Abstract
P-TEFb (CDK9/cyclin T) plays a central role in androgen receptor (AR)-mediated transactivation by phosphorylating both RNA polymerase 2 complex proteins and AR at S81. CDK9 dephosphorylation mobilizes P-TEFb from an inhibitory 7SK ribonucleoprotein complex, but mechanisms targeting phosphatases to P-TEFb are unclear. We show that AR recruits protein phosphatase 1α (PP1α), resulting in P-TEFb mobilization and CDK9-mediated AR S81 phosphorylation. This increased pS81 enhances p300 recruitment, histone acetylation, BRD4 binding and subsequent further recruitment of P-TEFb, generating a positive feedback loop that sustains transcription. AR S81 is also phosphorylated by CDK1, and blocking basal CDK1-mediated S81 phosphorylation markedly suppresses AR activity and initiation of this positive feedback loop. Finally, androgen-independent AR activity in castration-resistant prostate cancer (CRPC) cells is driven by increased CDK1-mediated S81 phosphorylation. Collectively these findings reveal a mechanism involving PP1α, CDK9 and CDK1 that is used by AR to initiate and sustain P-TEFb activity, which may be exploited to drive AR in CRPC.
INTRODUCTION
The androgen receptor (AR) plays a central role in prostate cancer (PCa) development, and most patients initially respond to androgen deprivation therapy (ADT, surgical or medical castration), but the tumors inevitably recur despite castrate levels of androgen (castration-resistant prostate cancer, CRPC). AR expression is generally increased in CRPC, and one mechanism further driving its activity is intratumoral synthesis of androgens from adrenal gland derived precursors or de novo from cholesterol (1,2). This activity can be suppressed by drugs including abiraterone (which inhibits the enzyme CYP17A1 required for androgen synthesis) or by the direct AR antagonist enzalutamide, and both abiraterone and enzalutamide are now FDA approved for treatment of CRPC. However, patients who respond to these agents generally relapse within 1–2 years and AR appears to still be contributing to the growth of these relapsed tumors in at least a substantial subset of patients. Therefore, there remains a critical need to further elucidate mechanisms contributing to AR activity and identify novel therapeutic targets.
AR activity can be modulated by phosphorylation at multiple sites (3), with serine 81 (S81) in the N-terminal domain being the major site that undergoes phosphorylation in response to androgen (4). S81 phosphorylation can enhance AR binding of the coactivator EP300 (p300), with subsequent acetylation of histones and possibly AR, and an S81A mutation can markedly impair AR activity (5–7). In androgen-dependent PCa cells, the androgen-stimulated phosphorylation of S81 increases gradually over about 8 h and appears to be dependent on AR binding to chromatin, as it can be prevented by a mutation in the AR DNA binding domain (4,8). This S81 phosphorylation is mediated primarily by CDK9, presumably in association with cyclin T (6). The CDK9/cyclin T complex (positive transcriptional elongation factor b, P-TEFb) is recruited to enhancer/promoter sites by BRD4, where its primary function is to phosphorylate NELF (Negative Elongation Factor) and serine 2 in the RNA polymerase 2 C-terminal domain (CTD), and thereby stimulate transcriptional elongation (9–11).
A large fraction of P-TEFb is held in an inactive state by association with a complex containing 7SK RNA and HEXIM1, and can be released from this complex through dephosphorylation of CDK9 by protein phosphatases, including protein phosphatase 1 (PP1) (12–17). There are three isoforms of the PP1 catalytic subunit, with PP1α being the major isoform. PP1α associates with a large number of regulatory proteins that determine its localization and activity (18). A regulatory protein targeting PP1 to chromatin is PNUTS (PP1 Nuclear Targeting Subunit), but additional mechanisms that may target PP1α to chromatin to drive CDK9 dephosphorylation and P-TEFb mobilization remain to be identified (19). We reported previously that AR could interact with PP1α, and that PP1α could enhance AR activity by dephosphorylating S650 in the AR hinge region and thereby prevent its nuclear export (20,21). We further found that PP1α could increase AR protein stability by suppressing the activity of ubiquitin ligases that target AR (22). In this study we show that AR also recruits PP1α to chromatin, where it mediates the dephosphorylation and mobilization of CDK9 to both drive transcriptional elongation and AR S81 phosphorylation, leading to further CDK9 recruitment via p300 and BRD4 to drive AR transcriptional activity.
While AR S81 phosphorylation in response to androgen stimulation is mediated primarily by CDK9, this site can also be phosphorylated by CDK1 (23). CDK1 activity is increased in CRPC (24), and we showed previously that increased CDK1 activity could sensitize PCa cells to low levels of androgen and may thereby contribute to AR activity in CRPC (23). In this study we show that basal (in steroid-depleted medium) AR S81 phosphorylation and transcriptional activity in CRPC cells are mediated by CDK1, and cannot be suppressed by AR antagonists. Moreover, we show that CDK1 inhibition in both androgen-dependent and independent PCa cells suppresses responses to androgen stimulation. Taken together our findings indicate that low levels of basal CDK1-mediated AR S81 phosphorylation are important to initiate transcription in response to androgen, which can then stimulate a positive feedback loop with recruitment PP1α and BRD4, mobilization and recruitment of P-TEFb, and subsequent further AR S81 phosphorylation. We hypothesize that increased CDK1 activity in CRPC acts through this positive feedback loop, and that therapies targeting components of this loop may be effective in CRPC.
MATERIALS AND METHODS
Reagents
Compounds and vendors are as follows: dihydrotestosterone (DHT) and nocodazole (Sigma-Aldrich); R1881 (PerkinElmer); RO-3306 (Alexis Biochemicals); CDK9 Inhibitor II (EMD Millipore); C646 (Xcess Biosciences); Dinaciclib (SCH-727965, Selleckchem); Tautomycin (Calbiochem and Enzo Life Science); Fostriecin, Roscovitine, CGP74514A, Olomoucine II, CDK1/2 inhibitor and CDK1/5 inhibitor (Calbiochem). The Pfizer CDK1 inhibitor (PF-00176275 or AG024322) was kindly provided by Pfizer Inc. The BRD4 antagonist JQ1 was kindly provided by Dr J. Bradner (DFCI, Boston, MA, USA). FBS (fetal bovine serum) and CDS (charcoal-dextran stripped serum) were from Hyclone. AR constructs and mutants, and the CDK1-AF and cyclin B1 (CCNB1) expression plasmids have been described previously (5,22,23). siRNAs are as below: control siRNA (Dharmacon, Cat. D-001810-01-05); CDK1 siRNAs (Dharmacon, Cat. M-003224-03; Cell Signaling, Cat. 3500S), PP1α siRNAs (Dharmacon, Cat. M-008927-00; Santa Cruz, Cat. Sc-36299); and AR siRNA (AR-1981, target sequence: AGGUUCUCUGCUAGACGACdTdT). The Flag-tagged AR-W741C-S650A(Ser650Ala) plasmid was generated based on a reported vector (5) using DNA mutagenesis.
Antibodies and immunoblotting
The sources for the antibodies and control IgGs were as following: AR (Santa Cruz, Cat. sc-816); pAR-S81 (EMD Millipore, Cat. 07–1375); pAR-S308 (Santa Cruz, Cat. sc-26406); pRNA Pol II Ser2 (Abcam, Cat., ab5095); pRNA Pol II Ser5 (Abcam, Cat. ab5131); CDK1 (Cell Signaling, Cat. 9112); pCDK1-T161 (Cell Signaling, Cat. 9114); CDK9 (Santa Cruz, Cat. sc-8338); Cyclin T1 (Santa Cruz, Cat. sc-10750); BRD4 (Bethyl, Cat. A301-985A); p300 (Santa Cruz, Cat. sc-585); Histone 3 (Abcam, Cat. ab1791); H3K27Ac (Abcam, Cat. ab4729); pH3-Ser10 (EMD Millipore, Cat. 06–570); FoxA1 (Abcam, Cat. Ab23738); PSA (Meridian Life Science, Cat. K92110R); Flag-M2 (Sigma-Aldrich, Cat. F3165); HA (Cell Signaling, Cat. 3724); β-Tubulin (EMD Millipore, Cat. MAB3408); β-Actin (Abcam, Cat. Ab6276); GAPDH (Abcam, Cat. Ab9485); Hsp90 (Santa Cruz, Cat. sc-69703), PP1 (Santa Cruz: Cat. sc-443; Cat. sc-6104; and Cat. sc-6105), and control IgGs (Santa Cruz: normal rabbit IgG, Cat. sc-2027; and normal goal IgG, Cat. sc-2028). Protein A and protein G was from Pierce (Cat. 20334 and Cat. 20399, respectively). The anti-flag M2 affinity gel was from Sigma-Aldrich (Cat. A2220). Western blots were developed using X-Ray film (Research Products International) and the Western Lightning Plus-ECL reagent (PerkinElmer). Images were acquired using a CanoScan LiDE 210 scanner.
Peptide competition assay
For peptides: three indicated peptides (AR-S81-P: Q Q Q E T [pSER] P R Q Q Q; AR-S81-C: Q Q Q E T S P R Q Q Q; AR-S650-P: A S S T T [pSER] P T E E T) were from Genscript USA. The peptides were suspended in solvent A (99.8% Water, 0.1% Acetonitrile and 0.1% Trifluoroacetic acid) and aliquots were stored at −80°C. For blocking, indicated folds (in molar concentration as compared to the primary antibody) of peptides were added to the primary antibody suspension, and then incubated by rotation at room temperature for 2 h, followed by proceeding to blotting.
Cell culture and transfection
LNCaP were grown in RPMI-1640 containing 10% FBS; the LNCaP-Abl cell line was maintained in phenol-red free RPMI-1640 medium (Gibco) containing 10% CDS; C4-2 and VCaP cells were grown in Dulbecco's modified Eagle's medium (DMEM) medium containing 10% FBS; and 293T cells were grown in DMEM medium with 5% FBS. Cell line identity was confirmed based on short tandem repeat testing. For androgen-starving conditions, cells were grown in medium containing CDS. Plasmids and siRNA transfections were carried out using the Lipofectamine 2000 (Invitrogen), following the manufacturer's directions.
Chromatin immunoprecipitation (ChIP) and qPCR
ChIP analysis was as previously described (25). Antibodies used were anti- AR (Santa Cruz, Cat. sc-816), anti-pAR-S81 (EMD Millipore, Cat. 07–1375), anti-pRNA Pol II Ser2 (Abcam, Cat., ab5095), anti-pRNA Pol II Ser5 (Abcam, Cat. ab5131), anti-CDK9 (Santa Cruz, Cat. sc-8338), anti-cyclin T1 (Santa Cruz, Cat. sc-10750), anti-BRD4 (Bethyl, Cat. A301-985A); anti-p300 (Santa Cruz, Cat. sc-585), anti-H3K27Ac (Abcam, Cat. ab4729), anti-PP1α (combined Santa Cruz sc-6104 and sc-6105). Primers for ChIP were: PSA-enhancer: Forward, 5΄-GCCTGGATCTGAGAGAGATATCATC-3΄; Reverse, 5΄-ACACCTTTTTTTTTCTGGATTGTTG-3΄; PSA-promoter: Forward, 5΄-TCCTGAGTGCTGGTGTCTTAG-3΄; Reverse, 5΄-CAGGATGAAACAGAAACAGGG-3΄; KLK2-Enhancer: Forward, 5΄-GCCTTTGCTCAGAAGACACA-3΄; Reverse, 5΄-ACAAGAGTGGAAGGCTCTGG -3΄; KLK2-Promoter: Forward, 5΄-CCTGTTGCTGTTCATCCTGA-3΄; Reverse, 5΄-CCTATGGATCATGGAGATGTGA-3΄.
RNA isolation and qRT-PCR analysis
RNA isolation was carried out using the TriZOL reagent (Ambion) and the qRT-PCR analysis on gene expression was performed with the TaqMan One-Step RT-PCR Master Mix Reagents (Applied Biosystems). The TaqMan primers were: KLK2 pre-mRNA: Forward, 5΄-GCCACAGCCCAGTTTTTCTC-3΄; Reverse, 5΄-TGGAGGCTCACACACTGAAGA-3΄; Probe, 5΄-FAM-ACCCATAGTCTTGCGCCCCAGGA-3΄. The TaqMan primer-probe sets for KLK2 and KLK3/PSA (FAM labeled) and the internal control GAPDH (VIC-TAMRA labeled) transcripts were purchased as inventoried mixes from Applied Biosystems.
Co-immunoprecipitations (Co-IP)
Cells were harvested in triton lysis buffer (TLB) containing protease inhibitors, as described previously (Chen et al. (20)). The lysates were incubated on ice for 20 min and centrifuged for 5 min at maximum speed. 20 μl of the supernatant was saved as input and the rest was mixed with anti-flag M2 beads, followed by incubation at 4°C on a rotator for 2 h. Then the input and washed (4×) beads were boiled in 60 μl of Laemmli buffer containing 5% β-mercaptoethanol.
Cellular fractionation and cell proliferation analysis
The cellular fractionation assay was performed with the Subcellular Protein Fractionation Kit (Pierce), following the manufacturer's direction. Cell proliferation assay was carried out as below: cells were cultured in 96-well plate for 2 days and then treated for 3 days as indicated. The cell counting analysis was performed with the CellTiter-Glo assay kit (Promega), following the manufactures’ manual.
Statistical analyses
Data in bar graphs represent mean ± SD of three technical replicates, and are representative of at least three independent experiments. Results for immunoblotting are representative of at least three independent experiments.
RESULTS
AR S81 phosphorylation by CDK9 is tightly linked to chromatin binding and transcriptional activation
Consistent with previous studies, androgen-dependent LNCaP PCa cells have very low levels of AR pS81 in the absence of exogenous androgens (cultured in medium with charcoal/dextran stripped serum, CSS medium), these levels can be increased by treatment with androgen (DHT) and this increase can be suppressed by cotreatment with direct AR antagonists (bicalutamide or enzalutamide) or by a CDK9 inhibitor (CDK9 inhibitor II, iII) (Figure 1A). The specificity of the AR pS81 antibody was supported by peptide blocking experiments (Supplementary Figure S1). These results support the conclusion that S81 phosphorylation of the chromatin-associated AR is mediated by CDK9, presumably in concert with cyclin T in the P-TEFb complex.
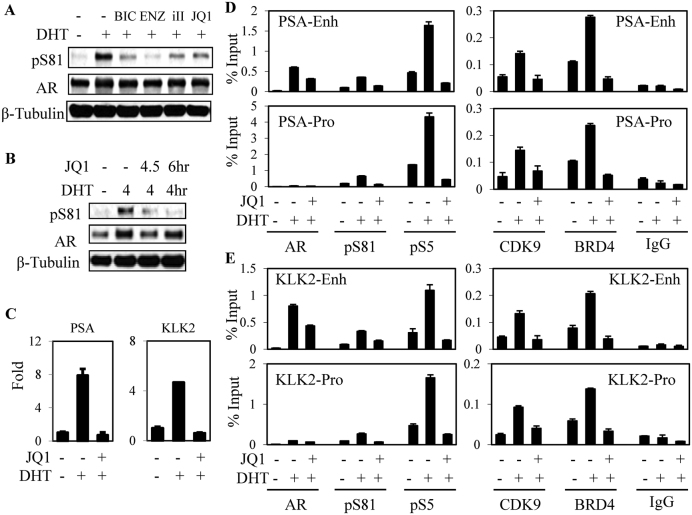
Figure 1.
Androgen-stimulated AR Ser81 phosphorylation is associated with chromatin occupancy of BRD4 and P-TEFb. (A) LNCaP cells in steroid-depleted medium (RPMI with charcoal/dextran stripped serum, CDS medium) were pre-treated for 30 min with AR antagonist (bicalutamide: BIC, 10 μM; or enzalutamide: ENZ, 10 μM), CDK9 inhibitor (CDK9 inhibitor II: iII, 50 μM) or JQ1 (500 nM) as indicated, and then treated for 4 h with 10 nM DHT followed by lysis and immunoblotting. (B and C) LNCaP cells in CDS medium were treated for 4 h with DHT and BRD4 antagonist JQ1 (500 nM) as indicated, followed by immunoblotting (B) or RNA isolation and qRT-PCR analysis (C). (D and E) LNCaP cells in CDS medium were treated for 4 h with DHT and JQ1 as indicated, followed by ChIP-qPCR for occupancy of AR, pS81 (S81 phosphorylated AR), pS5 (S5 phosphorylated RNA Pol II), CDK9 and BRD4 on AR-regulated elements (AREs) in the KLK3/PSA (D) and KLK2 (E) genes.
Recruitment of P-TEFb to chromatin can be mediated through an interaction with the bromodomain protein BRD4 (26,27), which binds to acetylated histones and has been reported to also interact directly with AR (28). Therefore, to further assess whether AR S81 phosphorylation is mediated by chromatin-associated P-TEFb, we used JQ1 (which blocks BRD4 binding to acetylated lysines) to suppress BRD4 binding to chromatin. JQ1 suppressed the DHT-stimulated increase in pS81 (Figure 1A and B) and, consistent with previous reports, prevented induction of the strongly AR regulated PSA and KLK2 genes (Figure 1C). Using ChIP-qPCR we further confirmed that JQ1 blocked the DHT-stimulated recruitment of BRD4 and CDK9 to the PSA enhancer and promoter (Figure 1D). AR binding was similarly decreased, and there was a marked decrease in levels of pS81 AR associated with these sites. Finally, binding of active (phosphorylation on S5) RNA polymerase 2 (pS5) was also decreased and similar effects were seen on the KLK2 enhancer and promoter (Figure 1E). These findings further establish that BRD4 mediates P-TEFb recruitment to these AR-regulated genes, and that chromatin-associated CDK9 mediates AR S81 phosphorylation.
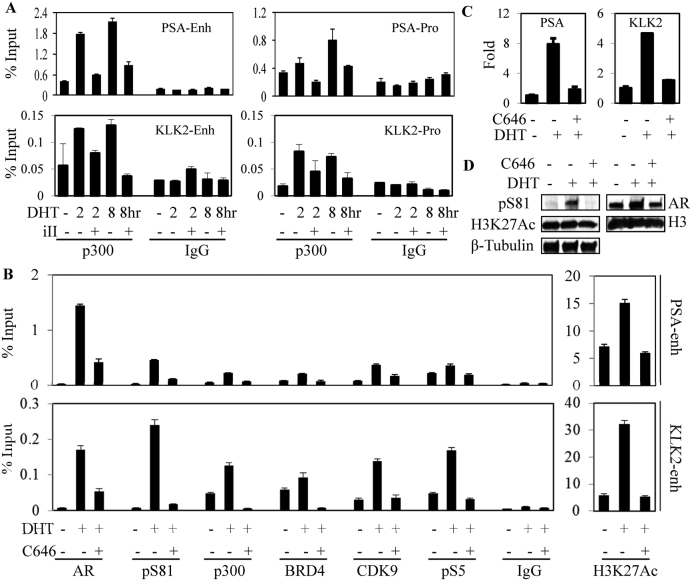
AR S81 phosphorylation has been reported to enhance binding of the histone acetyltransferase EP300 (p300) (7), suggesting that p300 binding and subsequent histone acetylation may drive a positive feedback loop through increased BRD4 recruitment, a subsequent increase in P-TEFb recruitment and further AR S81 phosphorylation. Consistent with pS81 enhancement of p300 binding, suppressing S81 phosphorylation by treatment with a CDK9 inhibitor (iII) decreased DHT stimulated binding of p300 to the PSA and KLK2 genes (Figure 2A). It should be noted that iII also impairs AR recruitment to chromatin (5), so the decrease in p300 binding may reflect both decreased total and S81 phosphorylated AR. We next used an inhibitor of p300 acetyltransferase activity (C646) to further dissect the role of p300 in AR mediated transcriptional activation. Treatment with C646 prevented DHT stimulated H3K27 acetylation on the PSA and KLK2 gene enhancers, and impaired the DHT stimulated binding of AR to these genes (Figure 2B). Significantly, C646 completely blocked the DHT-stimulated recruitment of BRD4, CDK9 and RNA polymerase 2, as well as the CDK9-mediated phosphorylation of AR S81. Binding of p300 was also abrogated, consistent with its binding being enhanced by AR S81 phosphorylation and by histone acetylation. As expected, C646 prevented the DHT stimulated induction PSA and KLK2 mRNA (Figure 2C). Finally, by immunoblotting we found that C646 globally suppressed DHT stimulated AR S81 phosphorylation (Figure 2D). It should be noted that while iII and JQ1 had no clear effects on levels of total AR, CDK9 or BRD4, treatment with C646 did decrease total AR, possibly due to decreased AR acetylation (7), which may contribute to decreased AR binding to chromatin (Supplementary Figure S2). Together these results support a positive feedback mechanism for androgen-stimulated AR activity whereby low levels of pS81 AR enhance recruitment of p300, with subsequent histone acetylation, increased recruitment of BRD4 and P-TEFb and subsequent further AR S81 phosphorylation.
Figure 2.
AR S81 phosphorylation and p300 recruitment coordinate a positive-feedback loop to sustain AR-mediated transactivation. (A) LNCaP cells in CDS medium were treated for 4 h with DHT (10 nM) and CDK9 antagonist (iII: CDK9 inhibitor II, 50μM), followed by ChIP-qPCR for p300 binding to the AREs in the KLK3/PSA and KLK2 genes. (B) LNCaP cells in CDS medium were treated with DHT and p300 antagonist (C646, 10 μM), followed by ChIP-qPCR for binding to AR-regulated PSA and KLK2 enhancers. H3K27Ac: Lysine 27 acetylated histone H3. (C and D) LNCaP cells in CDS medium were treated with DHT and C646, followed by RNA isolation for qRT-PCR (C) or immunoblotting (D).
PP1α is recruited to chromatin by AR to mobilize CDK9
A large fraction of P-TEFb is held in an inactive state by association with a complex containing 7SK RNA and HEXIM1, and can be released from this complex through dephosphorylation of CDK9 at S175 and T186 by phosphatases including PP1α (12–15,17,29). We reported previously that nuclear PP1α was increased in response to androgen, and that it associates with AR and dephosphorylates S650 in the AR hinge region, thereby enhancing AR nuclear retention (20). These findings suggested that PP1α also may be recruited to chromatin by AR, and could thereby enhance AR transcriptional activity through CDK9 dephosphorylation and subsequent mobilization of P-TEFb. To initially test this hypothesis, we used ChIP to determine whether PP1α was recruited in conjunction with AR to the AR regulated enhancer in the PSA gene. Significantly, we found that stimulation with DHT for 4 h resulted in recruitment of both AR and PP1α to the AR regulated enhancer in the PSA gene, and also to the AR regulated enhancers in the KLK2 and TMPRSS2 genes (Figure 3A). This recruitment of PP1α was also observed at shorter times and persisted for at least 8 h (Figure 3B). In an independent PCa cell line (VCaP), we similarly found that DHT treatment increased pS81, PSA and PP1α binding to the PSA and KLK2 enhancers (Supplementary Figure S3A–C).
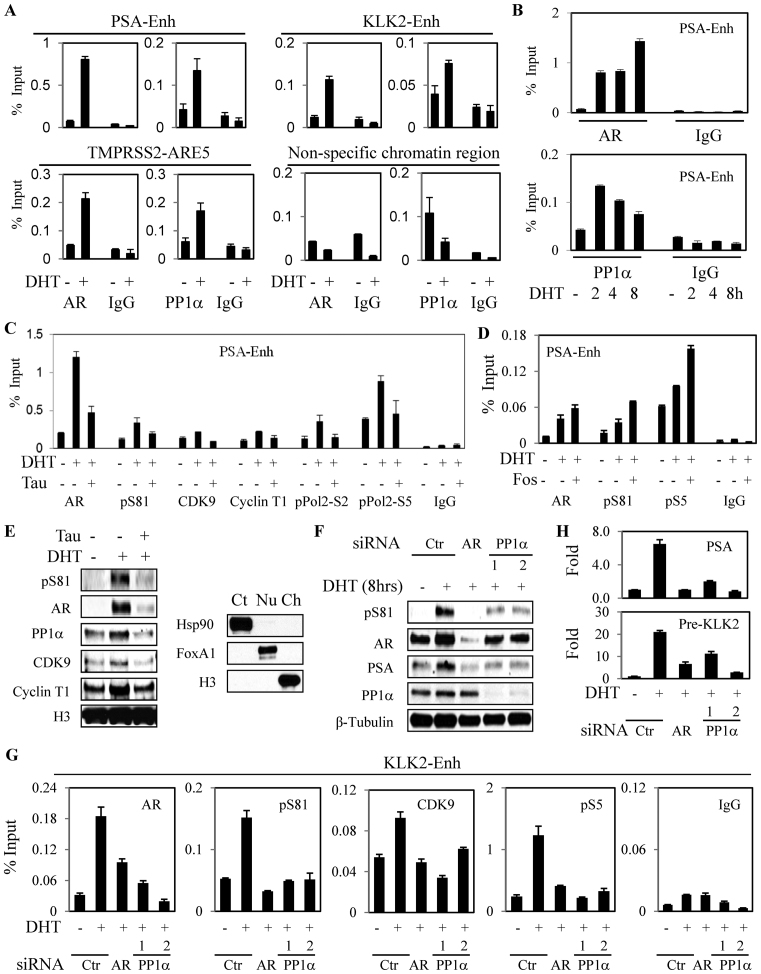
Figure 3.
AR stimulates PP1α recruitment and mobilization of P-TEFb. (A and B) LNCaP cells in CDS medium were treated for 2 h (A) or 2–8 h (B) with androgen (10 nM DHT) as indicated, followed by ChIP-qPCR for AR and PP1α binding to the indicted AREs. (C) LNCaP cells in CDS medium were treated for 4 h with PP1α inhibitor tautomycin (Tau, 200 nM) and DHT, followed by ChIP-qPCR. (D) LNCaP in CDS medium were treated overnight with PP2A-specific inhibitor fostriecin (Fos, 100 nM) and DHT, followed by ChIP-qPCR. (E) LNCaP cells in CDS medium were treated for 4 h with tautomycin and DHT as indicated, followed by isolation of cytoplasmic, nuclear and chromatin fractions, with HSP90, FOXA1 and H3 used as marker proteins for the cellular fractions, respectively (right panel). The chromatin fraction was then immunoblotted for the indicated proteins (left panel). (F–H) LNCaP cells in CDS medium were transfected with control siRNA, AR-specific siRNA or two independent PP1α siRNAs (20 nM) for 3 days, followed by treatment with DHT (10 nM). Total proteins were normalized and immunoblotted (F); cells were harvested for ChIP-qPCR analysis (G); or qRT-PCR analysis was carried out for PSA of KLK2 pre-mRNA (values normalized to the basal activity without DHT, set to 1) (H).
We next used ChIP to determine whether treatment with the PP1α inhibitor tautomycin had an effect on CDK9 activity at the AR regulated PSA enhancer. Indeed, tautomycin (Tau) prevented the DHT-stimulated increase in chromatin occupancy of P-TEFb (CDK9 and cyclin T1) and decreased binding of RNA polymerase II phosphorylated at serine 2 (site phosphorylated by CDK9) and 5 (Figure 3C). Consistent with the CDK9 mediated phosphorylation of AR at S81, tautomycin also decreased binding of S81 phosphorylated AR (Figure 3C). To address whether these effects of tautomycin may reflect increased S650 phosphorylation, we examined the behavior of a Flag-tagged S650A/W741C AR transiently expressed in LNCaP cells. Treatment with bicalutamide (which stimulates the W741C mutant but not endogenous AR in LNCaP cells) increased binding of the Flag-tagged AR to the KLK2 and KLK3 enhancers. Significantly, this binding was prevented by tautomycin, indicating that the loss of chromatin binding is not due to increased S650 phosphorylation (Supplementary Figure S4). We also confirmed that tautomycin did not decrease levels of total AR, CDK9 or BRD4 (see Supplementary Figure S2).
Importantly, while tautomycin is the most potent available inhibitor of PP1α, it is also a less potent inhibitor of PP2A. However, in contrast to tautomycin, the selective PP2A inhibitor fostreicin (Fos) enhanced AR and pS81 AR binding to chromatin (Figure 3D). This result is consistent with previous data showing PP2A can dephosphorylate pS81 and suppress AR activity (20,30,31). To broadly assess effects of tautomycin, we next isolated chromatin from LNCaP cells treated with DHT and tautomycin. As expected, in response to DHT there was increased total and pS81 AR in the chromatin fraction, as well as increased PP1α, and P-TEFb (CDK9 and cyclin T) (Figure 3E). Significantly, these increases were all prevented by tautomycin treatment. Treating with tautomycin similarly blocked the DHT-stimulated increases in chromatin bound AR (total and pS81), BRD4 and P-TEFb in VCaP cells (Supplementary Figure S3D).
To further establish that the effects of tautomycin were mediated through PP1α (the major PP1 catalytic subunit isoform), we examined the effects of transiently decreasing PP1α with two independent siRNA. Both siRNA decreased PP1α, and this was associated with marked decreases in DHT-induced AR S81 phosphorylation (Figure 3F). Moreover, as observed with tautomycin, PP1α siRNA impaired or prevented the DHT-stimulated recruitment of AR, pS81 AR, pRNA pol2 and CDK9 to the KLK2 enhancer (Figure 3G). Consistent with this result, PP1α siRNA also suppressed the DHT-stimulated transcription of KLK2 and KLK3 (Figure 3H). Together these findings demonstrate that PP1α is recruited to chromatin by AR and that its catalytic activity contributes to the recruitment of P-TEFb.
PP1α binding to AR is regulated by the AR N-C terminal interaction
We reported previously that PP1α binding to AR was not dependent on androgen (20). In further studies we have found that DHT treatment can decrease AR-PP1α binding (Figure 4A). This decrease appears related to the specific agonist conformation induced by DHT binding, as the AR antagonists bicalutamide or enzalutamide do not impair AR-PP1α interaction and instead can suppress the decrease in this interaction caused by DHT (Figure 4B). The loss of AR-PP1α association mediated by DHT could also be suppressed by the AR antagonists mifepristone (RU486), cyproterone acetate (CPA) and ARN-509 (ARN), with the latter purer antagonist being most effective (Figure 4C). To further confirm that PP1α binding is decreased when AR is in the agonist conformation, we examined the AR mutant W741C, which folds into the agonist conformation when bound to bicalutamide (32). As shown in Figure 4D, bicalutamide markedly decreased PP1α binding to the W741C mutant AR. Finally, we found that enzalutamide similarly decreased PP1α binding to the F876L mutant AR, for which enzalutamide acts as an agonist (Figure 4D) (33–35).
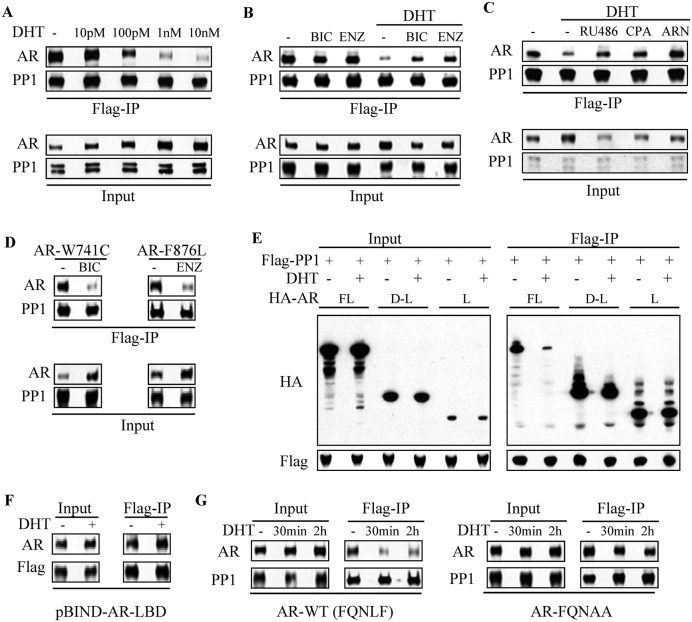
Figure 4.
PP1α-AR binding is disrupted by androgen-induced AR N/C interaction. (A) 293T cells transfected with Flag-PP1α and HA-AR full length were incubated overnight in CDS medium without or with indicated doses of androgen, followed by co-IP analysis. (B) 293T cells were transfected with Flag-PP1α and HA-AR. Cells were then incubated overnight in CDS medium with bicalutamide (BIC, 10 μM), enzalutamide (ENZ, 10 μM), DHT (10 nM) and combinations as indicated, followed by co-IP analysis. (C) 293T cells were transfected with Flag-PP1α and AR, followed by incubation overnight in CDS medium without or with DHT in combination with indicated AR antagonists (mifepristone: RU486; cyproterone acetate: CPA; and ARN509: ARN; 10 μM each), followed by co-IP analysis. (D) 293T cells were transfected with Flag-PP1α and AR W741C (bicalutamide-activated mutant) or F876L (enzalutamide-activated mutant) constructs. Cells were then incubated overnight in CDS medium without or with indicated ligands (BIC: 10 μM of bicalutamide and ENZ: 10 μM of enzalutamide), followed by co-IP analysis. (E) 293T cells were transfected with Flag-PP1α and indicated HA-AR constructs. Cells were then incubated overnight in CDS medium without or with DHT (10 nM) for co-IP analysis. (F) 293T cells were co-transfected of Flag-PP1α and pBIND-AR-LBD. Cells were then incubated overnight in CDS medium without or with 10 nM DHT as indicated, followed by co-IP analysis. (G) 293T cells were transfected with Flag-PP1α together with HA-AR wild-type (WT) versus its FQNLF motif mutant (FQNAA). The cells were then incubated overnight in CDS medium, followed by treatment with 10 nM DHT for 30 min or 2 h, followed by co-IP.
These results suggested that PP1α may be interacting with the AR ligand binding domain (LBD). To assess whether the PP1α-AR interaction was mediated by the AR LBD, we then compared PP1α binding to full length AR (FL) versus to constructs containing the AR DBD-LBD or only the LBD. As shown in Figure 4E, PP1α bound to the full length AR and this binding was decreased by DHT. Both the DBD-LBD and LBD constructs were also bound by PP1α, with binding to the LBD being the strongest, consistent with a binding site in the LBD. However, surprisingly, DHT did not clearly decrease PP1α binding to the DBD-LBD or LBD constructs. We also examined another construct containing the AR LBD fused to the yeast GAL4 DBD (pBIND-LBD), and similarly found that PP1α binding to pBIND-LBD was not decreased by DHT (Figure 4F).
Significantly, the DHT mediated conformational change in the LBD induces a strong interaction between an LXXLL-like motif in the AR NTD (FQNLF) and the coactivator binding site in the LBD (AR N-C interaction) (36–39). To determine whether this interaction interferes with PP1α binding, we introduced mutations into the FQNLF site that abrogate the N-C interaction (36). As shown in Figure 4G, DHT did not suppress PP1α binding to the FQNAA mutant AR, supporting the conclusion that PP1α binding to the LBD is negatively regulated by the N-C interaction. Importantly, while FRET studies have shown that the N–C interaction occurs rapidly after androgen stimulation, they also indicate that this interaction is disrupted when AR is on chromatin, which appears necessary to enhance binding of coactivator proteins through their LXXLL-motifs (40–43). Taken together these results indicate that AR recruits PP1α to chromatin through a site on the LBD, and that access to this site is regulated by the AR N–C interaction.
DHT-stimulated AR S81 phosphorylation and activity are dependent on basal S81 phosphorylation by CDK1
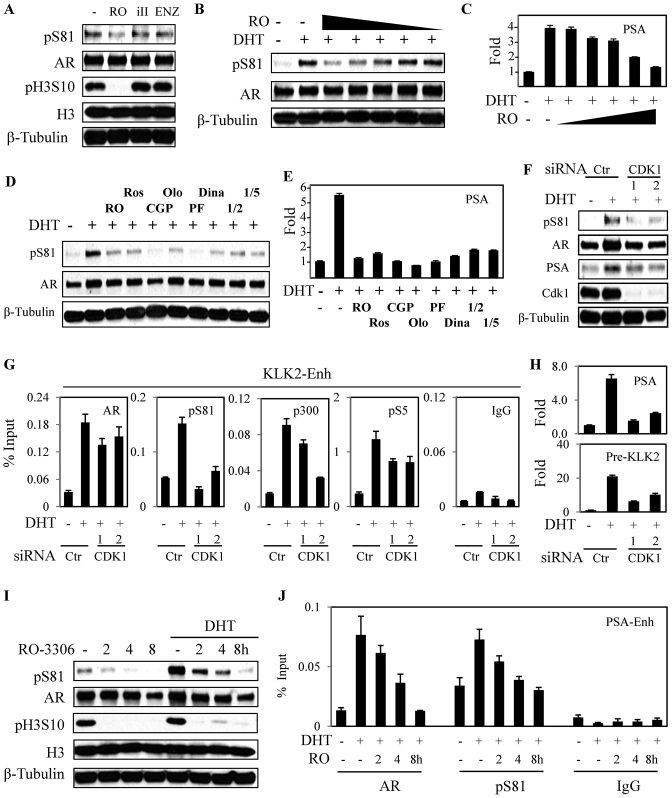
Although CDK9 is the dominant kinase mediating S81 phosphorylation in response to DHT, we reported previously that S81 could also be phosphorylated by CDK1 and that CDK1 activity could sensitize PCa cells to low levels of androgen (5,23). Consistent with these previous results, the low basal pS81 levels in LNCaP cells cultured in steroid-depleted medium could be further reduced by treatment with a CDK1 inhibitor (RO-3306) for as little as 2 h (Figure 5A). This treatment also reduced CDK1 mediated histone 3 S10 phosphorylation (H3S10) phosphorylation, with more modest effects of a CDK9 inhibitor (iII) or enzalutamide. Pretreatment with RO-3306 for 2 h also decreased S81 phosphorylation in response to a subsequent 4 h stimulation with DHT (Figure 5B), and this was associated with a decrease in DHT-stimulated PSA gene expression (Figure 5C). These effects of RO-3306 on pS81 were observed at concentrations at or below those required to decrease H3S10 phosphorylation (∼5 μM), supporting an on-target effect (Supplementary Figure S5A). Basal and DHT stimulated pS81 levels in LNCaP cells (Supplementary Figure S5B and Figure 5D, respectively) and PSA gene expression (Figure 5E), were similarly decreased by a series of other CDK1 inhibitors that have varying potency toward other CDKs.
Figure 5.
Basal CDK1 activity is required for AR-regulated locus transactivation. (A) LNCaP cells in CDS medium were treated for 2 h with CDK1 inhibitor RO-3306 (RO, 10 μM), CDK9 inhibitor (iII, 50 μM) and AR antagonist enzalutamide (ENZ, 10 μM). Total proteins were normalized for blotting. (B and C) LNCaP cells in CDS medium were pre-treated for 2 h with CDK1 inhibitor RO-3306 (0.5, 1, 2.5, 5 and 10 μM), followed by 4 h treatment with DHT (10 nM) as indicated. Total proteins were harvested for blotting (B) and total RNA was harvested for qRT-PCR (C) analyses, respectively. (D and E) LNCaP cells in CDS medium were pre-treated for 2 h with indicated inhibitors (RO-3306: RO, 10 μM; Roscovitine: Ros, 10 μM; CGP74514A: CGP, 10 μM; Olomoucine: Olo, 10 μM; PF: Pfizer compound AG024322, 10 μM; Dinaciclib: Dina, 1 μM; CDK1/2 inhibitor: 1/2, 20 μM; and CDK1/5 inhibitor: 1/5, 100 μM), followed by 4 h treatment with DHT (10 nM) as indicated. Total proteins were harvested for blotting (D) and total RNA was harvested for qRT-PCR analysis (E). (F–H) LNCaP cells in CDS medium were transfected with control siRNA or two independent CDK1 siRNAs (20 nM) for 3 days, followed by 4 h treatment with DHT (10 nM). Total proteins were normalized and assessed by immunblotting (F); cells were harvested for ChIP-qPCR analysis (G); or qRT-PCR analysis was carried out with genes of interest (normalized to the DHT negative samples being set as 1) (H). (I and J) LNCaP cells in CDS medium were pre-treated with CDK1 inhibitor RO-3306 (10 μM) for indicated time points, followed by 4 h treatment without or with DHT (10 nM) as indicated. Total proteins were harvested for blotting (F), or cells were subjected to ChIP for AR and pS81 AR (G).
Two independent CDK1 siRNA were next used to further assess the role of CDK1 in basal AR phosphorylation and activity. Both siRNA suppressed DHT-stimulated S81 phosphorylation and PSA expression (Figure 5F). ChIP-qPCR studies showed that the CDK1 siRNA also decreased DHT-stimulated recruitment of AR, pS81 AR, pRNA polymerase 2 and p300 to the KLK2 enhancer (Figure 5G). Consistent with this finding, the DHT-stimulated expression of KLK2 and KLK3 were also suppressed by the CDK1 siRNA (Figure 5H).
We next used RO-3306 to determine whether acute short-term CDK1 inhibition impairs DHT-stimulated AR binding to chromatin, and whether this is correlated with decreased S81 phosphorylation. As shown in Figure 5I, pretreatment with RO-3306 for 2–8 h progressively decreased basal and DHT-stimulated S81 phosphorylation. Significantly, the decreases in pS81 were associated with marked decreases in DHT-stimulated AR binding to chromatin (Figure 5J). Finally, we examined the effects of CDK1 inhibition with RO-3306 in VCaP cells. Consistent with the findings in LNCaP cells, RO-3306 decreased basal levels of pS81 and PSA expression, and suppressed the induction of S81 phosphorylation and PSA expression in response to DHT treatment (Supplementary Figure S6A–D). Together these results support a requirement for basal CDK1-mediated AR S81 phosphorylation, which may function to initiate a positive feedback loop through enhanced recruitment of p300, BRD4 and P-TEFb, with subsequent amplification of the response through CDK9 mediated S81 phosphorylation.
CDK1-mediated S81 phosphorylation drives AR antagonist-resistant AR activity in CRPC cells
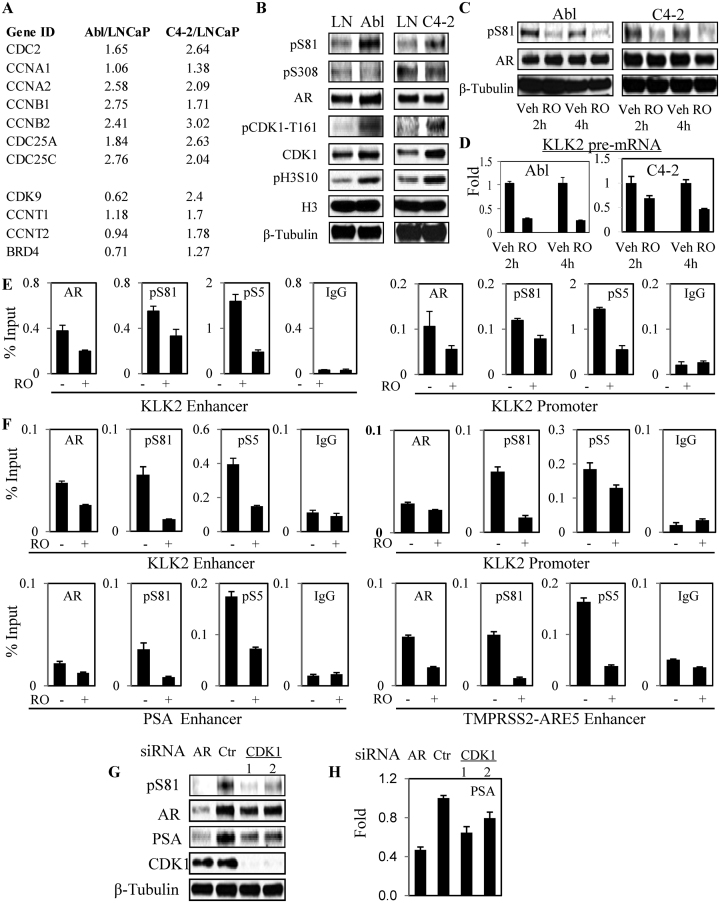
We reported previously that CDK1 activity was increased in CRPC clinical samples versus untreated primary PCa (24) (Supplementary Figure S7). The Abl and C4-2 cell lines are castration-resistant PCa models derived from LNCaP cells by in vitro culture in androgen-depleted medium (Abl) or by selection in castrated mice (C4-2) (44,45). Similarly to CRPC in patients, both Abl and C4-2 have high levels of AR that has basal transcriptional activity in the absence of exogenous androgen. Relative to LNCaP cells, both also have increased expression of mRNA encoding CDK1 and associated cyclins (Figure 6A). In contrast, P-TEFb and BRD4 are increased in C4-2, but not Abl. Consistent with increased CDK1 activity, both Abl and C4-2 have increased levels of total and T161 phosphorylated CDK1 and pH3S10, and have increased basal S81 phosphorylated AR (Figure 6B). CDK1 can also phosphorylate AR on S308 (46), but increased pS308 was not observed in the Abl or C4-2 versus LNCaP cells under these conditions. Significantly, inhibition of CDK1 with RO-3306 for 2–4 h substantially reduced basal pS81 in Abl and C4-2 cells (Figure 6C). A series of other CDK1 inhibitors (with varying effects on other CDKs) also markedly decreased this basal S81 phosphorylation (Supplementary Figure S8).
Figure 6.
Increased CDK1 activity drives basal AR activity in CRPC cells. (A) Expression of CDK1 (CDC2) and genes mediating CDK1 activation in CRPC Abl versus LNCaP (GSE11428) and between C4-2 versus LNCaP (GSE63479) under basal (CDS medium) growth conditions. (B) LNCaP, Abl and C4-2 cells in CDS medium were harvested and equal amounts of protein were analyzed by immunoblotting. (C and D) Abl and C4-2 cells in CDS medium were treated with 10 μM RO-3306 for 2 and 4 h, followed by blotting for AR and pS81 (C) and qRT-PCR analyses of KLK2 pre-mRNA (D) expression. (E and F) Abl (E) and C4-2 (F) cells in CDS medium were treated with RO-3306 for 2 h, followed by ChIP analysis of the indicated AR-regulated loci for occupancy by AR, pS81 and Ser5 phosphorylated RNA Pol II (pS5). (G and H) C4-2 cells in CDS medium were transfected with control siRNA, AR siRNA or two independent CDK1 siRNAs (20 nM) for 3 days. Total proteins were normalized and analyzed by immunoblotting (G); or qRT-PCR analysis was carried out for PSA mRNA (value normalized to control siRNA) (H).
We next asked whether this CDK1-dependent pS81was contributing to the basal AR activity in Abl or C4-2 cells. Abl cells do not express substantial levels of many typical AR target genes such as PSA (likely due to promoter methylation), but they do express KLK2. Therefore, to assess for rapid effects on transcription, we used qRT-PCR to quantify levels of unspliced KLK2 pre-mRNA. We found that RO-3306 treatment caused a rapid decline in KLK2 pre-mRNA in both Abl and C4-2 cells (Figure 6D). Examination of the KLK2 gene by ChIP in the Abl cells showed that the decrease in KLK2 pre-mRNA was associated with decreases in binding of AR (total and pS81) and of pS5-RNA polymerase 2 to the KLK2 enhancer and promoter (Figure 6E). Examination of the KLK2, PSA and TMPRSS2 genes by ChIP-qPCR in C4-2 yielded similar results (Figure 6F). Finally, using two independent CDK1 siRNA in C4-2 cells, we confirmed that CDK1 depletion decreased basal AR S81 phosphorylation and PSA expression (Figure 6G and H).
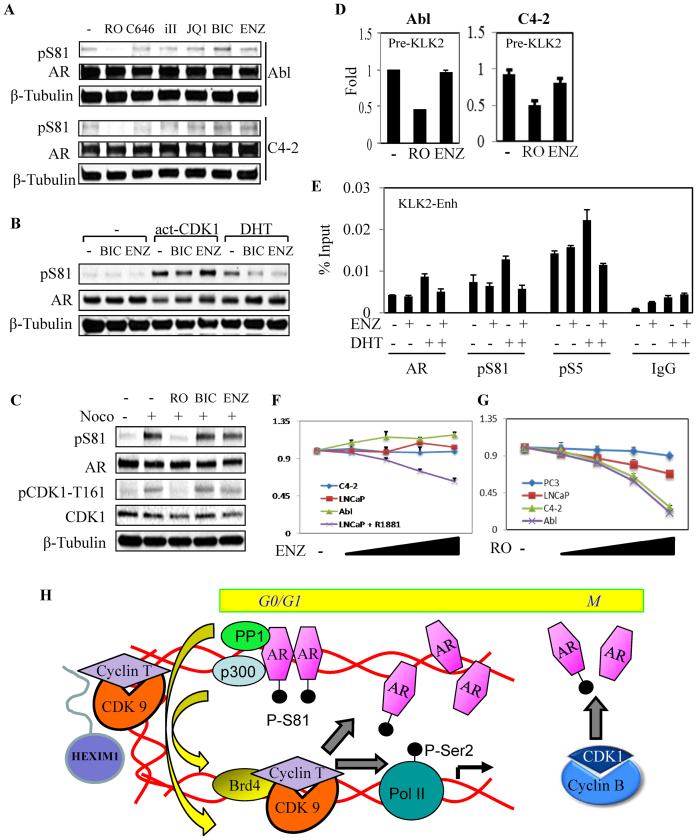
In contrast to CDK1 inhibition, basal S81 phosphorylation in Abl and C4-2 cells was not suppressed by inhibition of p300 (C646), CDK9 (iII), BRD4 (JQ1) or treatment with AR antagonists (bicalutamide or enzalutamide), indicating that it is independent of chromatin or ligand binding (Figure 7A). This inability of AR antagonists to suppress basal pS81 suggested that CDK1-mediated AR phosphorylation may be a mechanism contributing to AR antagonist resistance. Therefore, to directly address whether AR antagonists could suppress CDK1-mediated S81 phosphorylation, we transfected cells with expression vectors encoding AR and a constitutively active mutant CDK1 (act-CDK1). Significantly, bicalutamide or enzalutamide could block DHT-stimulated, but not CDK1-stimulated S81 phosphorylation (Figure 7B). We next used nocodozole to arrest LNCaP cells in mitosis and thereby further activate endogenous CDK1. As expected, pS81 levels were markedly increased by nocodozole. However, this increase was not prevented by bicalutamide or enzalutamide (Figure 7C). Consistent with these results, basal AR transcriptional activity in Abl and C4-2 cells was not suppressed by enzalutamide (Figure 7D), and enzalutamide did not decrease basal AR binding to the KLK2 enhancer in C4-2 cells (Figure 7E).
Figure 7.
CDK1-mediated basal pS81 and AR activation are not repressed by direct AR antagonists. (A) Abl and C4-2 cells in CDS medium were treated for 4 h with indicated compounds (CDK1 inhibitor RO-3306 (RO, 10 μM), p300 inhibitor (C646, 10 μM), CDK9 inhibitor (iII, 10 μM); BRD4 antagonist (JQ1, 500 nM), bicalutamide (BIC, 10 μM) and enzalutamide (ENZ, 10 μM) and total proteins were normalized for blotting. (B) 293T cells in CDS medium were co-transfected with AR, alone or with activated CDK1 (CDK-AF) and cyclin B1 vectors (23), followed by overnight treatments with bicalutamide (BIC, 10 μM), enzalutamide (ENZ, 10 μM) and DHT (10 nM) as indicated for blotting. (C) LNCaP cells in CDS medium were treated overnight with nocodazole (Noco, 50 ng/ml), followed by 4 h treatment with RO-3306, bicalutamide or enzalutamide (10 μM each) for blotting. (D) Abl and C4-2 cells in CDS medium were treated for 4 h with CDK1 inhibitor RO-3306 (10 μM) or enzalutamide (ENZ, 10 μM), followed by RNA isolation for qRT-PCR analysis of KLK2 pre-mRNA expression. (E) C4-2 cells in CDS medium were treated with enzalutamide (ENZ, 10 μM) and DHT (10 nM) as indicated for 4 h, followed by ChIP analysis. (F) LNCaP (without or with androgen as indicated), Abl and C4-2 cells in CDS medium were treated for 3 days with a range of doses of enzalutamide (0.4, 1, 2.5 and 10 μM), followed by cell proliferation analysis. The results were normalized to the untreated control that was set as 1. (G) Androgen-sensitive PCa cell line (LNCaP), CRPC cell lines (Abl and C4-2) and AR-negative PCa cell line (PC3) in CDS medium were treated for 3 days with a range of doses of RO-3306 (0.4, 1, 2.5 and 10 μM), followed by cell proliferation analysis. The results were normalized to the untreated control that was set as 1. (H) Proposed model for AR transcriptional activation. AR recruits PP1α to dephosphorylate and mobilize P-TEFb (CDK9/cyclin T) from an inhibitory 7SK complex, which can then phosphorylate RNA polymerase 2 (and associated proteins) for elongation and phosphorylate AR S81. The latter pS81 then enhances p300 binding, histone acetylation, BRD4 binding and further recruitment of P-TEFb to generate a positive feedback loop that sustains transcription of AR-regulated genes. CDK1 generates a basal pool of pSer81 that is needed to initiate this pathway, and increased CDK1-mediated pS81 phosphorylation may drive this pathway at very low androgen levels, or in the presence of AR antagonists, in CRPC.
As these data showed that CDK1 was critical for basal AR activity in Abl and C4-2 cells, we next assessed the effects of CDK1 inhibition on PCa cell growth under androgen-depleted conditions. As expected, enzalutamide had no effect on the growth of Abl, C4-2 or LNCaP cells in steroid-depleted medium, but did block the ability of androgen (R1881) to stimulate LNCaP growth (Figure 7F). In contrast, both Abl and C4-2 cells showed increased sensitivity to the growth suppressive effects of RO-3306 (Figure 7G), and this occurred at concentrations that suppressed basal S81 phosphorylation (5-10 μM, see Supplementary Figure S5A). While higher drug levels would clearly globally suppress growth of neoplastic and normal cells, these results suggest that effects on AR may provide a therapeutic index for CDK1 inhibitors in advanced CRPC.
DISCUSSION
AR stimulates transcription through the recruitment of multiple proteins, with CDK9 being one such key protein that is required for transcriptional elongation and for phosphorylation of AR at S81. The CDK9/cyclin T (P-TEFb) complex may be recruited by a direct interaction between AR and CDK9 (6,47). Alternatively, P-TEFb can be recruited by BRD4, which is in turn recruited through binding to acetylated histones and may also interact directly with AR (11,28). In any case, a large fraction of P-TEFb is held in an inactive complex that includes 7SK RNA and Hexim1 and must first be mobilized, with one mechanism for mobilization being through dephosphorylation of CDK9 mediated by protein phosphatases including PP1α (12–17). PP1α can be targeted to the nucleus by the protein PNUTS, and can be selectively targeted by the HIV-1 Tat protein to the HIV-1 promoter, but it has not been clear whether there are further mechanisms for the recruitment of PP1α to specific subsets of genes (16,19). In this study we show that AR mediates the recruitment of PP1α to a series of AR-regulated genes. We further show that the PP1α-AR association is mediated by the AR LBD, either directly or possibly via a yet to be identified regulatory protein and that this binding is regulated by the AR N/C interaction. Significantly, most current data indicate that the N/C interaction is disrupted upon chromatin binding, presumably in order to make the coactivator binding groove available for recruitment of coactivator proteins with LxxLL-motifs (40,41,43,48). Taken together, these findings reveal a mechanism for the selective recruitment of PP1α to AR target genes, with subsequent mobilization and recruitment of P-TEFb to initiate transcriptional elongation.
Additional mechanisms may be deployed by other transcription factors to mobilize and recruit P-TEFb. Significantly, while a large fraction of the inactive P-TEFb-7SK RNA complex is in the soluble nuclear fraction, recent data indicate that it is also positioned at many promoters prior to transcription factor binding (15,49–53). NFκB and the HIV transcription factor Tat can recruit the protein phosphatase PPM1G to then dephosphorylate and mobilize CDK9 (15), and Tat may also recruit PP1α for this purpose (12,16,17). In addition to dephosphorylation of CDK9, the helicase DDX21 and the splicing factor SRSF2 may release P-TEFb by other mechanisms (51,54). Finally, AR regulated enhancer RNAs were recently found to bind cyclin T and thereby compete it away from 7SK, similarly to HIV TAR RNA (55). Therefore, AR may employ more than one mechanism for P-TEFb recruitment.
In addition to phosphorylation of RNA polymerase 2, CDK9 mediates the phosphorylation of AR at S81 that occurs subsequent to androgen stimulation and chromatin binding (4,6,8). S81 phosphorylation enhances AR transcriptional activity (5,6), which may be mediated through increased binding of p300 and subsequent histone acetylation (7). Histone acetylation can then enhance binding of BRD4, with subsequent further recruitment of P-TEFb to drive transcriptional elongation. We propose that this further recruitment of P-TEFb also drives further S81 phosphorylation, resulting in a positive feedback loop to sustain high-level transcription of AR regulated genes (Figure 7G).
While CDK9 is the major kinase mediating S81 phosphorylation in response to androgen, this site can also be phosphorylated by CDK1 (23). We have hypothesized that CDK1-mediated S81 phosphorylation occurs physiologically during late G2 and M-phases of the cell cycle, and functions to generate a pool of hyperactive AR that serves to initiate the transcription of AR regulated genes when cells exit mitosis (5). We further propose that increased CDK1 activity in advanced CRPC (that persists to some degree throughout the cell cycle) increases the pool of hyperactive S81 phosphorylated AR, and that this mechanism contributes to AR activity and resistance to AR antagonists. A recent study found that CDK1 can also phosphorylate AR at S308 during G2/M phase, which appears to be a physiological mechanism to suppress AR chromatin binding during mitosis (46). Interestingly, we did not observe increased S308 phosphorylation in the CRPC cell lines C4-2 or Abl, suggesting there may be additional mechanisms in CRPC to selectively enhance S81 phosphorylation. Overall these studies indicate that PP1α, CDK9 and CDK1 cooperate to drive AR transcriptional activity through a positive feedback loop and that agents targeting components of this loop may be effective in CRPC.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Jay Bradner (DFCI, Boston, MA, USA) for JQ1 and Pfizer for the AG024322 compound. We also thank Dr Ankur Sharma (BIDMC, Boston, MA, USA) for help in the model and Drs Sen Chen, Ziyang Yu, ManLi Luo, YuMin Lin and Pengyu Huang (BIDMC, Boston, MA, USA) for reagents and technical assistance.
Footnotes
Present Address: Changmeng Cai, Center for Personalized Cancer Therapy, University of Massachusetts Boston, Boston, MA 02125, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NIH [K99/R00 CA135592]; DOD [W81XWH-14-1-0016 to SC]; NIH [P01 CA163227]; SPORE in Prostate Cancer P50 [CA090381 to S.P.B.]; Tongji Hospital, Tongji Medical School, HuaZhong University of Science and Technology (Wuhan, China) Scholarship (to X.L., in part); U.S. Department of Defense ['W81XWH-14-1-0016΄]. Funding for open access charge: NIH [P01 CA163227].
Conflict of interest statement. None declared.
REFERENCES
- 1. Yuan X., Cai C., Chen S., Chen S., Yu Z., Balk S.P.. Androgen receptor functions in castration-resistant prostate cancer and mechanisms of resistance to new agents targeting the androgen axis. Oncogene. 2014; 33:2815–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mostaghel E.A., Plymate S.R., Montgomery B.. Molecular pathways: targeting resistance in the androgen receptor for therapeutic benefit. Clin. Cancer Res. 2014; 20:791–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gioeli D., Paschal B.M.. Post-translational modification of the androgen receptor. Mol. Cell Endocrinol. 2012; 352:70–78. [DOI] [PubMed] [Google Scholar]
- 4. Gioeli D., Ficarro S.B., Kwiek J.J., Aaronson D., Hancock M., Catling A.D., White F.M., Christian R.E., Settlage R.E., Shabanowitz J. et al. Androgen receptor phosphorylation. Regulation and identification of the phosphorylation sites. J. Biol. Chem. 2002; 277:29304–29314. [DOI] [PubMed] [Google Scholar]
- 5. Chen S., Gulla S., Cai C., Balk S.P.. Androgen receptor serine 81 phosphorylation mediates chromatin binding and transcriptional activation. J. Biol. Chem. 2012; 287:8571–8583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gordon V., Bhadel S., Wunderlich W., Zhang J., Ficarro S.B., Mollah S.A., Shabanowitz J., Hunt D.F., Xenarios I., Hahn W.C. et al. CDK9 regulates AR promoter selectivity and cell growth through serine 81 phosphorylation. Mol. Endocrinol. 2010; 24:2267–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhong J., Ding L., Bohrer L.R., Pan Y., Liu P., Zhang J., Sebo T.J., Karnes R.J., Tindall D.J., van Deursen J. et al. p300 acetyltransferase regulates androgen receptor degradation and PTEN-deficient prostate tumorigenesis. Cancer Res. 2014; 74:1870–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Black B.E., Vitto M.J., Gioeli D., Spencer A., Afshar N., Conaway M.R., Weber M.J., Paschal B.M.. Transient, ligand-dependent arrest of the androgen receptor in subnuclear foci alters phosphorylation and coactivator interactions. Mol. Endocrinol. 2004; 18:834–850. [DOI] [PubMed] [Google Scholar]
- 9. Cho S., Schroeder S., Ott M.. CYCLINg through transcription: posttranslational modifications of P-TEFb regulate transcription elongation. Cell Cycle. 2010; 9:1697–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jonkers I., Lis J.T.. Getting up to speed with transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 2015; 16:167–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen R., Yik J.H., Lew Q.J., Chao S.H.. Brd4 and HEXIM1: multiple roles in P-TEFb regulation and cancer. Biomed. Res. Int. 2014; 2014:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ammosova T., Obukhov Y., Kotelkin A., Breuer D., Beullens M., Gordeuk V.R., Bollen M., Nekhai S.. Protein phosphatase-1 activates CDK9 by dephosphorylating Ser175. PLoS One. 2011; 6:e18985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen R., Liu M., Li H., Xue Y., Ramey W.N., He N., Ai N., Luo H., Zhu Y., Zhou N. et al. PP2B and PP1alpha cooperatively disrupt 7SK snRNP to release P-TEFb for transcription in response to Ca2+ signaling. Genes Dev. 2008; 22:1356–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yik J.H., Chen R., Nishimura R., Jennings J.L., Link A.J., Zhou Q.. Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol. Cell. 2003; 12:971–982. [DOI] [PubMed] [Google Scholar]
- 15. McNamara R.P., McCann J.L., Gudipaty S.A., D’Orso I.. Transcription factors mediate the enzymatic disassembly of promoter-bound 7SK snRNP to locally recruit P-TEFb for transcription elongation. Cell Rep. 2013; 5:1256–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ammosova T., Jerebtsova M., Beullens M., Lesage B., Jackson A., Kashanchi F., Southerland W., Gordeuk V.R., Bollen M., Nekhai S.. Nuclear targeting of protein phosphatase-1 by HIV-1 Tat protein. J. Biol. Chem. 2005; 280:36364–36371. [DOI] [PubMed] [Google Scholar]
- 17. Ammosova T., Washington K., Debebe Z., Brady J., Nekhai S.. Dephosphorylation of CDK9 by protein phosphatase 2A and protein phosphatase-1 in Tat-activated HIV-1 transcription. Retrovirology. 2005; 2:47–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Peti W., Nairn A.C., Page R.. Structural basis for protein phosphatase 1 regulation and specificity. FEBS J. 2013; 280:596–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ciurciu A., Duncalf L., Jonchere V., Lansdale N., Vasieva O., Glenday P., Rudenko A., Vissi E., Cobbe N., Alphey L. et al. PNUTS/PP1 regulates RNAPII-mediated gene expression and is necessary for developmental growth. PLoS Genet. 2013; 9:e1003885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen S., Kesler C.T., Paschal B.M., Balk S.P.. Androgen receptor phosphorylation and activity are regulated by an association with protein phosphatase 1. J. Biol. Chem. 2009; 284:25576–25584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gioeli D., Black B.E., Gordon V., Spencer A., Kesler C.T., Eblen S.T., Paschal B.M., Weber M.J.. Stress kinase signaling regulates androgen receptor phosphorylation, transcription, and localization. Mol. Endocrinol. 2006; 20:503–515. [DOI] [PubMed] [Google Scholar]
- 22. Liu X., Han W., Gulla S., Simon N.I., Gao Y., Cai C., Yang H., Zhang X., Liu J., Balk S.P. et al. Protein phosphatase 1 suppresses androgen receptor ubiquitylation and degradation. Oncotarget. 2016; 7:1754–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen S., Xu Y., Yuan X., Bubley G.J., Balk S.P.. Androgen receptor phosphorylation and stabilization in prostate cancer by cyclin-dependent kinase 1. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:15969–15974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stanbrough M., Bubley G.J., Ross K., Golub T.R., Rubin M.A., Penning T.M., Febbo P.G., Balk S.P.. Increased expression of genes converting adrenal androgens to testosterone in androgen-independent prostate cancer. Cancer Res. 2006; 66:2815–2825. [DOI] [PubMed] [Google Scholar]
- 25. Wang Q., Carroll J.S., Brown M.. Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol. Cell. 2005; 19:631–642. [DOI] [PubMed] [Google Scholar]
- 26. Jang M.K., Mochizuki K., Zhou M., Jeong H.S., Brady J.N., Ozato K.. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell. 2005; 19:523–534. [DOI] [PubMed] [Google Scholar]
- 27. Yang Z., Yik J.H., Chen R., He N., Jang M.K., Ozato K., Zhou Q.. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell. 2005; 19:535–545. [DOI] [PubMed] [Google Scholar]
- 28. Asangani I.A., Dommeti V.L., Wang X., Malik R., Cieslik M., Yang R., Escara-Wilke J., Wilder-Romans K., Dhanireddy S., Engelke C. et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature. 2014; 510:278–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang Y., Dow E.C., Liang Y.Y., Ramakrishnan R., Liu H., Sung T.L., Lin X., Rice A.P.. Phosphatase PPM1A regulates phosphorylation of Thr-186 in the Cdk9 T-loop. J. Biol. Chem. 2008; 283:33578–33584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yang C.S., Vitto M.J., Busby S.A., Garcia B.A., Kesler C.T., Gioeli D., Shabanowitz J., Hunt D.F., Rundell K., Brautigan D.L. et al. Simian virus 40 small t antigen mediates conformation-dependent transfer of protein phosphatase 2A onto the androgen receptor. Mol. Cell. Biol. 2005; 25:1298–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yang C.S., Xin H.W., Kelley J.B., Spencer A., Brautigan D.L., Paschal B.M.. Ligand binding to the androgen receptor induces conformational changes that regulate phosphatase interactions. Mol. Cell. Biol. 2007; 27:3390–3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hara T., Miyazaki J., Araki H., Yamaoka M., Kanzaki N., Kusaka M., Miyamoto M.. Novel mutations of androgen receptor: a possible mechanism of bicalutamide withdrawal syndrome. Cancer Res. 2003; 63:149–153. [PubMed] [Google Scholar]
- 33. Balbas M.D., Evans M.J., Hosfield D.J., Wongvipat J., Arora V.K., Watson P.A., Chen Y., Greene G.L., Shen Y., Sawyers C.L.. Overcoming mutation-based resistance to antiandrogens with rational drug design. Elife. 2013; 2:e00499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Joseph J.D., Lu N., Qian J., Sensintaffar J., Shao G., Brigham D., Moon M., Maneval E.C., Chen I., Darimont B. et al. A clinically relevant androgen receptor mutation confers resistance to second-generation antiandrogens enzalutamide and ARN-509. Cancer Discov. 2013; 3:1020–1029. [DOI] [PubMed] [Google Scholar]
- 35. Korpal M., Korn J.M., Gao X., Rakiec D.P., Ruddy D.A., Doshi S., Yuan J., Kovats S.G., Kim S., Cooke V.G. et al. An F876L mutation in androgen receptor confers genetic and phenotypic resistance to MDV3100 (enzalutamide). Cancer Discov. 2013; 3:1030–1043. [DOI] [PubMed] [Google Scholar]
- 36. He B., Kemppainen J.A., Wilson E.M.. FXXLF and WXXLF sequences mediate the NH2-terminal interaction with the ligand binding domain of the androgen receptor. J. Biol. Chem. 2000; 275:22986–22994. [DOI] [PubMed] [Google Scholar]
- 37. Askew E.B., Minges J.T., Hnat A.T., Wilson E.M.. Structural features discriminate androgen receptor N/C terminal and coactivator interactions. Mol. Cell. Endocrinol. 2012; 348:403–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hur E., Pfaff S.J., Payne E.S., Gron H., Buehrer B.M., Fletterick R.J.. Recognition and accommodation at the androgen receptor coactivator binding interface. PLoS Biol. 2004; 2:E274–E283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. He B., Gampe R.T. Jr, Kole A.J., Hnat A.T., Stanley T.B., An G., Stewart E.L., Kalman R.I., Minges J.T., Wilson E.M.. Structural basis for androgen receptor interdomain and coactivator interactions suggests a transition in nuclear receptor activation function dominance. Mol. Cell. 2004; 16:425–438. [DOI] [PubMed] [Google Scholar]
- 40. van Royen M.E., Cunha S.M., Brink M.C., Mattern K.A., Nigg A.L., Dubbink H.J., Verschure P.J., Trapman J., Houtsmuller A.B.. Compartmentalization of androgen receptor protein-protein interactions in living cells. J. Cell. Biol. 2007; 177:63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. van Royen M.E., van Cappellen W.A., de Vos C., Houtsmuller A.B., Trapman J.. Stepwise androgen receptor dimerization. J. Cell Sci. 2012; 125:1970–1979. [DOI] [PubMed] [Google Scholar]
- 42. Schaufele F., Carbonell X., Guerbadot M., Borngraeber S., Chapman M.S., Ma A.A., Miner J.N., Diamond M.I.. The structural basis of androgen receptor activation: intramolecular and intermolecular amino-carboxy interactions. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:9802–9807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Klokk T.I., Kurys P., Elbi C., Nagaich A.K., Hendarwanto A., Slagsvold T., Chang C.Y., Hager G.L., Saatcioglu F.. Ligand-specific dynamics of the androgen receptor at its response element in living cells. Mol. Cell. Biol. 2007; 27:1823–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Thalmann G.N., Anezinis P.E., Chang S.M., Zhau H.E., Kim E.E., Hopwood V.L., Pathak S., von Eschenbach A.C., Chung L.W.. Androgen-independent cancer progression and bone metastasis in the LNCaP model of human prostate cancer. Cancer Res. 1994; 54:2577–2581. [PubMed] [Google Scholar]
- 45. Pfeil K., Eder I.E., Putz T., Ramoner R., Culig Z., Ueberall F., Bartsch G., Klocker H.. Long-term androgen-ablation causes increased resistance to PI3K/Akt pathway inhibition in prostate cancer cells. Prostate. 2004; 58:259–268. [DOI] [PubMed] [Google Scholar]
- 46. Koryakina Y., Knudsen K.E., Gioeli D.. Cell-cycle-dependent regulation of androgen receptor function. Endocr. Relat. Cancer. 2015; 22:249–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lee D.K., Duan H.O., Chang C.. Androgen receptor interacts with the positive elongation factor P-TEFb and enhances the efficiency of transcriptional elongation. J. Biol. Chem. 2001; 276:9978–9984. [DOI] [PubMed] [Google Scholar]
- 48. Bai S., He B., Wilson E.M.. Melanoma antigen gene protein MAGE-11 regulates androgen receptor function by modulating the interdomain interaction. Mol. Cell. Biol. 2005; 25:1238–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cherrier T., Le Douce V., Eilebrecht S., Riclet R., Marban C., Dequiedt F., Goumon Y., Paillart J.C., Mericskay M., Parlakian A. et al. CTIP2 is a negative regulator of P-TEFb. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:12655–12660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. D’Orso I., Frankel A.D.. RNA-mediated displacement of an inhibitory snRNP complex activates transcription elongation. Nat. Struct. Mol. Biol. 2010; 17:815–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ji X., Zhou Y., Pandit S., Huang J., Li H., Lin C.Y., Xiao R., Burge C.B., Fu X.D.. SR proteins collaborate with 7SK and promoter-associated nascent RNA to release paused polymerase. Cell. 2013; 153:855–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. McNamara R.P., Reeder J.E., McMillan E.A., Bacon C.W., McCann J.L., D’Orso I.. KAP1 recruitment of the 7SK snRNP complex to promoters enables transcription elongation by RNA polymerase II. Mol. Cell. 2016; 61:39–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Flynn R.A., Do B.T., Rubin A.J., Calo E., Lee B., Kuchelmeister H., Rale M., Chu C., Kool E.T., Wysocka J. et al. 7SK-BAF axis controls pervasive transcription at enhancers. Nat. Struct. Mol. Biol. 2016; 23:231–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Calo E., Flynn R.A., Martin L., Spitale R.C., Chang H.Y., Wysocka J.. RNA helicase DDX21 coordinates transcription and ribosomal RNA processing. Nature. 2015; 518:249–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhao Y., Wang L., Ren S., Wang L., Blackburn P.R., McNulty M.S., Gao X., Qiao M., Vessella R.L., Kohli M. et al. Activation of P-TEFb by androgen receptor-regulated enhancer RNAs in castration-resistant prostate cancer. Cell Rep. 2016; 15:599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.