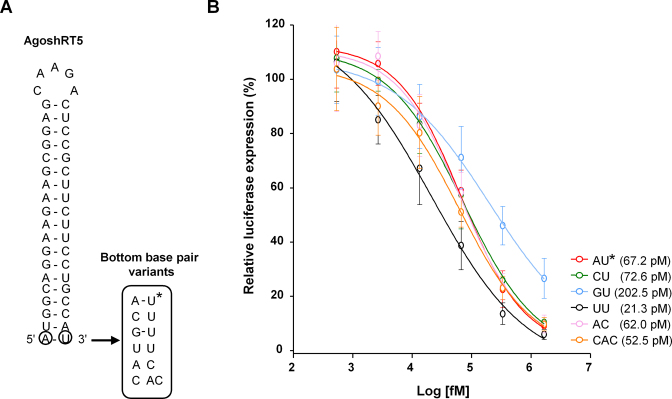
To test whether the modulating effect of the 5΄-terminal nt on AgoshRNA knockdown is based on altered promoter activity or on differential AgoshRNA processing/activity, we designed a set of 6 synthetic AgoshRNA molecules based on AgoshRT5 (Figure 8A). HEK293T cells were co-transfected with the synthetic AgoshRNA and the appropriate reporter plasmid. We included a control unrelated synthetic RNA that did not reduce Luciferase expression and this level of luciferase expression was set at 100%. A clear dose-dependent knockdown activity was measured for all synthetic AgoshRNA variants (Figure 8B). An RNA folding step before transfection into the cells was essential, consistent with the idea that these transcripts are active as structured hairpin molecules. All 6 synthetic AgoshRNAs exhibit roughly similar activities, which argues that the major difference observed among AgoshRNA constructs with 5΄ end variation is due to variation in promoter +1 usage. The UU variant exhibited a subtly improved knockdown activity (IC50: 21.3 pM) compared to the other variants (IC50: 5΄ end U < A < C < G). This observation contrasts with the results obtained for AgoshRNAs transcribed by the RNA polymerase III promoter, but is consistent with the fact that the MID domain of the human Ago2 protein has a binding preference for small RNAs with 5΄ U or A during RISC loading (36,37). These data confirm that the prominent changes in the transcription start site of the C and U constructs transcribed by the H1 promoter are linked to their reduced silencing activity. The chemically synthetized AU variant was slightly less active than the AC variant as observed previously. The CAC variant (with an additional C at the 5΄ end) is slightly more potent than the AC variant, consistent with previous studies based on chemically synthesized AgoshRNAs (38).
Figure 8.
Synthetic AgoshRT5 variants. (A) Synthetic AgoshRT5 molecules with variation in the bottom bp. The bottom bp (encircled) was mutated. (B) The knockdown activity of the different AgoshRT5 variants was determined by co-transfection with a luciferase reporter with the complementary target sequence. HEK293T cells were co-transfected with 100 ng of the respective firefly luciferase reporter plasmid, 1 ng renilla luciferase plasmid as internal control and increasing concentrations of the corresponding AgoshRT5 construct. An unrelated synthetic AgoshRNA was used as control, for which the activity was set at 100% luciferase expression. We performed three independent transfections, each in duplicate, and standard deviations were calculated. *: wild-type. To estimate the inhibitory concentration (IC50) of synthetic AgoshRNA constructs using nonlinear regression models (GraphPad Prism software). The response was used in the log form (log10(dose)).
DISCUSSION
Recent evidence indicates that one can create shRNAs with a small loop and short duplex length that avoid Dicer processing and instead are processed by Ago2 (13,15,16,18). This new design was termed AgoshRNA with the major advantage that no passenger strand is generated that can induce unwanted side effects. It is important to realize that siRNA design algorithms cannot be applied to shRNA and AgoshRNA design (39). It is therefore key to understand this novel processing route in mechanistic detail such that one can design the most effective and selective RNA reagents. In this study, we designed AgoshRNAs that are as potent as classical shRNAs, but that have little or no passenger strand activity.
There is accumulating evidence that the AgoshRNA pathway is very similar to that of miR-451. One of the hallmarks of miR-451 is that it starts with 5΄ A instead of 5΄ U that is more common in other miRNAs and this A remains unpaired at the bottom of the stem. A pilot study indicated that the identity of the bottom bp may be important for AgoshRNA activity (20). Therefore, we designed a complete set of AgoshRNA variants with all 16 dinucleotide combinations at the bottom of the stem to dissect general rules for AgoshRNA design. All four AN variants exhibited profound silencing activity followed by the GN variants, whereas the CN and UN variants exhibited little and no silencing activity, respectively. Thus, the identity of the +1 nt is indeed important. Subtly improved activity was scored for the unpaired nt combinations among the AN and GN variants. Consistent with the luciferase data, AN and GN variants exhibited the most abundant ∼30 nt Ago2 products on northern blot. Very similar results were obtained for a second AgoshRNA template and the same activity profiles were scored in an unrelated cell type. We thus conclude that AgoshRNA optimization is possible by introduction of a bottom mismatch with 5΄-terminal nt A or G.
The +1 position of the AgoshRNA transcript also represents the +1 position in the H1 promoter and thus may have multiple mechanistic implications at the DNA and RNA level. The +1 may have an effect on the H1 transcriptional activity and the actual choice of transcription start site. The transcript with +1 sequence variation may also be more stable or processed more efficiently. The bottom bp may also affect the stability of the folded transcript and affect the choice to be processed by Dicer or Ago2. One should also consider AgoshRNA binding to the Ago2 protein, which may favor certain 5΄-terminal nt. We will discuss these possibilities in more detail below.
An ‘early’ effect is possible at the transcriptional level due to the +1 change in the polymerase III H1 promoter. We measured a 2-fold higher transcription efficiency for the A and G constructs, which may thus partially explain the optimal AgoshRNA activity. The same purine preference was scored for the U6 promoter. In fact, we observed that the H1 promoter starts transcription at multiple sites, which results in considerable variation at the 5΄ end of the transcript. These results are consistent with a previous study in which the polymerase III promoters (H1 and U6) were used to express small RNAs (35). They investigated the effect of mutations around the +1 position on transcription initiation and described that the sequence from position −3 to +4 affects the precision and efficiency of transcription initiation. It was suggested that A or G can be used for initiation when present in the −1/+2 window. However, it is still unclear if this ‘A/G rule’ applies to transcripts with an unrelated sequence. Therefore, a more complete analysis may still be required that includes transcripts with different −3/+4 sequences. We demonstrated that the +1 identity contributes strongly to the accuracy of transcription initiation by the H1 and U6 promoters. The human H1 WT promoter (+1A) yields a fairly discrete transcript and the G mutant is only a bit less precise, but the product variation is significantly increased for the C and U constructs, consistent with the activity profile.
In this study, we show that H1 transcription starts most frequently at the -1 position as previously reported (27). Thus, the optimized AgoshRNA molecule will in fact have a 2-nt 5΄ overhang (CN+1). The 3΄ overhang will read NUU, the latter 2-nt from the transcription terminal signal. By comparison, the similarly processed miR-451 has a 3΄-terminal CUC overhang created by Drosha processing. The 3΄ nt overhang is not likely to sterically hinder RNA uptake by the Ago2 enzyme and the silencing activity as we confirmed with the chemically synthesized CAC AgoshRNA. It has also been reported that synthetic molecules mimicking miR-451 (agsiRNA) are slightly more potent with an additional A at the 5΄ end (38). The 2-nt 5΄ overhang may facilitate anchorage to the MID domain of Ago2 or binding into the Ago2 groove to trigger Ago2 slicer activity, but this has not been studied yet.
Another possibility is that a strong bp at the bottom of the hairpin affects the stability of the folded transcript to cause a shift from Ago2 to Dicer processing as observed for the GC variants. We did not find a clear pattern among the other variants. The MID domain of the human Ago2 protein has a binding preference for small RNAs with 5΄ U or A during RISC loading (36,37). However, we scored superior AgoshRNA activity for the AN and GN variants, which argues against this scenario.
Combined with our previous findings, we have defined the parameters for the design of optimized AgoshRNA molecules: a small 5 nt loop (CAAGA) and a duplex length of 18 bp with a bottom mismatch. A is recommended at position +1 as 5΄-terminal nt, at least when the RNA polymerase III H1 promoter is used for expression of the RNA molecule. The optimized AgoshRNA molecules are as potent as regular shRNAs, but lack the passenger strand activity and thus reduce the risk of adverse effects. We previously listed other putative advantages of the AgoshRNA design (15,18). Ago2-mediated processing yields more precise RNA molecules than Dicer cleavage, which is known to create imprecise ends (27,40). The shorter duplex of AgoshRNAs may exhibit an improved safety profile because innate immunity sensor like interferon will be triggered less likely by shorter RNA duplexes (41). AgoshRNAs may possibly mimic miR-451 in causing exclusive loading into Ago2, thus avoiding off targeting via Ago1, 3 and 4 (42). Another potential advantage is that AgoshRNAs are active in Dicer-deficient cells, e.g. monocytes that lack Dicer expression or cells that lack the RNAi machinery (15,17). Therefore, the AgoshRNA design may constitute a new platform for gene silencing that outperforms current miRNA and shRNA technology.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Nederlandse Organisatie voor Wetenschappelijk Onderzoek – Chemische Wetenschappen (NWO-CW, Top Grant); Zorg Onderzoek Nederland – Medische Wetenschappen (ZonMw, Translational Gene Therapy Grant). Zongl-liang Gao is recipient of a fellowship of the China Scholarship Council (CSC). The open access publication charge for this paper has been waived by Oxford University Press – NAR Editorial Board members are entitled to one free paper per year in recognition of their work on behalf of the journal.
Conflict of interest statement. None declared.
REFERENCES
- 1. Napoli C., Lemieux C., Jorgensen R.. Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell. 1990; 2:279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C.. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998; 391:806–811. [DOI] [PubMed] [Google Scholar]
- 3. Khvorova A., Reynolds A., Jayasena S.D.. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003; 115:209–216. [DOI] [PubMed] [Google Scholar]
- 4. Schwarz D.S., Hutvagner G., Du T., Xu Z., Aronin N., Zamore P.D.. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003; 115:199–208. [DOI] [PubMed] [Google Scholar]
- 5. Brummelkamp T.R., Bernards R., Agami R.. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002; 296:550–553. [DOI] [PubMed] [Google Scholar]
- 6. Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T.. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001; 411:494–498. [DOI] [PubMed] [Google Scholar]
- 7. Hammond S.M., Bernstein E., Beach D., Hannon G.J.. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000; 404:293–296. [DOI] [PubMed] [Google Scholar]
- 8. Tam O.H., Aravin A.A., Stein P., Girard A., Murchison E.P., Cheloufi S., Hodges E., Anger M., Sachidanandam R., Schultz R.M. et al. . Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008; 453:534–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cheloufi S., Dos Santos C.O., Chong M.M., Hannon G.J.. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010; 465:584–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cifuentes D., Xue H., Taylor D.W., Patnode H., Mishima Y., Cheloufi S., Ma E., Mane S., Hannon G.J., Lawson N.D. et al. . A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010; 328:1694–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yang J.S., Maurin T., Robine N., Rasmussen K.D., Jeffrey K.L., Chandwani R., Papapetrou E.P., Sadelain M., O’Carroll D., Lai E.C.. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:15163–15168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yoda M., Cifuentes D., Izumi N., Sakaguchi Y., Suzuki T., Giraldez A.J., Tomari Y.. Poly(A)-specific ribonuclease mediates 3΄-end trimming of Argonaute2-cleaved precursor microRNAs. Cell Rep. 2013; 5:715–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dallas A., Ilves H., Ge Q., Kumar P., Shorenstein J., Kazakov S.A., Cuellar T.L., McManus M.T., Behlke M.A., Johnston B.H.. Right- and left-loop short shRNAs have distinct and unusual mechanisms of gene silencing. Nucleic Acids Res. 2012; 40:9255–9271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ge Q., Ilves H., Dallas A., Kumar P., Shorenstein J., Kazakov S.A., Johnston B.H.. Minimal-length short hairpin RNAs: the relationship of structure and RNAi activity. RNA. 2010; 16:106–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu Y.P., Schopman N.C., Berkhout B.. Dicer-independent processing of short hairpin RNAs. Nucleic Acids Res. 2013; 41:3723–3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu Y.P., Karg M., Harwig A., Herrera-Carrillo E., Jongejan A., van kampen A., Berkhout B.. Mechanistic insights on the Dicer-independent AGO2-mediated processing of AgoshRNAs. RNA. Biol. 2014; 12:92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Herrera-Carrillo E., Harwig A., Liu Y.P., Berkhout B.. Probing the shRNA characteristics that hinder Dicer recognition and consequently allow Ago-mediated processing and AgoshRNA activity. RNA. 2014; 20:1410–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Berkhout B., Liu Y.P.. Towards improved shRNA and miRNA reagents as inhibitors of HIV-1 replication. Future Microbiol. 2014; 9:561–571. [DOI] [PubMed] [Google Scholar]
- 19. Yang J.S., Maurin T., Lai E.C.. Functional parameters of Dicer-independent microRNA biogenesis. RNA. 2012; 18:945–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Herrera-Carrillo E., Harwig A., Berkhout B.. Towards optimization of AgoshRNA molecules that use a non-canonical RNAi pathway: variations in the top and bottom base pairs. RNAr Biol. 2015; 12:447–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schopman N.C., Liu Y.P., Konstantinova P., Ter Brake O., Berkhout B.. Optimization of shRNA inhibitors by variation of the terminal loop sequence. Antiviral Res. 2010; 86:204–211. [DOI] [PubMed] [Google Scholar]
- 22. Ter Brake O., Konstantinova P., Ceylan M., Berkhout B.. Silencing of HIV-1 with RNA interference: a multiple shRNA approach. Mol. Ther. 2006; 14:883–892. [DOI] [PubMed] [Google Scholar]
- 23. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003; 31:3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Westerhout E.M., Ooms M., Vink M., Das A.T., Berkhout B.. HIV-1 can escape from RNA interference by evolving an alternative structure in its RNA genome. Nucleic Acids Res. 2005; 33:796–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ruijter J.M., Thygesen H.H., Schoneveld O.J., Das A.T., Berkhout B., Lamers W.H.. Factor correction as a tool to eliminate between-session variation in replicate experiments: application to molecular biology and retrovirology. Retrovirology. 2006; 3:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ratkowsky D.A. Principles of Nonlinear-Regression Modeling. J. Ind. Microbiol. 1993; 12:195–199. [Google Scholar]
- 27. Harwig A., Herrera-Carrillo E., Jongejan A., van Kampen A.H., Berkhout B.. Deep sequence analysis of AgoshRNA processing reveals 3΄ A addition and trimming. Mol. Ther. Nucleic Acids. 2015; 4:e247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu Y.P., Haasnoot J., Ter Brake O., Berkhout B., Konstantinova P.. Inhibition of HIV-1 by multiple siRNAs expressed from a single microRNA polycistron. Nucleic Acids Res. 2008; 36:2811–2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sun G., Li H., Rossi J.J.. Cloning and detecting signature microRNAs from mammalian cells. Methods Enzymol. 2007; 427:123–138. [DOI] [PubMed] [Google Scholar]
- 30. Lim J., Ha M., Chang H., Kwon S.C., Simanshu D.K., Patel D.J., Kim V.N.. Uridylation by TUT4 and TUT7 marks mRNA for degradation. Cell. 2014; 159:1365–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shen B., Goodman H.M.. Uridine addition after microRNA-directed cleavage. Science. 2004; 306:997. [DOI] [PubMed] [Google Scholar]
- 32. Rollins J., Veras I., Cabarcas S., Willis I., Schramm L.. Human Maf1 negatively regulates RNA polymerase III transcription via the TFIIB family members Brf1 and Brf2. Int. J. Biol. Sci. 2007; 3:292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rumi M., Ishihara S., Aziz M., Kazumori H., Ishimura N., Yuki T., Kadota C., Kadowaki Y., Kinoshita Y.. RNA polymerase II mediated transcription from the polymerase III promoters in short hairpin RNA expression vector. Biochem. Biophys. Res. Commun. 2006; 339:540–547. [DOI] [PubMed] [Google Scholar]
- 34. Ma H., Zhang J., Wu H.. Designing Ago2-specific siRNA/shRNA to avoid competition with endogenous miRNAs. Mol. Ther. Nucleic Acids. 2014; 3:e176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ma H., Wu Y., Dang Y., Choi J.G., Zhang J., Wu H.. Pol III promoters to express small RNAs: delineation of transcription initiation. Mol. Ther. Nucleic Acids. 2014; 3:e161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Frank F., Sonenberg N., Nagar B.. Structural basis for 5΄-nucleotide base-specific recognition of guide RNA by human AGO2. Nature. 2010; 465:818–822. [DOI] [PubMed] [Google Scholar]
- 37. Hu H.Y., Yan Z., Xu Y., Hu H., Menzel C., Zhou Y.H., Chen W., Khaitovich P.. Sequence features associated with microRNA strand selection in humans and flies. BMC Genomics. 2009; 10:413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sun G., Yeh S.Y., Yuan C.W., Chiu M.J., Yung B.S., Yen Y.. Molecular properties, functional mechanisms, and applications of sliced siRNA. Mol. Ther. Nucleic Acids. 2015; 4:e221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Taxman D.J., Livingstone L.R., Zhang J., Conti B.J., Iocca H.A., Williams K.L., Lich J.D., Ting J.P., Reed W.. Criteria for effective design, construction, and gene knockdown by shRNA vectors. BMC. Biotechnol. 2006; 6:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gu S., Jin L., Zhang Y., Huang Y., Zhang F., Valdmanis P.N., Kay M.A.. The loop position of shRNAs and pre-miRNAs is critical for the accuracy of dicer processing in vivo. Cell. 2012; 151:900–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bridge A.J., Pebernard S., Ducraux A., Nicoulaz A.L., Iggo R.. Induction of an interferon response by RNAi vectors in mammalian cells. Nat. Genet. 2003; 34:263–264. [DOI] [PubMed] [Google Scholar]
- 42. Dueck A., Ziegler C., Eichner A., Berezikov E., Meister G.. microRNAs associated with the different human Argonaute proteins. Nucleic Acids Res. 2012; 40:9850–9862. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.