Abstract
Adenosine deaminases acting on RNA (ADARs) catalyze the editing of adenosine residues to inosine (A-to-I) within RNA sequences, mostly in the introns and UTRs (un-translated regions). The significance of editing within non-coding regions of RNA is poorly understood. Here, we demonstrate that association of ADAR2 with RNA stabilizes a subset of transcripts. ADAR2 interacts with and edits the 3΄UTR of nuclear-retained Cat2 transcribed nuclear RNA (Ctn RNA). In absence of ADAR2, the abundance and half-life of Ctn RNA are significantly reduced. Furthermore, ADAR2-mediated stabilization of Ctn RNA occurred in an editing-independent manner. Unedited Ctn RNA shows enhanced interaction with the RNA-binding proteins HuR and PARN [Poly(A) specific ribonuclease deadenylase]. HuR and PARN destabilize Ctn RNA in absence of ADAR2, indicating that ADAR2 stabilizes Ctn RNA by antagonizing its degradation by PARN and HuR. Transcriptomic analysis identified other RNAs that are regulated by a similar mechanism. In summary, we identify a regulatory mechanism whereby ADAR2 enhances target RNA stability by limiting the interaction of RNA-destabilizing proteins with their cognate substrates.
INTRODUCTION
Post-transcriptional RNA processing is essential for regulation of cellular gene expression, and is mediated by a large class of RNA-binding proteins (RBPs). RNA editing is one such widespread, post-transcriptional process that introduces changes in the sequence of the RNA transcript. Adenosine deaminases acting on RNA (ADARs) are double-stranded RBPs that catalyze the hydrolytic deamination of adenosine residues to inosine, a process referred to as A-to-I editing (1,2). Adenosine base pairs with uridine; in contrast, inosine base pairs with cytidine. Thus, this substitution alters the RNA sequence. Three types of ADAR enzymes have been identified to-date, ADAR1, ADAR2, ADAR3, but only ADAR1 and ADAR2 exhibit recognizable editing activity (1–4). Both ADAR1 and ADAR2 are known to be essential in mammals (3–5). While a few mechanistic details of ADAR function are known, more insights are required in order to understand the molecular basis of their physiological and pathological effects (6–8).
Transcriptome-wide studies have demonstrated that majority of A-to-I editing events occur within Alu repetitive sequences of short interspersed element (SINE) origin, which are primarily located within introns or 3΄UTRs (untranslated regions) of RNAs (9–13). Alu elements form long intra-molecular RNA duplexes with closely lying inverted Alu repeat sequences (IRAlus), and are recognized by ADARs for A‐to‐I editing (2,9,14,15). While majority of editing occurs in the non-coding parts of the transcriptome, only a handful of studies have described the role of editing within non-coding regions (16–19). In general, the 3΄UTRs of mRNAs regulate RNA localization, stability and translation. Therefore, it is possible that A-to-I editing within the 3΄UTRs of mRNAs could play a role in regulating these processes. A few studies utilizing reporter and endogenous mRNA have suggested that editing within the 3΄UTR could influence gene expression, by restricting the nuclear export of hyper-edited RNA (16,20,21). It has been suggested that the association of A-to-I hyper-edited transcripts with p54nrb/NonO, a component of the paraspeckle, is responsible for the nuclear retention of edited transcripts (20,22). However, other studies have shown that mRNAs with hyper-edited 3΄UTRs localize to the cytoplasm, indicating that editing alone is not enough to restrict the export of transcripts (23,24). Recent studies have shown that A-to-I editing of the 3΄UTR could influence the binding of microRNAs (miRNAs) to transcripts (25). Except for these limited examples, the precise functional significance of A-to-I editing within the 3΄UTR is largely unknown, and demands further investigation. Furthermore, apart from editing-related functions of ADARs, a handful of recent studies have also demonstrated editing-independent roles of ADARs (8,26–28).
In the present study, we used nuclear-retained Ctn RNA as a model system to gain insights into the biological significance of ADAR associations within 3΄UTRs of RNAs. We have previously demonstrated that Ctn RNA regulates the expression of its protein-coding partner, mCat2 (mouse cationic amino acid transporter 2) (21). Both Ctn RNA and mCat2 mRNA are transcribed from the same gene, however, due to alternative poly(A) site selection, Ctn RNA has a longer 3΄ end (hereafter named ‘3΄UTR’, given its shared sequence with mCat2 mRNA). mCAT2 facilitates the cellular uptake of L-arginine, which is utilized as a substrate for the synthesis of nitric oxide in the cell. Ctn RNA is an abundant and very stable transcript, and is also induced as part of the antiviral response (21). While SINE repeats within the 3΄UTR of Ctn RNA are known to be A-to-I edited, how such editing affects the properties of Ctn RNA is not known. Our findings reveal that ADAR2 association with the 3΄UTR promotes the stability of Ctn RNA by limiting its association with two RNA-destabilizing proteins, HuR and PARN [poly(A)-specific ribonuclease] deadenylase, capable of destabilizing Ctn RNA. Furthermore, transcriptomic analysis indicates that in addition to Ctn RNA, ADAR2 also stabilizes other classes of RNAs by limiting the binding of HuR and PARN. Our studies provide mechanistic insights into the role of ADAR2 in regulating the stability of RNA through its association with non-coding regions of transcripts.
MATERIALS AND METHODS
RNA editing
To measure A-to-I editing levels, FwR and IR2 repeat sequences were amplified using repeat-specific primers by RT-PCR and the products (of expected size) were sequenced. In the electropherograms, editing sites appeared as mixed A and G peaks. Editing levels were determined by measuring peak heights using Bioedit software and were calculated as a ratio of G-peak height to the A+G-peak height (G/G+A).
Ribonucleoprotein immunoprecipitation (RIP)
Ribonucleoprotein immunoprecipitation (RIP) was performed using an established protocol (29). Wild-type (WT) and Adar2-KO cells (1 × 107) were used for RNA immunoprecipitation utilizing reversible chemical crosslinking of RNA–protein interactions by formaldehyde followed by immunoprecipitation using Anti-FLAG (F1804, Sigma, USA) or PARN antibody (sc-135242; Santa Cruz Biotechnology, USA), or anti-ADAR2 antibody (HPA018277, Sigma, USA). For ADAR2 RIP in mouse embryonic fibroblasts (MEFs), following reversible chemical crosslinking of RNA–protein interactions by formaldehyde, cells were lysed in 0.1% NP-40 and 1 mM ethylenediaminetetraacetic acid (EDTA). Immunoprecipitation was performed in buffer containing 0.05% NP-40 and 1 mM EDTA. Following IP, extracts were reverse cross-linked and total RNA was extracted using Trizol LS (Invitrogen, USA). Extracted RNA was treated with RNase-free DNase I (Sigma, USA), and RT was conducted using random-hexamer primers as per the manufacturer's instructions (Applied Biosystems, USA). qPCR analysis was performed using gene-specific primers.
For native RIP analysis of endogenous RNP complexes from whole-cell extracts, cells were lysed in 20 mM Tris-HCl at pH 7.5, 100 mM KCl, 5 mM MgCl2 and 0.5% NP-40 for 10 min on ice and centrifuged at 10 000 × g for 15 min at 4°C (30). The supernatants were incubated with protein A-Sepharose beads coated with antibodies that recognized HuR (sc-5261, Santa Cruz Biotechnology) or with control IgG for 1 h at 4°C. After the beads were washed with NT2 buffer (50 mM Tris-HCl at pH 7.5, 150 mM NaCl, 1 mM MgCl2 and 0.05% NP-40), the complexes were incubated with 20 units of RNase-free DNase I (15 min at 37°C) and further incubated with 0.1% sodium dodecyl sulphate/0.5 mg/ml Proteinase K (15 min at 55°C) to remove DNA and proteins, respectively. Acidic phenol (Ambion) was used to extract RNA for RIP analysis. Reverse transcription (RT) was performed using random hexamers and reverse transcriptase (Maxima, Thermo Scientific) and RT-qPCR using gene-specific primers, and SYBR green master mix (Kapa Biosystems), using an Applied Biosystems 7300 instrument. Normalization of RIP results was carried out by quantifying in parallel the relative levels of Gapdh mRNA in each IP sample. These abundant RNAs are non-specific contaminants present in the IP components.
RNA stability assay
To measure RNA stability, 5 μg/ml Actinomycin D (Sigma-aldrich, USA) or Flavopiridol (1 µM) was added to cells and incubated for different time durations as indicated. At each time point, total RNA was harvested using Trizol and used for RT-qPCR analysis. RNA decay rate was quantified by fitting an exponential curve to the data points (y = a * e∧-bt), whereby y is the (relative) amount of RNA and t is time. The half-life was then calculated using: t(1/2) = ln2 / b (31). The half-life was calculated from each experiment and the average half-life along with standard deviations shown in the graph. Anti-HA (Covance, USA), Anti-FLAG (F1804, Sigma, USA), beta-Actin (ab6276, Abcam, USA) was used to measure protein levels in western blots.
Nuclear and cytoplasmic fractionation
WT and Adar2-KO (1 × 106) cells were used for fractionation. Cells were washed with PBS and resuspended in RSB buffer (10 mM Tris-HCl pH7.4, 100 mM NaCl, 2.5 mM MgCl2, RNase Inhibitor) and lysed in RSB buffer containing Digitonin (8 μg/ml) (D141-100MG, Sigma-Aldrich, USA) for 10 min on ice. Cells were centrifuged (2000 rpm, 4°C, 10 min) and the supernatant (cytoplasmic fraction) collected. The pellet (nuclear fraction) was washed with RSB + digitonin by the procedure described above. Trizol LS (10296-028, Invitrogen, USA) was added to the cytoplasmic fraction while Trizol was added to the nuclear fraction. Neat1 and Actin were used to confirm the purity of the nuclear and cytoplasmic fractions, respectively. Ct values of nuclear or cytoplasmic fractions were normalized to total RNA.
RNA-FISH
To detect Ctn RNA, RNA-FISH analysis was performed as previously described (29). Ctn RNA localization to paraspeckles was increased during transcriptional reactivation. Therefore, for Ctn RNA FISH, cells were treated with the transcriptional inhibitor DRB (5,6-Dichloro-1-beta-D-ribofuranosylbenzimidazole) followed by reactivation of transcription by removal of inhibitor with medium for 3 h. Immunofluorescence staining of HuR using HuR antibody (1:100 for 2 h at room temperature; sc-5261, Santa Cruz Biotechnology) in WT and Adar2-KO MEFs was performed as previously described (29).
RESULTS
ADAR2 edits specific sites within the 3΄UTR of Ctn RNA
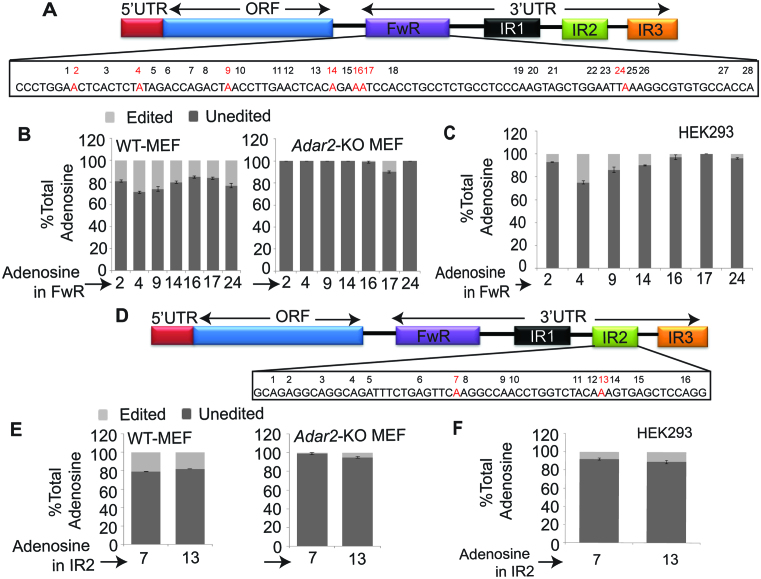
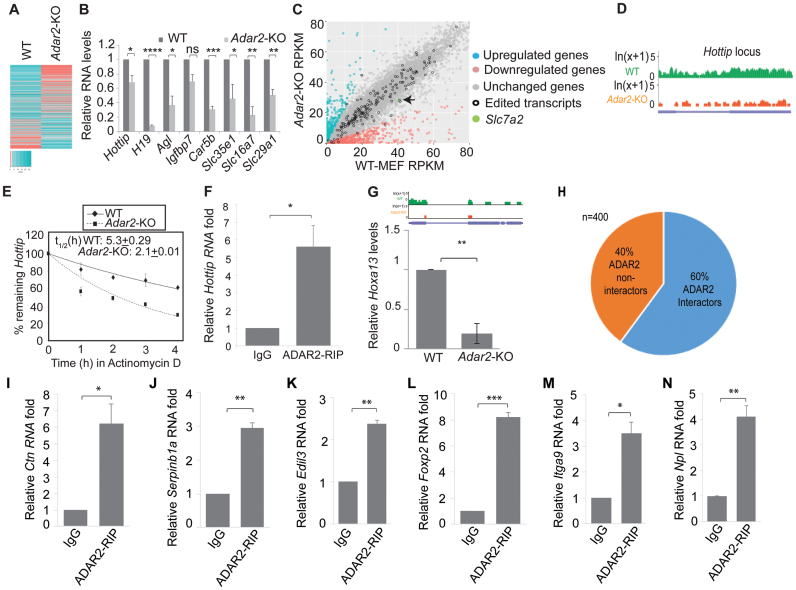
The 3΄UTR of Ctn RNA harbors three repeat elements (IR1, IR2 and IR3) of SINE origin that are inverted with respect to the forward repeat (FwR) (Figure 1A). In mouse tissues, several of the adenosines within the FwR and IR2 (inverted repeat 2) are A-to-I edited (21). ADAR2 RIP from lysates prepared from mouse brain, followed by microarray analyses (Mouse Genome 430A 2.0 micro array), confirmed the interaction between ADAR2 and Ctn RNA (Supplementary Table S1) (32). To determine if ADAR2 edits Ctn RNA, we analyzed the editing profile of endogenous Ctn RNA (in FwR and IR2) in Adar2-KO MEFs and compared it to that of WT MEFs (Figure 1A, B, D and E; Supplementary Figure S1Aa and B). In WT, the editing efficiency of several of the adenosines within the FwR and IR2 was found to be ∼20–30% (Figure 1B and E; Supplementary Figure S1B). In contrast, we observed a significant reduction in Ctn RNA editing in Adar2-KO MEFs (Figure 1B and E; Supplementary Figure S1B).
Figure 1.
ADAR2 edits SINE elements in the 3΄UTR of Ctn RNA. (A and D) Schematic of transcript organization of Ctn RNA, indicating edited adenosines (red) in (A) forward repeat (FwR) and (D) inverted repeat 2 (IR2) respectively. Numbers above each Adenosine indicate its position in the sequence. (B and E) Relative % of unedited versus edited adenosines at each site in (B) FwR and (E) IR2 in WT MEFs and Adar2-KO MEFs. (C and F) Relative % of unedited versus edited adenosines at each site in the (C) FwR and (F) IR2 of transiently expressed Ctn RNA along with ADAR2 (24 h after transfection) in HEK293 cells (in cells where only Ctn RNA was transfected, no editing was observed in majority of sites). X-axis shows the relative position of individual adenosines within the FwR or IR2. Error bars in (B and C) and (E and F) represent the means ± SD of three independent experiments (biological replicates).
To provide further evidence of the involvement of ADAR2 in editing Ctn RNA, we transiently transfected Ctn RNA, alone or in combination with Flag-tagged Adar2, into HEK293 cells (Supplementary Figure S1C). Despite the presence of normal background levels of ADARs, HEK293 cells display low intrinsic A-to-I editing activity and thus have been previously used as a system to study editing of substrates by exogenously expressed ADAR isoforms (Supplementary Figure S1Ab and D) (33–36). We observed that most of the adenosines within the repeats of Ctn RNA (except site 17 in FwR) that were A-to-I edited in WT-MEFs were also edited in Adar2 exogenously expressed HEK293 cells (Figure 1C and F; Supplementary Figure S1D). Together, these results suggest that ADAR2 interacts with and edits adenosines within the 3΄UTR of Ctn RNA.
ADAR2 promotes the stability of Ctn RNA
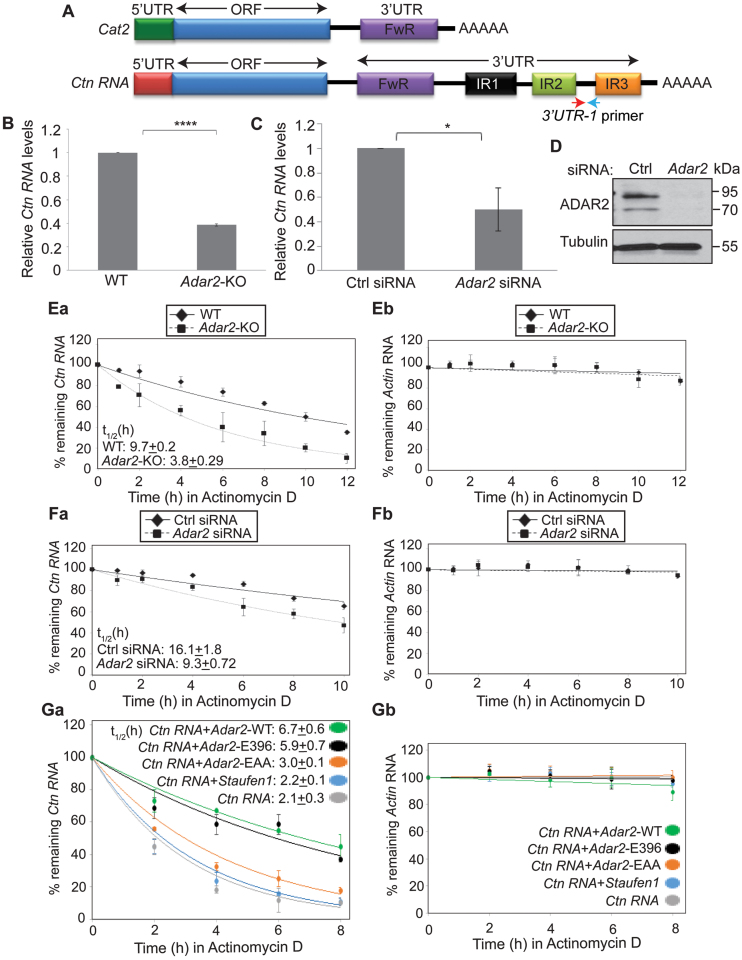
Next, we wanted to determine the functional significance of ADAR2 interaction with the 3΄UTR of Ctn RNA. To this end, we determined if ADAR2 influences the cellular abundance of Ctn RNA. We measured the total levels of Ctn RNA in WT and Adar2-KO MEFs using a primer pair unique to the long 3΄UTR of Ctn RNA (Figure 2A and Supplementary Figure S2). RT-qPCR results revealed that Ctn RNA levels were significantly reduced in Adar2-KO MEFs than in WT (Figure 2B). However, the total levels of a large number of other transcripts, including Actin and Gapdh remained unchanged in Adar2-KO cells (Supplementary Table S4). Consistent with these observations, Ctn RNA levels were significantly lowered in ADAR2-depleted, transformed MEFs than in control siRNA-treated cells (Figure 2C and D). We also measured mCat2 RNA levels in WT and Adar2-KO MEFs by qRT-PCR using a primer from the ORF (open reading frame) (Supplementary Figure S3A and B). We observed a mild reduction of mCat2 levels in Adar2-KO MEFs. However, please note, that the ORF primer is common to both Ctn RNA and mCat2 and the observed reduction may be due to lower levels of Ctn RNA levels and not mCat2 transcripts. RNA-FISH and nucleo-cytoplasmic fractionation analyses also demonstrated that majority of Ctn RNA in both WT and Adar2-KO MEFs retained in the nucleus (Supplementary Figure S3C and E). Therefore, we conclude that neither Ctn RNA localization nor mCat2 mRNA levels significantly change in Adar2-depleted cells. Our earlier studies have shown that mCat2 could also be transcribed from an alternative promoter (21). It is possible that a significant fraction of mCat2 present in Adar2-KO cells could be transcribed from the alternate promoter.
Figure 2.
ADAR2 promotes the stability of Ctn RNA. (A) Schematic of mCat2 and Ctn RNA. 3΄UTR-1 primer pair is specific to Ctn RNA, as it has been designed from a region in between IR2 and IR3, which lies downstream the predicted poly(A) site of Cat2 (Please see Supplementary Figure S2 for details of primer location). Arrows indicate approximate primer positions. (B) Relative levels of Ctn RNA in WT and Adar2-KO MEFs, determined by RT-qPCR using 3΄UTR-1 primer. (C) Relative Ctn RNA levels in Ctrl and Adar2 siRNA treated (48 h) transformed MEFs determined by RT-qPCR using 3΄UTR-1 primer. (D) Western blot analysis of ADAR2 in Ctrl and Adar2 siRNA treated (48 h) transformed MEFs respectively. Tubulin is used as a loading control. Measurement of the stability of (Ea and Fa) Ctn RNA and (Eb and Fb) Actin mRNA by RT-qPCR in WT, Adar2-KO MEFs (E) and Adar2 siRNA-treated transformed MEFs (F) in presence of the transcriptional inhibitor Actinomycin D at indicated time points. Half-life of Ctn RNA (t1/2) in both cells has been indicated. Please note that Actin mRNA does not show significant degradation in the duration of the experiment. Measurement of stability of (Ga) exogenously expressed Ctn RNA in HEK293 cells co-expressed with vector or human Adar2-WT or E396A deaminase-dead mutant of rat Adar2 (38) or RNA binding dead mutant of ADAR2-EAA or Staufen1 and (Gb) Actin mRNA was determined by Actinomycin D chase (after 48 h of transfection) followed by RT-qPCR. To normalize for transfection efficiency, Ctn RNA levels were normalized to GFP mRNA from GFP plasmid, which is co-transfected along with Ctn RNA in all of the experiments. In all RT-qPCR experiments Gapdh mRNA (GAPDH mRNA in HEK293 cells) was used as a normalization control as it did not show gene expression changes with any treatment. Error bars in (B, C, E, F and G) represent mean ± SD of three independent experiments (biological replicates). *P < 0.05, ****P < 0.0001, using Student's t-test.
We next determined the half-life (t1/2) of Ctn RNA in WT and Adar2-KO MEFs by incubating cells with Actinomycin D to block transcription and then measuring the time required until Ctn RNA levels were reduced to 50% of the original abundance. The results showed that in Adar2-KO primary MEFs, Ctn RNA showed reduced stability (Figure 2E). We also observed similar results upon treatment with another transcriptional inhibitor—flavopiridol (Supplementary Figure S4A). Additionally, we measured the stability of EU (5-ethynyl uridine)-labelled nascent RNA and observed that nascent Ctn RNA exhibited lower stability in Adar2-KO MEFs (Supplementary Figure S4B and C). We also observed ∼2-fold reduction in the stability of Ctn RNA in ADAR2-depleted, transformed MEFs when compared to control siRNA-treated cells (Figure 2F). Altogether these results indicate that Adar2 influences the stability of Ctn RNA.
To further corroborate ADAR2-mediated stabilization of Ctn RNA, we determined the half-life of exogenously expressed Ctn RNA, alone or co-transfected with full length Adar2 in HEK293 cells. Consistent with the results obtained in MEFs, Ctn RNA showed higher stability in the presence of ADAR2 (Figure 2G). To determine whether ADAR2 regulates the stability of Ctn RNA via A-to-I editing, we examined the stability of Ctn RNA reporter in HEK293 cells that were co-transfected with an inactive deaminase-domain containing mutant of ADAR2 (E396A) Supplementary Figure S5A and B). (37,38). Our results showed that even the deaminase-dead mutant of ADAR2 could enhance the stability of Ctn RNA (Figure 2G; Supplementary Figure S5A and B). Cells co-expressed with Ctn RNA and an Adar2 RNA-binding dead mutant (ADAR2-EAA, where both dsRBD1 and dsRBD2 have been mutated) (39) showed only a mild increase in Ctn RNA stability (Figure 2G and Supplementary Figure S5C). However, the rescue did not occur to the same extent as observed in case of ADAR2-WT and ADAR2-E396. Similarly, HEK293 cells overexpressing Staufen1, another ADAR2-unrelated double-stranded RNA binding protein, did not rescue the stability of Ctn RNA reporter RNA (40) (Figure 2G and Supplementary Figure S5D). Collectively, these results demonstrate that the association of ADAR2 with Ctn RNA, and not necessarily its editing activity that contributes to Ctn RNA stability. Ctn RNA is a nuclear RNA that is mobilized/cleaved at the 3΄UTR during cellular stress to rapidly generate mCat2-like mRNA. Our present results suggest a potential mechanism through which Ctn RNA maintains a stable nuclear reserve.
ADAR2 negatively regulates the association of HuR to Ctn RNA
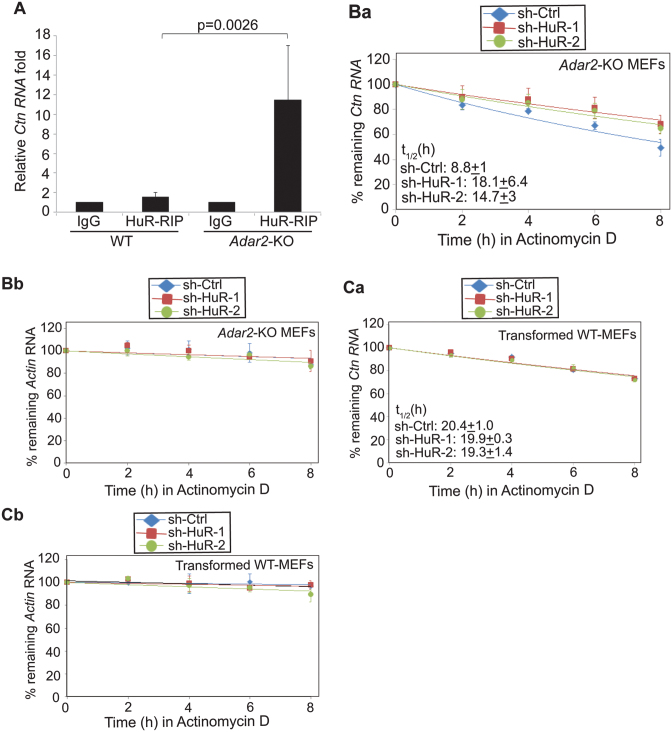
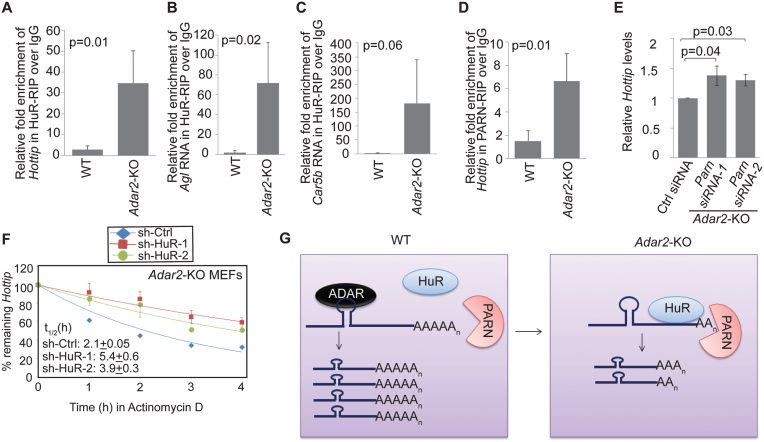
Next, we investigated the mechanism of ADAR2-dependent regulation of Ctn RNA stability. Previous studies have suggested that ADAR1 enhances the stability of mRNAs by facilitating the recruitment of HuR/ELAVL1 to these mRNAs (8,41). HuR/ELAVL1 is a U-/AU-rich element binding protein, is an established regulator of mRNA stability and potentially binds to ∼26 000 sites across the transcriptome (42–44). We identified potential binding sites of HuR using the HuR binding motif in the 3΄UTR of Ctn RNA (RBPmap—http://rbpmap.technion.ac.il/ and Supplementary Table S2A) (45). In addition, a published CLIP study also reported HuR binding sites in the 3΄UTR of human SLC7A2 (Supplementary Table S2B) (44). Since HuR is an established regulator of RNA stability, we investigated if HuR is involved in mediating ADAR2-dependent changes in Ctn RNA stability. To this end, we examined the status of HuR interaction with Ctn RNA in the presence or absence of ADAR2. RIP studies using an anti-HuR antibody indicated that HuR did not interact with Ctn RNA in WT MEF cell extracts (Figure 3A). Surprisingly, however, HuR displayed robust interaction with Ctn RNA in Adar2-KO MEFs (Figure 3A). This dramatic difference in the association of HuR with Ctn RNA was not due to differences in the total levels or localization of HuR between WT and Adar2-KO cells (Supplementary Figure S6A and B). Instead, the data suggest that ADAR2 negatively regulates HuR interaction with Ctn RNA.
Figure 3.
HuR destabilizes Ctn RNA in absence of ADAR2. (A) RIP using HuR antibody followed by RT-qPCR to detect Ctn RNA in WT and Adar2-KO MEFs. Normalization of RIP results was carried out by quantifying in parallel the relative levels of Gapdh mRNA in each IP sample. Stability of (Ba) Ctn RNA and (Bb) Actin mRNA in sh-Ctrl and sh-HuR expressed Adar2-KO MEFs. Stability of (Ca) Ctn RNA and (Cb) Actin mRNA in sh-Ctrl and sh-HuR expressed WT MEFs. In all RT-qPCR experiments Gapdh mRNA was used as a normalization control as it did not show changes in its levels with any treatment. Error bars represent mean ± SD of six (A) and three (B and C) independent experiments (biological replicates). Student's t-test was performed for determining statistical significance.
Even though HuR is known to increase the stability of mRNAs, several recent studies have described the role of HuR in destabilizing specific set of RNAs (30,46). Thus, we examined whether HuR is involved in destabilizing Ctn RNA. To test this possibility, we depleted HuR in Adar2-KO primary MEFs and determined the stability of Ctn RNA in these cells. In comparison to control cells, HuR-depleted Adar2-KO MEFs showed increased stability of Ctn RNA (Figure 3B and Supplementary Figure S6C). However, HuR depletion in WT MEFs did not alter Ctn RNA stability (Figure 3C). These results suggest that in the absence of ADAR2, the increased interaction of HuR with Ctn RNA reduces its stability.
HuR destabilizes Ctn RNA by facilitating the interaction between Ctn RNA and PARN deadenylase
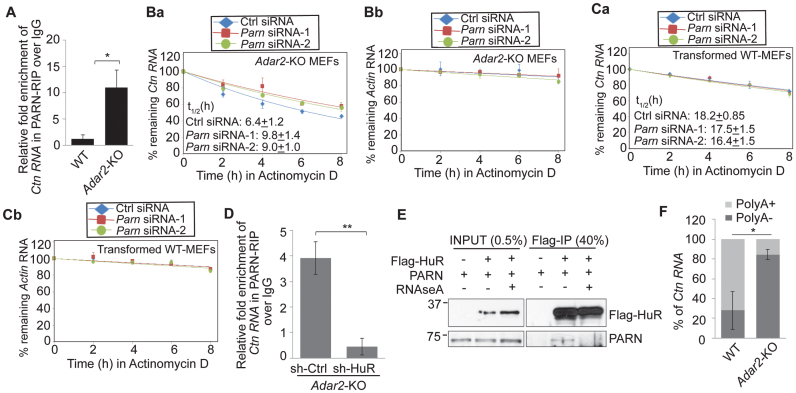
Next, we investigated the mechanism of HuR-mediated destabilization of Ctn RNA in the absence of ADAR2. Previous studies have shown that AU-binding proteins including HuR destabilize specific mRNAs by facilitating the recruitment of PARN (an exoribonuclease that shortens the poly(A) tail and thus, destabilizes RNA) and exosome components to mRNAs (44,46,47). Thus, we wondered if HuR plays a similar role in destabilizing Ctn RNA, by facilitating the recruitment of PARN. To test this possibility, we determined the interaction of Ctn RNA with PARN deadenylase in Adar2-KO MEFs by RIP analysis. The results indicate that like HuR, PARN also showed increased association with Ctn RNA, specifically in Adar2-KO extracts (Figure 4A; Supplementary Figure S7A and B). Finally, Ctn RNA displayed increased stability in Adar2-KO cells depleted of Parn (Figure 4B; Supplementary Figure S7C and D), indicating that PARN destabilizes Ctn RNA in the absence of ADAR2. However, similar to what was observed in case of HuR, Parn depletion did not affect Ctn RNA stability in WT cells (Figure 4C). This result indicates that in Adar2-KO cells, the increased interaction between PARN and Ctn RNA resulted in the PARN-mediated degradation of Ctn RNA.
Figure 4.
HuR facilitates the interaction between PARN and Ctn RNA in absence of ADAR2. (A) PARN RIP followed by Ctn RNA RT-qPCR in WT and Adar2-KO MEFs. Normalization of RIP results was carried out by quantifying in parallel the relative levels of Gapdh mRNA in each RIP sample. Decay curves of (Ba) Ctn RNA and (Bb) Actin mRNA in Ctrl and Parn-depleted Adar2-KO MEFs. Decay curves of (Ca) Ctn RNA and (Cb) Actin mRNA in Ctrl and Parn-depleted WT MEFs. (D) PARN RIP followed by RT-qPCR of Ctn RNA in sh-Ctrl and sh-HuR transfected Adar2-KO MEFs. Normalization of RIP results was carried out by quantifying in parallel the relative levels of Gapdh mRNA in each RIP sample. (E) Co-IP of FLAG-HuR and endogenous PARN in MEFs in presence and absence of RNase A. (F) % Ctn RNA levels measured in the PolyA+ and PolyA- fractions of WT and Adar2-KO MEFs by RT-qPCR using 3΄UTR-1 primer. In all RT-qPCR experiments Gapdh mRNA was used as a normalization control as it did not show gene expression changes with any treatment. Error bars represent mean ± SD of three (A–E) and five (F) independent experiments (biological replicates). *P <0.05, **P <0.01 using Student's t-test.
In our study, we observed that when compared to uninfected cells (Figure 2E), cells that are infected with a control shRNA lentivirus (Figure 3B) or treated with control siRNAs (Figure 4B), displayed an increase in the stability of Ctn RNA (Supplementary Figure S8). Ctn RNA levels are elevated as part of the antiviral response, including response to treatment of cells with IFNγ (interferon-γ) (21). We believe that the above-mentioned treatments could also lead to similar cellular responses, resulting in the enhanced stability of Ctn RNA. In any case, however, HuR- or PARN-depleted cells displayed enhanced stability for Ctn RNA when compared to control vector-infected or scrambled-siRNA-transfected cells, implying that these proteins indeed destabilize Ctn RNA.
To test the potential involvement of HuR in regulating the association of PARN with Ctn RNA, we measured the interaction between PARN and Ctn RNA in control versus HuR-depleted Adar2-KO cells, by PARN RIP followed by RT-qPCR analysis (Figure 4D and Supplementary Figure S7E). The interaction of PARN with Ctn RNA in Adar2-KO cells was significantly reduced in the absence of HuR (Figure 4D). These results support the model that HuR facilitates the interaction between PARN and Ctn RNA in the absence of ADAR2. Co-immunoprecipitation of FLAG-HuR with PARN in HEK293 cells showed that a small but significant fraction of PARN interacted with HuR in an RNA-dependent manner (Figure 4E). HuR has been previously shown to alter local mRNA secondary structure to facilitate binding of the miRISC complex to mRNA (48). By interacting with Ctn RNA, HuR could alter the local RNA secondary structure, which could facilitate PARN binding to Ctn RNA. Thus, our studies indicate that HuR serves as a co-factor that is required for the association of PARN-containing complex to RNA, in order to negatively regulate RNA stability.
Since the shortening of the poly(A) tail is a vital step for PARN-mediated mRNA decay, we asked if the decreased stability of Ctn RNA upon ADAR-depletion is associated with shortening of its poly(A) tail. To determine any change in the polyadenylation status of Ctn RNA, we measured the total levels of polyadenylated Ctn RNA in WT and Adar2-KO MEFs. We observed that levels of polyadenylated Ctn RNA were significantly reduced in Adar2-KO MEFs than in WT MEFs (Figure 4F).
ADAR2 influences the expression/stability of a large number of RNAs, including nuclear lncRNA Hottip
To determine if ADAR2 regulates the expression and/or stability of transcripts other than Ctn RNA, we performed polyA+ RNA sequencing of WT and Adar2-KO MEFs. As observed for Ctn RNA, several RNAs show reduced editing in Adar2-KO than in WT MEFs (Supplementary Table S3). In addition, we observed that ∼1300 genes were differentially expressed (>2-fold difference) between WT and Adar2-KO MEFs (Figure 5A and Supplementary Table S4). Of these, ∼800 genes were downregulated and another ∼500 genes were upregulated in Adar2-KO MEFs (Supplementary Tables S5 and S6). A large number of genes did not show significant change in their expression in WT and Adar2-KO cells (Supplementary Table S4). These data suggest that ADAR2 influences the cellular levels of a significant fraction of polyadenylated mRNA, including several of the known A-to-I-edited transcripts like Agl and Car5b mRNAs. We validated the RNA-seq results by RT-qPCR analysis for some of the downregulated genes (Figure 5B). Our results revealed that the expression and/or stability of several classes of RNA (long non-coding and protein-coding RNA) were reduced in Adar2-KO cells (Figure 5B). As seen in case of Ctn RNA, the ADAR2-mediated effects on RNA abundance were predominantly A-to-I editing-independent (Figure 5C).
Figure 5.
ADAR2 regulates the stability of various RNAs, including lncRNA Hottip. (A) Heat map of PolyA+ RNA-sequencing of WT and Adar2-KO showing differential gene expression (>2-fold difference). (B) RT-qPCR analysis of representative transcripts downregulated in Adar2-KO MEFs. (C) Plot comparing RNA levels, including that of edited transcripts between WT and Adar2-KO MEFs. The position of Slc7a2 (encodes Ctn RNA and mCat2 mRNA) has been marked in green and indicated by an arrow. (D) Levels of Hottip RNA measured by RNA-seq in WT and Adar2-KO MEFs. (E) Decay rates of Hottip in WT and Adar2-KO MEFs. (F) ADAR2 RIP followed by RT-qPCR to detect interaction between ADAR2 and Hottip in WT MEFs. Normalization of RIP results was carried out by quantifying in parallel the relative levels of Gapdh mRNA in each RIP sample. (G) Levels of Hoxa13 (Hottip target gene) measured by RNA-seq (upper panel) and RT-qPCR (lower panel) in WT and Adar2-KO MEFs (H) Pie chart showing percentage of ADAR2-interacting and non-interacting RNAs (based on ADAR2 RIP in mouse brain cells) (32) that are downregulated in Adar2-KO MEFs. (I–N) ADAR2 RIP followed by RT-qPCR of potential ADAR2-interacting mRNAs in transformed WT MEFs. Normalization of RIP results was carried out by quantifying in parallel the relative levels of Gapdh mRNA in each RIP sample. In all RT-qPCR experiments Gapdh was used as a normalization control as it did not show gene expression changes with any treatment. Error bars represent mean ± SD of three independent experiments (biological replicates). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, ns, not significant using Student's t-test.
Next, we determined if the stability of some of the downregulated transcripts was compromised in Adar2-KO cells. We observed that the gene expression of Hottip, a nuclear long non-coding RNA was reduced in Adar2-KO MEFs (Figure 5B and D). Furthermore, like Ctn RNA, Hottip RNA too exhibited a shorter half-life in Adar2-KO MEFs (2.1 h) in comparison to WT MEFs (5.3 h) (Figure 5E). RIP analysis in WT MEFs showed that ADAR2 interacted with Hottip RNA (Figure 5F). Since Hottip has been shown to activate the transcription of genes in the HoxA cluster, we measured the gene expression of HoxA13, which lies most proximal to Hottip (49), in WT and Adar2-KO cells. RNA-seq and RT-qPCR results showed that HoxA13 showed significant downregulation in Adar2-KO MEFs (Figure 5G). Together, these results show that like Ctn RNA, Hottip RNA is also stabilized by ADAR2. Apart from Ctn RNA and Hottip, we observed that among the validated genes, the stability of Agl and Car5b mRNAs was also reduced in Adar2-KO MEFs (Supplementary Figure S9A and B). Both Agl and Car5b mRNAs are edited transcripts that have structured 3΄UTRs, and harbor inverted SINE elements in their 3΄UTRs (Supplementary Table S3, Supplementary Figure S9C and F). It is possible that several of the other downregulated transcripts in Adar2-KO cells are regulated at the level of transcription, and further studies are needed to determine the potential mechanism(s) by which ADAR2 regulates the expression of these genes.
To determine the significance of ADAR2-mediated downregulation of gene expression, we performed gene ontology (GO) analysis of the genes that are downregulated in Adar2-KO MEFs (Supplementary Figure S10A). GO analysis indicated that ADAR2 influences the expression of genes involved in several vital cellular processes. For example, ADAR2-depleted cells showed downregulation of several immune response genes, including various chemokines and Toll-like receptors (TLRs). We validated the expression of several of these genes by RT-qPCR analysis (Supplementary Figure S10B). Recent studies have shown that the levels of several of the immune response gene mRNAs are found to be elevated in Adar1-KO mouse embryos, indicating that ADAR1 could negatively regulate the expression of immune response genes (6,50). However, our study revealed that unlike ADAR1, ADAR2 positively regulates the expression of a subset of immune-response genes (such as chemokines and TLRs), as Adar2-KO MEFs showed lower expression of these genes. Such effects could possibly be due to increased interaction of ADAR1 with some of these transcripts in the absence of ADAR2. On the other hand, both ADAR1 and ADAR2 could be regulating opposite functions under physiological or pathological conditions, as observed in the case of certain cancers (51). In the present scenario, while ADAR1 is required to suppress aberrant immune responses, ADAR2 could be required to ensure proper expression of immune response genes.
Next, we investigated if ADAR2 could interact with many of the downregulated transcripts. To identify ADAR2-interacting transcripts, we compared the list of genes that showed downregulation in Adar2-KO MEFs in the RNA-seq (in the present study) with the previously published ADAR2 RIP-microarray dataset in mouse brain (32) (Supplementary Table S7 and S8). Among the ∼400 mRNAs showing significant levels of expression in the brain, ADAR2 RIP microarray data showed that ∼60% (∼240) of them interacted with ADAR2 (Figure 5H). ADAR2 RIP followed by RT-qPCR analysis in WT MEFs as well as RT-qPCR analysis in WT and Adar2-KO cells revealed that several of the transcripts (five out of seven mRNAs tested) that were downregulated in Adar2-KO cells interacted with ADAR2 in WT cells (Figure 5H–N and Supplementary Figure S11).
ADAR2 stabilizes a subset of RNA by limiting the binding of RNA-destabilizing proteins HuR and PARN
Next, we investigated if, like Ctn RNA, ADAR2 also stabilized other transcripts by antagonizing the binding of HuR and PARN. To test this possibility, we determined the interaction of HuR with four of the RNAs –Hottip, Agl, Car5b and H19– those were downregulated in Adar2-KO MEFs, in WT and Adar2-KO MEFs. HuR-RIP revealed that Hottip, Agl and Car5b showed increased interaction with HuR specifically in Adar2-KO MEFs (Figure 6A–C). However, H19 (which is downregulated in Adar2-KO MEFs) showed similar interaction with HuR in both WT and Adar2-KO cells, suggesting that HuR is not involved in destabilizing H19 in the Adar2-KO MEFs (Supplementary Figure S12). In the case of Hottip, we further investigated the involvement of ADAR2 in the association of PARN with RNA. We observed increased association of PARN with Hottip in the Adar2-KO MEFs (Figure 6D). Finally, like Ctn RNA, Hottip RNA also showed increased cellular levels and stability upon depletion of PARN and HuR in Adar2-KO MEFs (Figure 6E and F). Based on all these results, we conclude that ADAR2 antagonizes the binding of HuR and PARN to its cognate RNA substrates, such as Ctn RNA and Hottip RNA, in order to stabilize them (Figure 6G).
Figure 6.
ADAR2 stabilizes RNAs by limiting the binding of RNA destabilizing proteins—HuR and PARN. (A–C) HuR RIP or (D) PARN RIP followed by RT-qPCR to measure the interaction of HuR with Hottip, Agl and Car5b and (D) PARN with Hottip RNAs in WT and Adar2–KO MEFs. Normalization of RIP results was carried out by quantifying in parallel the relative levels of Gapdh mRNA in each RIP sample. (E) RT-qPCR to detect Hottip levels in Ctrl and Parn-siRNA treated Adar2-KO MEFs. (F) Decay curve of Hottip RNA in Ctrl and HuR-depleted Adar2-KO MEFs. (G) Model depicting potential role of ADAR2 in stabilizing a sub-set of RNA by preventing the association of RNA-destabilizing factors to these RNAs. In all RT-qPCR experiments Gapdh was used as a normalization control as it did not show gene expression changes with any treatment. Error bars represent mean ± SD of three independent experiments (biological replicates). Statistical significance was determined by Student's t-test.
DISCUSSION
Only a limited number of studies have provided insights into the function of ADAR association with non-coding parts of the transcriptome, such as the 3΄UTR (16,17). In the present study, we focused on A-to-I-edited Ctn RNA to investigate the functional significance of ADAR interaction with its 3΄UTR. Our findings reveal that ADAR2 promotes the stability of Ctn RNA. RNA-seq analysis in Adar2-KO cells indicated potential involvement of ADAR2 in regulating the expression and/or stability of a large number of cellular transcripts. In addition, we have provided evidence for editing independent role of ADAR2 in regulating the expression and/or stability of transcripts. Several recent studies suggested editing-independent roles of ADARs (8,26–28,52). For example, ADARs are known to influence cellular abundance of miRNAs, potentially through editing-independent activities (27,28). An earlier study reported that ADAR1 by interacting with NF90 (nuclear factor 90) family of proteins influences NF90-mediated gene expression, in an editing-independent manner (26).
Besides the present study, a few other studies have attempted genome-wide transcriptome analyses in WT and Adar-KO or knockdown (KD) cells to determine the role of ADARs in gene expression (5,8). In one of the studies, by performing microarray analysis in WT and Adar2-KO brain cells, authors identified ∼80 transcripts that were upregulated upon Adar2 deletion. We believe that with the enhanced depth and coverage of RNA sequencing technology, our study provides a more comprehensive analysis of gene expression changes in WT and Adar2-KO MEFs (5). More recently, Wang et al. (8), have performed RNA-seq analysis of ADAR1 and ADAR2 KD B-cells (8). Their study indicated that ADAR1 or ADAR2 control the expression of a large number of genes. For example, ADAR1-depelted B-cells showed significant changes in the expression of ∼1238 genes. Similarly, ∼5000 genes showed altered expression in ADAR2-depleted B-cells (including 4154 transcripts increased by 2-fold, and those of 872 transcripts decreased by 2-fold). Thus, ADAR2 displays broader effect on gene expression compared to ADAR1. Furthermore, in the case of ADAR1, authors reported that the changes in gene expression observed in ADAR1-depleted cells did not correlate with the deamination activity of ADAR1 (8). Interestingly, in B-cells ADAR1 is shown to increase the stability of a subset of transcripts by facilitating the association of HuR to these mRNAs. However, in the present study we found that in MEFs, ADAR2 stabilizes several RNAs by preventing the association of HuR with mRNA. Thus, ADARs impact both the RNA stabilizing and destabilizing functions of HuR. The differences observed in these two studies could be attributed to cell type-specific differences in ADAR function. Alternatively, both ADAR1 and ADAR2 could perform opposite functions by differentially regulating the activities of RBPs such as HuR. ADAR1 and ADAR2 have been shown to have opposing functions in cancer, possibly by partnering with different proteins or differentially regulating the activities the same RBP (51). In the case of Ctn RNA, ADAR2 could recruit another yet unknown RBP to Ctn RNA, which then prevents the interaction of HuR with Ctn RNA. Future studies will be focused on identifying the mechanism by which ADAR2 prevents the interaction of HuR with Ctn RNA.
RBPs modulate the stability of RNAs by regulating the accessibility of RNAs to various stabilizing or destabilizing factors. ADAR2 regulates the stability of Ctn RNA and Hottip by preventing the association of HuR and PARN deadenylase to these RNAs. While HuR predominantly stabilizes RNA, other reports have described an RNA-destabilizing activity for HuR. For example, HuR uses AUF1 and Ago2 as co-factors in promoting the decay of p16(INK4) mRNA (53). HuR represses MYC mRNA levels in a let-7-dependent manner (42). HuR promotes the early steps of myogenesis by destabilizing nucleophosmin/NPM mRNA (46). HuR also promotes the decay of HOTAIR and linc-p21 lncRNAs by recruiting the Ago2/let7 complex (30,54). While neuronal ELAVL (which belongs to the same family of proteins as HuR/ELAVL1) stabilized ∼68% of its interacting target mRNAs, it also destabilized ∼7% of its interacting mRNAs (55). Collectively, these studies indicate proteins like HuR/ELAV though mostly involved in stabilizing RNAs, could also destabilize RNAs in a context-dependent manner. Both ADAR2 and HuR are known to interact with the 3΄UTR of several RNAs. A previous study showed the enrichment of HuR binding motifs near A-to-I edited regions (8). HuR consensus binding sites are found to be enriched ∼100 nt upstream and downstream of A-to-I edited sites. In the case of Ctn RNA too, we observed potential HuR binding sites near the repeat sequences (or the edited regions). Instead of direct competition, it is possible that in presence of ADAR2, HuR is unable to access its interaction sites in the 3΄UTR of Ctn RNA or other tested RNAs. Previous studies have suggested that ADARs could influence the binding of other RNA binding proteins to mRNA sequences that are located in close proximity. A recent ADAR1-CLIP-seq study has shown that ADAR1 precludes the binding of other 3΄UTR interacting RNA binding proteins such as CFIm68 and CstF64 even though they are not competing for the same binding sites (56). In this study, authors showed that CFIm68 or CstF64 binding motifs do not overlap with ADAR1 interacting sequences within the 3΄UTRs. Another study has shown that ADAR1 competes with Staufen-1 for binding to double-stranded regions in the 3΄UTR and that in turn affects the nuclear retention of transcripts (40).
Deadenylases such as PARN modulate the poly(A) tail length and thus, are important regulators of RNA stability (57). Deadenylation is a highly regulated process and the recruitment of deadenylase complexes to mRNA is regulated by several RBPs such as KSRP, CUG-BP and AU-rich element (ARE)-binding proteins such as HuR (46,58). Our results strongly support a role for HuR as a co-factor that is required for the association of PARN-containing complex to Ctn RNA, in order to negatively regulate RNA stability. Even though most of HuR is localized in the nucleus, most of the HuR-related functions have been attributed to the small cytoplasmic pool of HuR (59,60). Since HuR, PARN and ADAR2 localize in the nucleus, it is possible that they could compete or cooperate with each other in order to influence the stability of nuclear pool of RNAs (61,62).
Our studies highlight a novel mechanism whereby ADAR2 regulates RNA stability by limiting the interaction of RNAs with the decay-promoting proteins HuR and PARN deadenylase. Collectively, these studies imply that ADARs perform editing-dependent and -independent roles, and also influence the interaction of other RBPs to RNA, thereby controlling vital processes such as RNA stability. In addition, our study underscores the importance of the interaction of ADARs and/or editing within the non-coding regions of hundreds of RNAs in the mammalian transcriptome.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Drs K Nishikura (Wistar Institute, USA, [FLAG-ADAR2 plasmid]), C Wilusz (Colorado State University, USA, [Parn plasmid]), L Maquat (University of Rochester Medical Center, USA, [Staufen1 plasmid]) for the gift of reagents. We would also like to thank Prasanth lab members and Drs A Kalsotra (UIUC), A Lal (NCI, NIH) for critical reading of this manuscript.
Footnotes
Present address: Vidisha Tripathi, National Center for Cell Science, Ganeshkhind, Pune 411007, India.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institute of Health [1RO1GM088252 to K.V.P., 1RO1GM099669 to S.G.P.]; American Cancer Society [RSG-11-174-01-RMC to K.V.P.]; National Science Foundation career [1243372 to S.G.P.]; National Institute on Aging—Intramural Research Program, National Institute of Health [Z01-AG000511 to J.H.Y., M.G.]. Funding for open access charge: NIGMS, NIH [RO1GM088252].
Conflict of interest statement. None declared.
REFERENCES
- 1. Hundley H.A., Bass B.L.. ADAR editing in double-stranded UTRs and other noncoding RNA sequences. Trends Biochem. Sci. 2010; 35:377–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 2010; 79:321–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hartner J.C., Schmittwolf C., Kispert A., Muller A.M., Higuchi M., Seeburg P.H.. Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J. Biol. Chem. 2004; 279:4894–4902. [DOI] [PubMed] [Google Scholar]
- 4. Higuchi M., Maas S., Single F.N., Hartner J., Rozov A., Burnashev N., Feldmeyer D., Sprengel R., Seeburg P.H.. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000; 406:78–81. [DOI] [PubMed] [Google Scholar]
- 5. Horsch M., Seeburg P.H., Adler T., Aguilar-Pimentel J.A., Becker L., Calzada-Wack J., Garrett L., Gotz A., Hans W., Higuchi M. et al. Requirement of the RNA-editing enzyme ADAR2 for normal physiology in mice. J. Biol. Chem. 2011; 286:18614–18622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liddicoat B.J., Piskol R., Chalk A.M., Ramaswami G., Higuchi M., Hartner J.C., Li J.B., Seeburg P.H., Walkley C.R.. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science. 2015; 349:1115–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pestal K., Funk C.C., Snyder J.M., Price N.D., Treuting P.M., Stetson D.B.. Isoforms of RNA-editing enzyme ADAR1 independently control nucleic acid sensor MDA5-driven autoimmunity and multi-organ development. Immunity. 2015; 43:933–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang I.X., So E., Devlin J.L., Zhao Y., Wu M., Cheung V.G.. ADAR regulates RNA editing, transcript stability, and gene expression. Cell Rep. 2013; 5:849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bazak L., Levanon E.Y., Eisenberg E.. Genome-wide analysis of Alu editability. Nucleic Acids Res. 2014; 42:6876–6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim D.D., Kim T.T., Walsh T., Kobayashi Y., Matise T.C., Buyske S., Gabriel A.. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res. 2004; 14:1719–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Levanon E.Y., Eisenberg E., Yelin R., Nemzer S., Hallegger M., Shemesh R., Fligelman Z.Y., Shoshan A., Pollock S.R., Sztybel D. et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 2004; 22:1001–1005. [DOI] [PubMed] [Google Scholar]
- 12. Peng Z., Cheng Y., Tan B.C., Kang L., Tian Z., Zhu Y., Zhang W., Liang Y., Hu X., Tan X. et al. Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nat. Biotechnol. 2012; 30:253–260. [DOI] [PubMed] [Google Scholar]
- 13. Ramaswami G., Lin W., Piskol R., Tan M.H., Davis C., Li J.B.. Accurate identification of human Alu and non-Alu RNA editing sites. Nat. Methods. 2012; 9:579–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mallela A., Nishikura K.. A-to-I editing of protein coding and noncoding RNAs. Crit. Rev. Biochem. Mol. Biol. 2012; 47:493–501. [DOI] [PubMed] [Google Scholar]
- 15. Ulbricht R.J., Emeson R.B.. One hundred million adenosine-to-inosine RNA editing sites: hearing through the noise. Bioessays. 2014; 36:730–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen L.L., Carmichael G.G.. Nuclear editing of mRNA 3΄-UTRs. Curr. Top. Microbiol. Immunol. 2012; 353:111–121. [DOI] [PubMed] [Google Scholar]
- 17. Daniel C., Veno M.T., Ekdahl Y., Kjems J., Ohman M.. A distant cis acting intronic element induces site-selective RNA editing. Nucleic Acids Res. 2012; 40:9876–9886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ivanov A., Memczak S., Wyler E., Torti F., Porath H.T., Orejuela M.R., Piechotta M., Levanon E.Y., Landthaler M., Dieterich C. et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015; 10:170–177. [DOI] [PubMed] [Google Scholar]
- 19. Rueter S.M., Dawson T.R., Emeson R.B.. Regulation of alternative splicing by RNA editing. Nature. 1999; 399:75–80. [DOI] [PubMed] [Google Scholar]
- 20. Chen L.L., DeCerbo J.N., Carmichael G.G.. Alu element-mediated gene silencing. EMBO J. 2008; 27:1694–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Prasanth K.V., Prasanth S.G., Xuan Z., Hearn S., Freier S.M., Bennett C.F., Zhang M.Q., Spector D.L.. Regulating gene expression through RNA nuclear retention. Cell. 2005; 123:249–263. [DOI] [PubMed] [Google Scholar]
- 22. Hu S.B., Xiang J.F., Li X., Xu Y., Xue W., Huang M., Wong C.C., Sagum C.A., Bedford M.T., Yang L. et al. Protein arginine methyltransferase CARM1 attenuates the paraspeckle-mediated nuclear retention of mRNAs containing IRAlus. Genes Dev. 2015; 29:630–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Capshew C.R., Dusenbury K.L., Hundley H.A.. Inverted Alu dsRNA structures do not affect localization but can alter translation efficiency of human mRNAs independent of RNA editing. Nucleic Acids Res. 2012; 40:8637–8645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hundley H.A., Krauchuk A.A., Bass B.L.. C. elegans and H. sapiens mRNAs with edited 3΄ UTRs are present on polysomes. RNA. 2008; 14:2050–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Soundararajan R., Stearns T.M., Griswold A.L., Mehta A., Czachor A., Fukumoto J., Lockey R.F., King B.L., Kolliputi N.. Detection of canonical A-to-G editing events at 3΄ UTRs and microRNA target sites in human lungs using next-generation sequencing. Oncotarget. 2015; 6:35726–35736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nie Y., Ding L., Kao P.N., Braun R., Yang J.H.. ADAR1 interacts with NF90 through double-stranded RNA and regulates NF90-mediated gene expression independently of RNA editing. Mol. Cell. Biol. 2005; 25:6956–6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vesely C., Tauber S., Sedlazeck F.J., Tajaddod M., von Haeseler A., Jantsch M.F.. ADAR2 induces reproducible changes in sequence and abundance of mature microRNAs in the mouse brain. Nucleic Acids Res. 2014; 42:12155–12168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vesely C., Tauber S., Sedlazeck F.J., von Haeseler A., Jantsch M.F.. Adenosine deaminases that act on RNA induce reproducible changes in abundance and sequence of embryonic miRNAs. Genome Res. 2012; 22:1468–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tripathi V., Ellis J.D., Shen Z., Song D.Y., Pan Q., Watt A.T., Freier S.M., Bennett C.F., Sharma A., Bubulya P.A. et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell. 2010; 39:925–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yoon J.H., Abdelmohsen K., Srikantan S., Yang X., Martindale J.L., De S., Huarte M., Zhan M., Becker K.G., Gorospe M.. LincRNA-p21 suppresses target mRNA translation. Mol. Cell. 2012; 47:648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Conrad N.K., Mili S., Marshall E.L., Shu M.D., Steitz J.A.. Identification of a rapid mammalian deadenylation-dependent decay pathway and its inhibition by a viral RNA element. Mol. Cell. 2006; 24:943–953. [DOI] [PubMed] [Google Scholar]
- 32. Ohlson J., Enstero M., Sjoberg B.M., Ohman M.. A method to find tissue-specific novel sites of selective adenosine deamination. Nucleic Acids Res. 2005; 33:e167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Herb A., Higuchi M., Sprengel R., Seeburg P.H.. Q/R site editing in kainate receptor GluR5 and GluR6 pre-mRNAs requires distant intronic sequences. Proc. Natl. Acad. Sci. U.S.A. 1996; 93:1875–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Melcher T., Maas S., Herb A., Sprengel R., Higuchi M., Seeburg P.H.. RED2, a brain-specific member of the RNA-specific adenosine deaminase family. J. Biol. Chem. 1996; 271:31795–31798. [DOI] [PubMed] [Google Scholar]
- 35. Melcher T., Maas S., Herb A., Sprengel R., Seeburg P.H., Higuchi M.. A mammalian RNA editing enzyme. Nature. 1996; 379:460–464. [DOI] [PubMed] [Google Scholar]
- 36. Ohlson J., Pedersen J.S., Haussler D., Ohman M.. Editing modifies the GABA(A) receptor subunit alpha3. RNA. 2007; 13:698–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Macbeth M.R., Schubert H.L., Vandemark A.P., Lingam A.T., Hill C.P., Bass B.L.. Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing. Science. 2005; 309:1534–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ryman K., Fong N., Bratt E., Bentley D.L., Ohman M.. The C-terminal domain of RNA Pol II helps ensure that editing precedes splicing of the GluR-B transcript. RNA. 2007; 13:1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Valente L., Nishikura K.. RNA binding-independent dimerization of adenosine deaminases acting on RNA and dominant negative effects of nonfunctional subunits on dimer functions. J. Biol. Chem. 2007; 282:16054–16061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Elbarbary R.A., Li W., Tian B., Maquat L.E.. STAU1 binding 3΄ UTR IRAlus complements nuclear retention to protect cells from PKR-mediated translational shutdown. Genes Dev. 2013; 27:1495–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stellos K., Gatsiou A., Stamatelopoulos K., Perisic Matic L., John D., Lunella F.F., Jae N., Rossbach O., Amrhein C., Sigala F. et al. Adenosine-to-inosine RNA editing controls cathepsin S expression in atherosclerosis by enabling HuR-mediated post-transcriptional regulation. Nat. Med. 2016; 22:1140–1150. [DOI] [PubMed] [Google Scholar]
- 42. Kim H.H., Kuwano Y., Srikantan S., Lee E.K., Martindale J.L., Gorospe M.. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009; 23:1743–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lebedeva S., Jens M., Theil K., Schwanhausser B., Selbach M., Landthaler M., Rajewsky N.. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol. Cell. 2011; 43:340–352. [DOI] [PubMed] [Google Scholar]
- 44. Mukherjee N., Corcoran D.L., Nusbaum J.D., Reid D.W., Georgiev S., Hafner M., Ascano M. Jr, Tuschl T., Ohler U., Keene J.D.. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol. Cell. 2011; 43:327–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Paz I., Kosti I., Ares M. Jr, Cline M., Mandel-Gutfreund Y.. RBPmap: a web server for mapping binding sites of RNA-binding proteins. Nucleic Acids Res. 2014; 42:W361–W367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cammas A., Sanchez B.J., Lian X.J., Dormoy-Raclet V., van der Giessen K., Lopez de Silanes I., Ma J., Wilusz C., Richardson J., Gorospe M. et al. Destabilization of nucleophosmin mRNA by the HuR/KSRP complex is required for muscle fibre formation. Nat. Commun. 2014; 5:4190 (1–16). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lee J.E., Lee J.Y., Trembly J., Wilusz J., Tian B., Wilusz C.J.. The PARN deadenylase targets a discrete set of mRNAs for decay and regulates cell motility in mouse myoblasts. PLoS Genet. 2012; 8:e1002901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kundu P., Fabian M.R., Sonenberg N., Bhattacharyya S.N., Filipowicz W.. HuR protein attenuates miRNA-mediated repression by promoting miRISC dissociation from the target RNA. Nucleic Acids Res. 2012; 40:5088–5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang K.C., Yang Y.W., Liu B., Sanyal A., Corces-Zimmerman R., Chen Y., Lajoie B.R., Protacio A., Flynn R.A., Gupta R.A. et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011; 472:120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mannion N.M., Greenwood S.M., Young R., Cox S., Brindle J., Read D., Nellaker C., Vesely C., Ponting C.P., McLaughlin P.J. et al. The RNA-editing enzyme ADAR1 controls innate immune responses to RNA. Cell Rep. 2014; 9:1482–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chan T.H., Lin C.H., Qi L., Fei J., Li Y., Yong K.J., Liu M., Song Y., Chow R.K., Ng V.H. et al. A disrupted RNA editing balance mediated by ADARs (Adenosine DeAminases that act on RNA) in human hepatocellular carcinoma. Gut. 2014; 63:832–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Heale B.S., Keegan L.P., McGurk L., Michlewski G., Brindle J., Stanton C.M., Caceres J.F., O’Connell M.A.. Editing independent effects of ADARs on the miRNA/siRNA pathways. EMBO J. 2009; 28:3145–3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chang N., Yi J., Guo G., Liu X., Shang Y., Tong T., Cui Q., Zhan M., Gorospe M., Wang W.. HuR uses AUF1 as a cofactor to promote p16INK4 mRNA decay. Mol. Cell. Biol. 2010; 30:3875–3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yoon J.H., Abdelmohsen K., Kim J., Yang X., Martindale J.L., Tominaga-Yamanaka K., White E.J., Orjalo A.V., Rinn J.L., Kreft S.G. et al. Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nat. Commun. 2013; 4:2939 (1–14). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Scheckel C., Drapeau E., Frias M.A., Park C.Y., Fak J., Zucker-Scharff I., Kou Y., Haroutunian V., Ma’ayan A., Buxbaum J.D. et al. Regulatory consequences of neuronal ELAV-like protein binding to coding and non-coding RNAs in human brain. Elife. 2016; 5:1–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bahn J.H., Ahn J., Lin X., Zhang Q., Lee J.H., Civelek M., Xiao X.. Genomic analysis of ADAR1 binding and its involvement in multiple RNA processing pathways. Nat. Commun. 2015; 6:6355 (1–13). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Goldstrohm A.C., Wickens M.. Multifunctional deadenylase complexes diversify mRNA control. Nat. Rev. Mol. Cell Biol. 2008; 9:337–344. [DOI] [PubMed] [Google Scholar]
- 58. Tran H., Schilling M., Wirbelauer C., Hess D., Nagamine Y.. Facilitation of mRNA deadenylation and decay by the exosome-bound, DExH protein RHAU. Mol. Cell. 2004; 13:101–111. [DOI] [PubMed] [Google Scholar]
- 59. Atasoy U., Watson J., Patel D., Keene J.D.. ELAV protein HuA (HuR) can redistribute between nucleus and cytoplasm and is upregulated during serum stimulation and T cell activation. J. Cell Sci. 1998; 111:3145–3156. [DOI] [PubMed] [Google Scholar]
- 60. Nowotarski S.L., Shantz L.M.. Cytoplasmic accumulation of the RNA-binding protein HuR stabilizes the ornithine decarboxylase transcript in a murine nonmelanoma skin cancer model. J. Biol. Chem. 2010; 285:31885–31894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Berndt H., Harnisch C., Rammelt C., Stohr N., Zirkel A., Dohm J.C., Himmelbauer H., Tavanez J.P., Huttelmaier S., Wahle E.. Maturation of mammalian H/ACA box snoRNAs: PAPD5-dependent adenylation and PARN-dependent trimming. RNA. 2012; 18:958–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Desterro J.M., Keegan L.P., Lafarga M., Berciano M.T., O’Connell M., Carmo-Fonseca M.. Dynamic association of RNA-editing enzymes with the nucleolus. J. Cell Sci. 2003; 116:1805–1818. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.