Abstract
Super-enhancers are established through the interactions of several enhancers and a large number of proteins, including transcription factors and co-regulators; however, the formation of these interactions is poorly understood. By re-analysing previously published estrogen receptor alpha (ERα) ChIP-seq data sets derived from the MCF-7 cell line, we observed that in the absence of stimulation, future super-enhancers are represented by one or a few transcription factor binding event(s) and these extraordinary enhancers possess a response element largely specific to the ERα dimer. Upon hormonal stimulation, these primary binding sites are surrounded by a large amount of ERα and the critical components of active enhancers, such as P300 and MED1, and together with neighbouring sites bound by newly recruited ERα, they generate the functional super-enhancers. To further validate the role of canonical elements in super-enhancer formation, we investigated some additional signal-dependent transcription factors, confirming that certain, distinguished binding elements have a general organizer function. These results suggest that certain signal-specific transcription factors guide super-enhancer formation upon binding to strong response elements. These findings may reshape the current understanding of how these regulatory units assemble, highlighting the involvement of DNA elements instead of protein–protein interactions.
INTRODUCTION
Estrogen receptor alpha (ERα) is a well-studied member of the nuclear receptor (NR) superfamily and is a key hormone-regulated transcription factor (TF) in three-quarters of breast cancer cases (1). The activation of ERα through 17β-estradiol (referred to as estradiol or E2) results in the translocation of the receptor from the cytoplasm to the nucleus and strengthens the binding of the receptor and its collaborating factors to regulatory regions of DNA. These events contribute to the altered, particularly increased expression of the specific target genes (2). ERα primarily binds the estrogen response element (ERE), initially described as a conserved regulatory element of the promoter of Xenopus and chicken vitellogenin genes in 1986 (3). ERE is typically an inverted repeat of the NR half site (AGGTCA) that is separated by a 3-bp-long spacer (IR3). There are several variations of this element, which also binds the ligand-induced ER dimer (4).
The most widely used human cell line to study the behavior of ERα is MCF-7. MCF-7 is a well-established in vitro model for the investigation of estrogen-dependent biological processes of breast cancer development, as this model is an ER+ breast cancer-derived cell line isolated from the pleural effusion of a patient with metastatic breast cancer (5,6).
There are several putative ERα transcription factor binding sites (TFBSs) in the reference human genome, but only a portion of these sites is functionally relevant. Using general bioinformatic motif scans, more than 1 million of these putative sites were identified, while the number of optimal or near-optimal sequence motifs was predicted to be approximately 230 000 according to previous studies (7,8). Among these, only a small fraction, 5105 elements, was bound by ERα in MCF-7 cells, while approximately 11 000 additional ERα binding events were identified without the presence of any ERE in the DNA. The discrepancy between the putative ERα binding sites and those identified in experimental models may reflect the accessibility of the sites. Chromatin accessibility has been suggested to drive, in general, the site selection of TFs and chromatin regions with increased DNase I hypersensitivity were indeed enriched in cell line- and tissue-specific TFBSs (9). The emergence of these accessible sites may be primed by cell line-specific factors. Moreover, priming events at the chromatin level further prepare the conformational context for subsequent protein–DNA and protein–protein binding events that ultimately lead to transcriptional activation.
The major collaborative factors of ERα are Forkhead box protein A1 (FoxA1) and activator protein 2 gamma (AP2γ) (10,11). FoxA1 plays a role as a pioneer factor, and it has been proposed that FoxA1 binds ∼50% of the regions occupied by ERα, even in the absence of estradiol (12–14). Moreover, FoxA1 is indispensable for any ERα recruitment (15). AP2γ also joins to ER binding sites and may be involved in stabilizing ERα-chromatin interactions (10).
Recent studies have suggested that enhancers may form a higher-order structure within regulatory units, also called super-enhancers (SEs) (16). Several reports have suggested that the expression of cell type-specific genes is controlled through these markedly active enhancer groups. Thus, relevant genes have been published with respect to different types of cells, with genes defining the nature of a given cell (17). In cancer cells, SEs are associated with key oncogenes, such as c-MYC (16,18) and TAL1 (19); therefore, the genome-wide determination of SE-associated genes would be useful for mapping additional determinative genes that may be involved in tumor pathogenesis (20). Thus, an understanding of how these SEs work would be useful for bridging the gaps in cell differentiation and tumorigenesis. Moreover, an understanding of the mechanisms underlying the manner in which SEs influence the genetic programmes of a cell could bridge the phenotype–genotype gap as the foundation for the missing heritability.
According to a current approach of HOMER bioinformatic analysis suite (21), active SEs are defined based on the elevated local sequence read density of such genomic regions where the individual ChIP-seq (chromatin immunoprecipitation followed by high-throughput DNA sequencing) peaks of the key TFs are closer to each other than 12.5 kb. Each ChIP-seq peak or group of peaks is given a read density score, sorted based on score values and those that are located on the region of the graph with a slope >1 are considered SEs.
Although relevant data have accumulated concerning how these SEs function, little is known regarding the detailed mechanisms that drive the activities of these complexes. SEs are bound by master regulators (17,22), and their prediction can also be done based on the simultaneous presence of BRD4 and/or the active histone mark H3K27ac (23,24).
The Mediator complex, which is a key component of transcription initiation, is involved in the looping of enhancers to transcription start sites (25,26). The largest member of the Mediator complex, MED1, binds to several nuclear receptors, such as thyroid hormone receptor (TR), vitamin D receptor (VDR), peroxisome proliferator-activated receptor gamma (PPARγ), hepatocyte nuclear factor 4 alpha (HNF4α), ER and glucocorticoid receptor (GR), and it is also a co-regulator of SEs (17,27). Another subunit of the complex, MED14, also interacts with GR, HNF4α and PPARγ (27). Previous studies have demonstrated that the reduced level of Mediator members also affects the expression of lineage-specific genes, reflecting the absence of major TFs (28). This effect is consistent with the role of MED1 in forming a bridge between collaborating promoters and enhancers through DNA loop formation (29).
In the present study, using previously published and publicly available ChIP-seq data sets, we conducted a detailed examination of how ERα-driven SEs are formed. We compared the sites with the highest level of ERα binding that upon estradiol treatment, together with the newly activated regions, were organized into SEs. Unexpectedly, we identified a subset of the enhancers within SE clusters that plays distinct roles in the organization of these functional units, and only these enhancers possess canonical EREs. Moreover, these enhancers have a different profile of protein–protein interactions upon ligand treatment. Further examination of the interdependence of ERα, FoxA1 and AP2γ showed that different TFs have specific subsets of SEs, in which the cooperative binding events are incidental.
To further validate the role of canonical elements in SE formation, we investigated several additional NRs and TFs, confirming the pivotal role of strong binding sites in SE formation.
MATERIALS AND METHODS
Data selection
We investigated seven TFs in five different human and mouse cell types: ERα, FoxA1 and AP2γ in the MCF-7 cell line (10,30,31), androgen receptor (AR) in the prostate cancer-derived LNCaP cell line (32), JUNB in primary bone marrow-derived macrophages (BMDM) of the C57BL/6 mouse strain (33), VDR in mouse intestinal epithelial cells and RAR in the F9 mouse embryonic testis carcinoma cell line (34). Control and ligand-treated ChIP-seq samples for the above-mentioned TFs were selected from the publicly available gene expression omnibus database (35) (Supplementary Table S1).
We selected seven additional E2-treated ERα ChIP-seq samples (8 in total, from which at least 10 000 ERα TFBSs were predicted) for further comparison (Supplementary Table S2) (30,36–40). P300, BRD4, MED1 and H3K27ac ChIP-seq and DNase-seq experiments were also performed as follows (Supplementary Table S3) (41–43). For strengthening our model, several further sets samples of ERα ChIP-seq experiments were included in our analysis that cover: vehicle-treated, untreated samples, vehicle- and E2-treated FoxA1 knock-down samples, tamoxifen- and fulvestrant-treated samples as well as a time-course experiment of E2-treatment (Supplementary Table S3) (8,31,44–47).
Data processing
Raw sequence files of the selected ChIP-seq samples were downloaded and processed using a previously published computational pipeline with the hg19 and mm10 reference genomes (48). ChIP-seq peaks were predicted using MACS2 (49), and SEs were predicted from the ligand-treated samples using HOMER findPeaks (21). Artifacts, based on the blacklisted genomic regions of the Encyclopedia of DNA Elements (50), were removed from the peak sets using BEDTools (51).
Reads per kilobase per million mapped reads (RPKM) values for both the control and ligand-treated samples were calculated on the summit ±50 bp region of the peaks determined from the corresponding ligand-treated samples. Peaks with the highest read density (referred to as ‘mother’ peaks) of each future SE were selected from the control samples. The emerging SE peaks (referred to as ‘daughter’ peaks) were identified in the ligand-treated samples in a subsequent analysis. The detailed information about the sites of predicted SEs, mother and daughter enhancers was presented in Additional material (http://dx.doi.org/10.13140/RG.2.2.22940.49286).
Motif enrichment analysis was carried out by findMotifsGenome.pl and was performed on the −50, 50 bp flanking region of the peak summits. The individual and average read density of the different peak sets was determined by annotatePeaks.pl (21). Read distribution and average density heat maps were displayed by Java TreeView (52). Histograms and box plots were performed using GraphPad Prism version 6.00 for Windows (GraphPad Software, La Jolla, CA, USA, www.graphpad.com).
RESULTS
Putative ERα binding sites in MCF-7 cells
To determine the ERα cistrome, we collected public ChIP-seq data sets that included ligand-treated samples with the highest IP efficiency available. By re-analysing the top eight estradiol-treated MCF-7 samples derived from six laboratories (Supplementary Table S2), we identified a surprisingly large number of ERα binding sites (∼189 000 in total), although the consistency of these sites was rather low; specifically, in at least two samples, only 56 931 ERα TFBSs could be predicted (Supplementary Figure S1A and B). Among these, only 4387 TFBSs were present in all of the investigated samples, and we named these sites the ‘consensus peak set’ (the consensus ERα TFBSs were listed in Additional material). The observed heterogeneity may reflect genetic alterations, the number of cell passages, the estradiol concentration used in the specific experiments, the induction time of the estradiol response or other technical differences, e.g. the number of cells or the protocol used for ChIP. Measuring DNase I hypersensitivity to identify chromatin regions accessible for TFs revealed the consensus peak set with the highest chromatin accessibility (Supplementary Figure S1C).
While the majority of the detected ERα binding sites are variegated, ERα SEs showed a high degree of consistency within the reported experiments, highlighting the importance of these regulatory regions within the genome. Notably, more than 90% of the SEs contained at least one peak from the consensus set (Supplementary Table S2), and, interestingly, these peaks were typically present also in the vehicle-treated samples (Figure 1A).
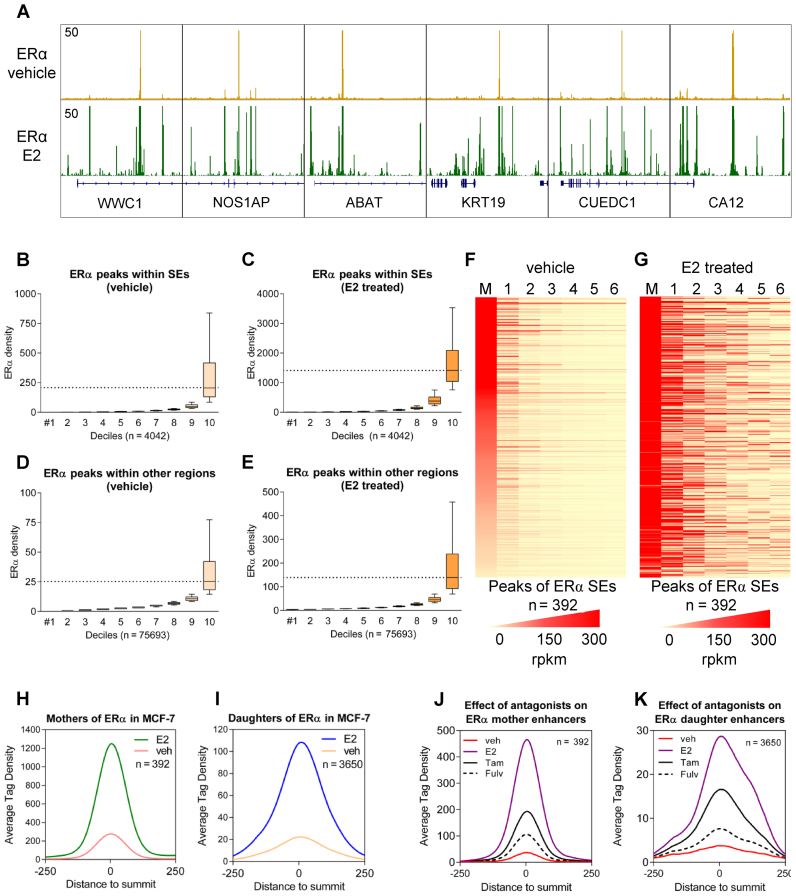
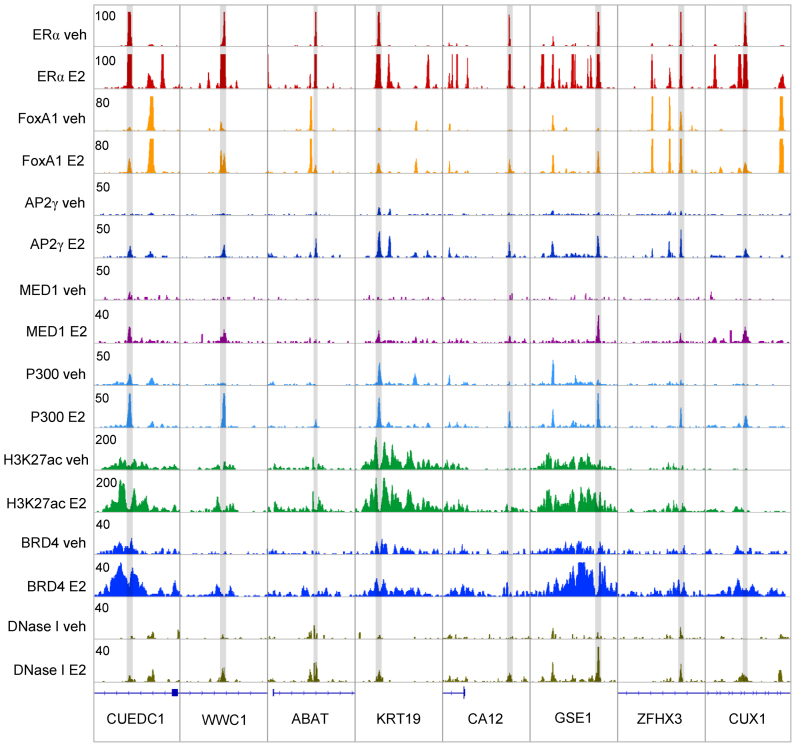
Figure 1.
MCF-7-specific estrogen receptor alpha (ERα) super-enhancers are represented by one (or a few) enhancer(s) even in the absence of estradiol treatment. (A) Integrative Genomics Viewer (IGV) snapshot of ERα ChIP-seq coverage representing six ERα super-enhancers (SEs) upon vehicle and estradiol (E2) treatment. The interval scale is 50 in both cases. (B and C) ERα tag density upon vehicle or E2 treatment in the deciles determined based on ERα recruitment at peaks within SEs. (D and E) ERα tag density upon vehicle or E2 treatment in deciles determined based on ERα recruitment at peaks not overlapping with SEs. The boxes represent the first and third quartiles, the horizontal lines indicate the median reads per kilobase per million mapped reads (RPKM) values, and the whiskers indicate the lower and upper extremes per decile. (F and G) ERα tag densities of the mother (M) and the top 6 daughter enhancers (1–6) within the same super-enhancer region in vehicle- and E2-treated MCF-7 cells. Enhancers were vertically sorted based on the RPKM values of the mother enhancers (in the first column), and the individual enhancers within a SE region were subsequently horizontally aligned based on the read enrichment of the vehicle-treated samples. (H and I) Histograms show the average tag density of mother (392) and daughter enhancers (3650) in the presence or absence of E2. (J and K) Histograms show the average tag density of mother (392) and daughter enhancers (3650) in the presence vehicle- or E2-treatment and upon tamoxifen- and fulvestrant-treatment. Samples derived from the same experiment.
Primary enhancers designate future super-enhancers
A comparison of the vehicle- and estradiol-treated samples (GSM614610 and GSM614611 ERα ChIP-seq samples were used during the main analysis) revealed that in the absence of ligand, each SE was represented by one or a few peak(s) and ligand-induced SEs formed around these activated genomic regions. In the selected sample, 392 highly covered regulatory regions could be predicted by the SE finder of HOMER (Supplementary Figure S1D).
Figure 1A concentrates on the new binding events but cannot emphasize the increase on pre-occupied regions; the exact coverage of the examples is shown on Supplementary Figure S1E. Notably, these individual peaks could not be predicted as SEs in the vehicle-treated samples, as the currently used prediction criteria rely mainly on the presence of clusters of enhancers. These peaks are individual or a few separate peaks, but upon ERα activation, a cluster of peaks assembles and is clearly identified as SEs. Based on this observation, we considered that these preferential primary enhancers may play a key role in the formation of SEs, such as regulatory units; therefore, we further investigated these genomic regions.
We examined the size distribution of ERα peaks (4042 peaks were investigated), plotting each member of ERα SEs based on their tag density (in deciles) (Figure 1B and C). As a control, we used other ERα TFBSs that do not cluster in SEs (75 693 peaks) (Figure 1D and E). The top 10% of SE peaks had an extremely high tag density compared with the other sites in both vehicle- and estradiol-treated samples (Figure 1B and C). Although the distribution of the tag density was similar under both conditions, a more than 5-fold increase in the median tag density was observed upon ligand treatment. To exclude technical issues as the cause of this observation, we performed the same analysis using another set of ERα ChIP-seq samples and observed the same results (Supplementary Figure S1F and G). Plotting the ERα enrichment of all ERα-bound single enhancers that are not members of a SE revealed one-tenth of the tag density of these sites compared with the peaks within SEs. These results suggest that the binding events outside of SEs are less pronounced (Figure 1D and E).
According to the assumption that primary enhancers form the basis of SEs, we referred to these elements as ‘mother enhancers’, and the subsequently appearing secondary enhancers were referred to as ‘daughter enhancers’. The determination of mother and daughter enhancers is described in the data processing description in the Materials and Methods section.
To globally determine whether the binding of ERα to the so-called mother enhancers precedes binding to the subsequent so-called daughter enhancers within the SEs, we sorted all ERα SEs based on the RPKM values of the mother enhancers (calculated from the vehicle-treated samples). In Figure 1F, mother enhancers were well separated, and a relatively low coverage of the daughter enhancers was observed. However, upon ligand treatment, the existence of enhancers was detected on the majority of ERα SEs (Figure 1F and G, Supplementary Figure S2A and B). We also plotted the average tag density of the ERα SE peaks, indicating that mother enhancers show even higher signals in the vehicle-treated samples than daughter enhancers upon estradiol treatment (Figure 1H and I, Supplementary Figure S3A and B).
To validate the precedence of mother enhancer occupancy, we compared vehicle-treated and untreated ERα ChIP-seq samples (Supplementary Figure S4A). These data suggest that vehicle treatment of MCF-7 cells changing the ERα binding neither on mother nor on daughter enhancer regions. The processed time-course experiment also suggested that mother enhancers are indeed occupied prior to daughter enhancers, and show a prolonged binding at later time points, too (Supplementary Figure S4B–D). Thus, there is a difference between the dynamics of the occupancy of the distinct SE peak sets. Importantly, the presence of ERα on mother enhancers is remarkable after ERα antagonist treatments such as tamoxifen and fulvestrant (Figure 1J and K). Moreover, in FoxA1-depleted state, as shown on Supplementary Figure S4E–G, the binding of ERα is reduced, but not abolished neither on mother nor on daughter enhancers.
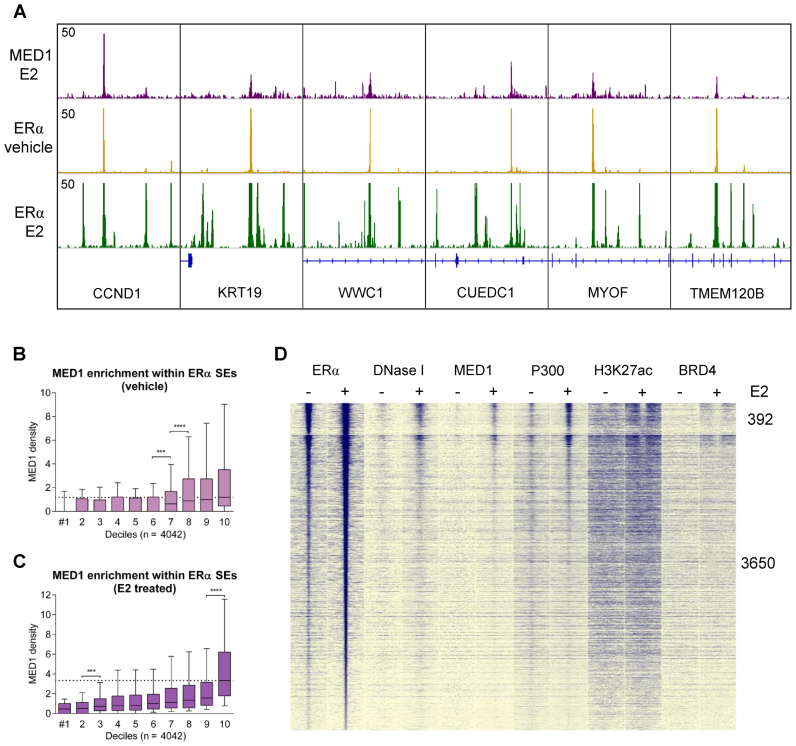
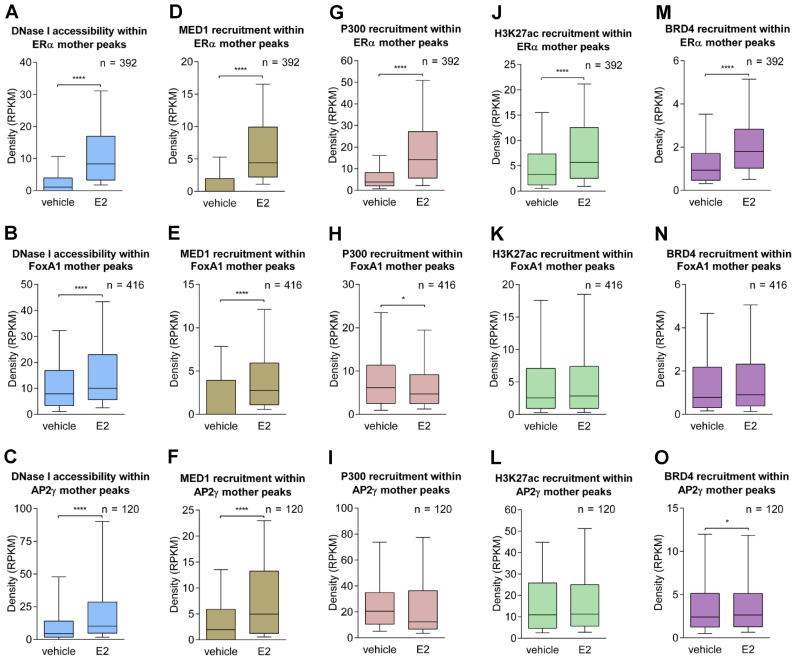
As MED1 was reported to be a key component of the Mediator complex bridging SEs with transcription start sites, we visualized the presence of this component within the ERα SEs. MED1 is recruited to ERα-bound sites with a high binding affinity to the buds or initiator(s) of SEs, namely, mother enhancers, upon estradiol treatment (Figure 2A). We also investigated the MED1 densities based on the previously defined and sorted deciles of ERα peaks, and as shown in Figure 2B and C, Mediator is preferentially recruited to ERα peaks with higher tag densities, particularly in estradiol-treated samples (Figure 2C). Collectively, we observed collaborating enhancers clustered in SEs, showing a much higher ERα occupancy than single enhancers, and even more prominent enrichments accompanied by Mediator binding were observed within SEs.
Figure 2.
MED1 is recruited to ERα-bound sites with an extremely high binding affinity. (A) IGV snapshot of MED1 and ERα ChIP-seq coverage, representing six ERα SEs upon vehicle and estradiol (E2) treatment. The interval scale is 50 in both cases. (B and C) MED1 tag density upon vehicle or E2 treatment in deciles determined based on ERα recruitment at peaks within SEs. The boxes represent the first and third quartiles, horizontal lines indicate the median RPKM values, and whiskers indicate the lower and upper extremes per decile. Paired t-test, * significant at P < 0.05; ** at P < 0.01; *** at P < 0.001; **** at P < 0.0001. (D) Read distribution plot of ERα, DNase I, MED1, P300, H3K27ac and BRD4 upon vehicle or E2 treatment, relative to SE peaks in 2-kb frames. Mother (392 in total) and daughter (3650 in total) peaks are sorted according to ERα tag density.
The read distribution plot around the individual enhancers demonstrated that mother enhancers are located in the most accessible chromatin regions with high levels of DNase I signal and show the highest MED1, P300, H3K27ac and BRD4 coverage upon induction, suggesting that the top ERα enhancers (392 mother enhancers in total) represent the most active regulatory regions, while the daughter enhancers (3650 in total) did not show this pattern (Figure 2D, Supplementary Table S3).
Based on the previous observation, namely, that each super-enhancer region is indicated by a markedly active region, we assessed whether mother and daughter enhancers were observed for other TFs and cell types. First, we examined the other two lineage-determining factors associated with MCF-7 cells, namely, FoxA1 and AP2γ (Supplementary Figure S2C–F). Second, we extended these investigations to other NRs in different cell types (AR in LNCaP cells, RAR in mouse F9 cells and VDR in mouse intestinal epithelial cells) and to JUNB in mouse BMDMs (Supplementary Figure S2G–N). In all experimental systems investigated (except for FoxA1, as described later in this manuscript), patterns similar to those for ERα in MCF-7 cells were observed, indicating that a more general phenomenon occurs in the presence of several TFs (Supplementary Figure S2A–N, Figure S3A–N).
Canonical elements drive transcription factors
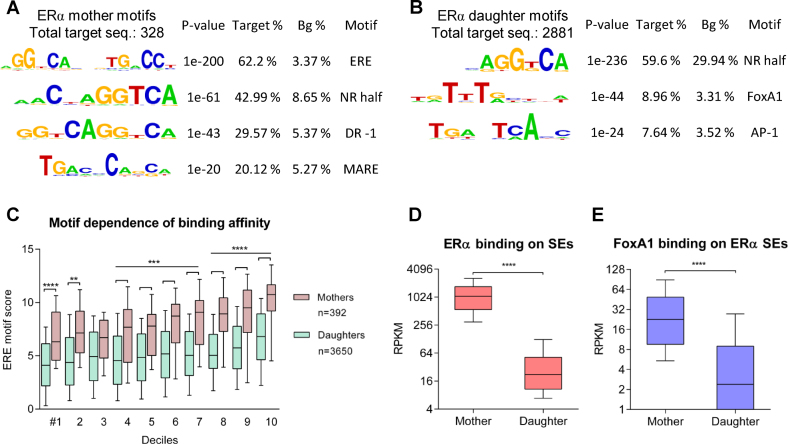
To better understand the factors that discriminate ERα mother and daughter enhancers, we applied a motif enrichment analysis for these regions. While mother enhancers showed strong canonical ERE motif enrichment from a relatively low number of target sequences, daughter enhancers were not enriched for the ERα dimer-specific elements but were enriched for NR half sites and FoxA1 motifs (Figure 3A and B and Supplementary Figure S5A and B). Interestingly, at the sites of mother enhancers, a unique NR direct repeat element with overlapping half sites was also identified. Considering the strong P-value (1e-200) and the high enrichment of ERE (62.2%) (compared with the background, which was 3.37%), despite the small number of target sequences (n = 328), we concluded that the high level of ERα recruitment at mother enhancers (even in the absence of estradiol) reflects strong canonical DNA elements. However, for the emergence of daughter peaks, other TFs, such as FoxA1 and activator protein 1 (AP-1), act in concert with the increased level of ERα after estradiol treatment.
Figure 3.
Canonical elements provide higher DNA-binding affinity than non-canonical elements. (A and B) Motif enrichments of mother and daughter enhancers. The P-value and target and background (Bg) percentages are included for each motif. (C) Estrogen response element (ERE) motif scores in the deciles determined based on ERα recruitment upon vehicle (in the case of mother enhancers) or E2 treatment (in the case of daughter enhancers) within SEs. (D and E) RPKM values of ERα and FoxA1 coverage at ERα mother and daughter enhancers. In the case of panels C, D and E, the boxes represent the first and third quartiles, the horizontal lines indicate the median RPKM values, and the whiskers indicate the 10th to 90th percentile ranges per decile. Paired t-test, * significant at P < 0.05, ** at P < 0.01, *** at P < 0.001, **** at P < 0.0001.
To validate these observations, we examined the motif dependence of binding frequency. We plotted the motif scores of the remapped EREs as a function of the deciles derived from the vehicle-treated peak size distribution in the case of mother enhancers and from the estradiol-treated peak size distribution in the case of daughter enhancers and observed that in all cases, the extremely high mother peaks had significantly stronger binding elements than the daughter enhancers (deciles 8–10 have P < 0.0001) (Figure 3C). This finding suggests that the strongest elements of accessible euchromatic regions irresistibly attract the ERα dimers. To determine whether this effect is also observed for the binding of other TFs, we investigated these correlations for other types of SEs.
Motif enrichment analysis at the sites of the mother and daughter enhancers predicted from other investigated samples showed the same phenomenon. For example, we detected better motif enrichment of mother enhancers in MCF-7 for FoxA1 than that of their daughters, and this effect was also observed for AP2γ (Supplementary Figure S5C–F). Interestingly, AP2γ daughter enhancers harboured the motifs of the main collaborating partners, FoxA1, ERα and AP-1, together with the CCCTC-binding factor (CTCF). In the case of AR in the LNCaP cell line, FoxA1 is needed for the establishment of mother enhancers, as shown by the extremely high motif enrichment in the mother enhancers (Supplementary Figure S5G and H). Importantly, daughter enhancers showed lower IR3/ARE (ARE: androgen response element) and FoxA1 motif enrichment, but a composite element of FoxA1 and AR was also identified, indicating a tight collaboration between these TFs, which may be less affected by other regulators.
In the case of JUNB, mother enhancers of mouse BMDM cells showed all the main specific motif enrichments, namely, AP-1, AP-1::IRF (IRF: interferon regulatory factor) composite element (AICE), cAMP response element and the motif of the lineage-determining CCAAT/enhancer-binding protein (C/EBP). These motifs showed weaker enrichment in daughter enhancers, and further motifs of collaborating TFs, namely, purine-rich box-1 (PU.1), nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) and signal transducer and activator of transcription (STAT) appeared (Supplementary Figure S5I and J). The RAR and VDR mother enhancers of mouse F9 and epithelial cells, respectively, also showed stronger specific motif enrichment than the corresponding daughter peaks, and GATA protein(s) may also occupy VDR mother enhancers. However, daughter enhancers also showed putative AP-1 and Krüppel-like factor (KLF) binding sites (Supplementary Figure S5K–N).
All examined TFs playing roles in SE formation showed a phenomenon similar to that of ERα, namely, these primary regulatory regions possessed canonical elements specific to dominant TF(s), while the further occupied regions had fewer specific elements together with their collaborative factors. The binding of canonical elements precedes, and upon treatment, likely facilitates the occupation of the nearby regions by dominant TFs and interacting partners.
In summary, the primary ChIP-seq peaks, which were present prior to estradiol stimulation, had significantly stronger binding elements than the activated peaks, suggesting that certain elements have high DNA–protein interaction affinities and that there is no need for cooperative binding with other factors, e.g. FoxA1 or AP2γ. Therefore, we examined the differentially enriched ERα and FoxA1 binding on ERα SEs. As shown in Figure 3D and E, the occurrence of ERα on the mother enhancers within ERα SEs is a predominant event (P < 0.0001), but interestingly, FoxA1 also shows a similar pattern (P < 0.0001). The results suggested that the binding affinity of ERα was definitely higher at the sites of mother enhancers, and FoxA1 showed a stronger binding at these sites despite the lack of its binding elements that might mean the importance of protein–protein interactions, namely the binding to ERα.
ERα, FoxA1 and AP2γ form distinct SEs
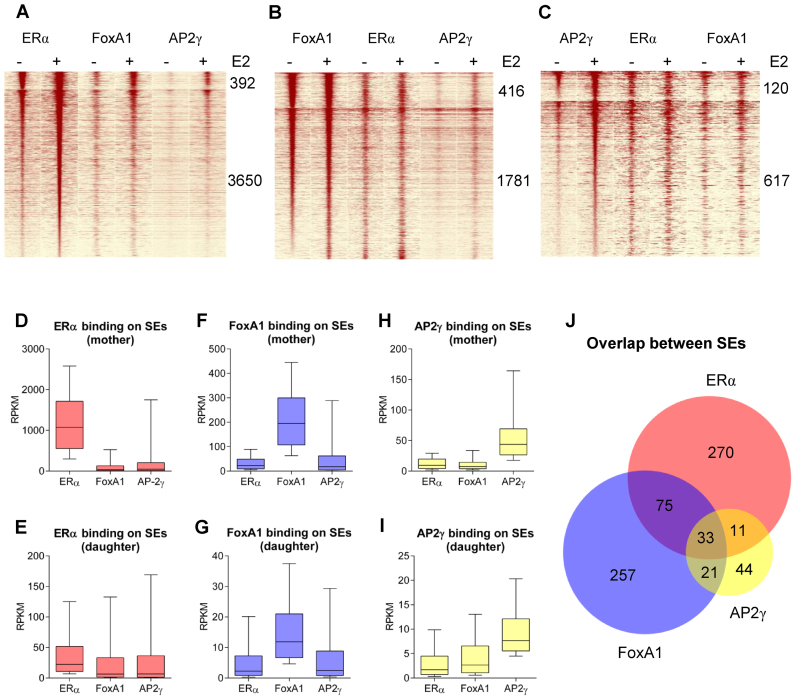
Previous studies have suggested that interactions with collaborating pioneer factors are necessary for ERα function; therefore, we conducted a detailed investigation of how ERα, FoxA1 and AP2γ recruitment correlates with each other at the SE regions and examined the extent of the overlap between the SEs of these MCF-7-specific TFs. For this purpose, we plotted the FoxA1 and AP2γ tag density of mother and daughter ERα enhancers (Figure 4A), and the resulting read distribution heat map showed a positive correlation between the three TFs following estradiol treatment, but the recruitment of the collaborating TFs was much lower than that of ERα. Examining the mother and daughter FoxA1 peaks revealed an increase in AP2γ binding but not in FoxA1 and ERα binding (Figure 4B), and we consistently obtained a similar result for AP2γ SE peaks (Figure 4C). Measuring the tag density of the three TFs on their SEs showed that ERα is typically much less frequently recruited at the mother (and daughter) peaks of the other two TFs (Figure 4D and E) compared with the recruitment of FoxA1 and AP2γ at the sites of ERα SEs. This effect was also observed for the recruitment of FoxA1 and AP2γ at each other's SEs (Figure 4F–I), suggesting that SEs are typically dominated by a single TF and that other TFs are likely collaborating partners in their activity. TF specificity was also confirmed by the overlap of SEs determined for ERα, FoxA1 and AP2γ (Figure 4J). The Venn diagram shows that of these three proteins, ERα and FoxA1 have a more distinct SE profile, while AP2γ has its own, less prominent, SE profile. This finding is consistent with the coverage results shown in Figure 4A–C, as AP2γ was induced at all SEs examined. The observed dominances may reflect the canonical elements because not only the ERα-bound mother enhancers but also those of FoxA1 and AP2γ showed stronger specific motif enrichment compared with that in the corresponding secondary regions (Figure 4A and B, Supplementary Figure S5A–F). On FoxA1- and AP2γ-driven SEs FoxA1 showed no notable change upon E2 treatment, but interestingly, it did show higher density on ERα-dominated SEs (Figure 4A–C). The former is not surprising as no significant change could previously been observed in FoxA1 binding upon E2 stimulation (46), but the latter is striking as we found a set of genomic regions, where E2 could affect FoxA1 binding.
Figure 4.
ERα, FoxA1 and AP2γ form distinct super-enhancers. (A–C) Read distribution plot of ERα, FoxA1 and AP2γ upon vehicle or E2 treatment, relative to each other's SE peaks in 2-kb frames. The number of mother and daughter peaks, sorted based on tag density, is indicated. (D–I) ERα, FoxA1 and AP2γ tag density of each other's mother and daughter enhancers in E2-treated MCF-7 cells. (J) Area-proportional Venn diagram illustrates the overlaps between the ERα, FoxA1 and AP2γ SEs.
Focusing on some ERα-driven SEs, we observed that there is typically at least one FoxA1-bound enhancer per SE in the absence of estradiol; however, these enhancers rather occasionally overlap with mother enhancers (Figure 5 and Supplementary Figure S6A and B). In contrast, AP2γ did not show enrichment prior to treatment and emerged together with ERα, typically at mother enhancers and specific daughter enhancers occupied by FoxA1 (Figure 5 and Supplementary Figure S6C and D). There is a similar enrichment at ERα mother enhancers as of AP2γ, which suggests an ERE/ERα-dependent FoxA1 and AP2γ recruitment by protein–protein interactions. Importantly, MED1 is specific only to mother enhancers, while P300 may also be recruited to regions bound by FoxA1. H3K27ac and BRD4, together with the DNA accessibility, also followed these tendencies, thus showing elevated activity upon treatment (Figures 5, 6A, D, G, J, M).
Figure 5.
Recruitment of FoxA1 and AP2γ at ERα super-enhancers. IGV snapshot of ERα ChIP-seq coverage, representing eight ERα SEs upon vehicle (veh) or E2 treatment and the simultaneous presence of FoxA1, AP2γ, MED1, P300, H3K27ac, BRD4 and DNase I upon vehicle and E2 treatment. The interval scales are indicated in the upper left corners. Peaks, highlighted in grey, represent the sites of ERα mother enhancers, while dashed lines indicate the location of FoxA1 peaks.
Figure 6.
FoxA1 and AP2γ mother enhancers are located in active regions but are not inducible compared with ERα mother enhancers. The box plots demonstrate the accessibility of DNase I and the recruitment of MED1, P300, H3K27ac and BRD4 within the (A, D, G, J and M) ERα mother enhancers, (B, E, H, K, N) FoxA1 mother enhancers and (C, F, I, L and O) AP2γ mother enhancers. The boxes represent the first and third quartiles, the horizontal lines indicate the median RPKM values and the whiskers indicate the 10th to 90th percentile ranges per decile. Paired t-test, * significant at P < 0.05, ** at P < 0.01, *** at P < 0.001, **** at P < 0.0001.
By examining the presence of DNase I, MED1, P300, H3K27ac, BRD4 and collaborating TFs on FoxA1- and AP2γ-driven SEs, we observed a striking phenomenon: as ERα recruitment is neither induced at these sites, P300, H3K27ac and BRD4 nor show enrichment, while DNase I signal and MED1 binding is induced upon treatment (Figure 6B and C, E and F, H and I, K and L, N and O, Supplementary Figure S7A and B). Notably, according to the enrichment of H3K27ac and BRD4, the slight decrease in P300 recruitment did not affect these regions, as they were already at least as active in the untreated state as the induced ERα SEs, and this activation did not change (Figure 6G–I). These results strongly suggest ERα-dependent MED1/P300 recruitment, followed by histone acetylation and BRD4 binding (Figure 6A, D, G, J, M), while at FoxA1- and AP2γ-dominated SEs, only MED1 recruitment and elevated chromatin accessibility were observed upon estradiol treatment (Figure 6E and F). On FoxA1- and AP2γ-dominated SEs estradiol treatment results in MED1 binding. However, because P300 is not further recruited, neither histone acetylation nor the reading module of the acetylated histones, namely BRD4 will be further recruited to these sites (Figure 6E, F, H, I, K, L, N, O and Supplementary Figure S7A and B). These events suggested that in the case of TF-specific SE recruitment and the increase in MED1 binding, an increase in DNase I hypersensitivity is not sufficient for the initiation of these events. However, it is important to note that AP2γ-dominated SEs showed the most enriched active enhancer marks (Figure 6C, F, I, L, O), while those of FoxA1 were the least active (Figure 6B, E, H, K, N). Moreover, ERα bound in a scattered manner to FoxA1- and AP2γ-specific SEs (Supplementary Figure S6E–L).
DISCUSSION
In this study we first describe ligand-inducible super-enhancers. Due to the almost prompt biological response (Supplementary Figure S4A–D) and the large variety of available (NGS) data (Supplementary Tables S1–S3), activation of ERα in MCF-7 is an appropriate system to model the formation of SEs, and our basic findings might be true for the establishment of any cluster of enhancers in any cell type. Using several cell types we tested further inducible TFs (Supplementary Table S1) and found that typically there is one (or a few) high-affinity response element (Figure 3A–C, Supplementary Figure S5) that forms the core of the developing SEs (Figures 1A, F and G and 2A). Inducible SEs possess the same phenomena as previously described for ‘steady-state’ SEs, the enrichment for Mediator proteins, co-regulators and active histone marks (17) (Figures 2D and 6), however, the large extent of protein–protein interactions between TFs was not as pronounced as previously described. In contrast, the examined SEs are driven rather by one single TF (Figure 4), which then recruited the further proteins (Figure 7).
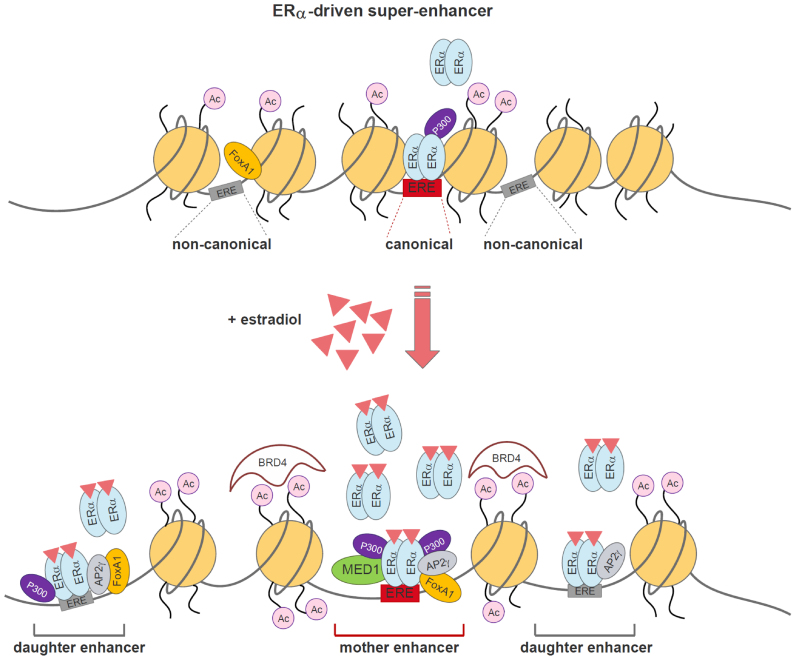
Figure 7.
Working model of an ERα-dominated SE in the presence or absence of estradiol. The upper, closed chromatin region demonstrates the presence of EREs within a SE, and one of these regions provides a canonical DNA-binding element (red box) for mother enhancers in the absence of treatment. Upon E2 treatment, the resulting SE region becomes more opened and E2 treatment recruits several ERα molecules, which may occupy the surrounding non-canonical EREs. The existence of a canonical element provides competition between ERs, and the attracted ERs likely bind to neighbouring non-canonical EREs rather than to similar non-canonical EREs in the distal regions of the genome. The SE region becomes more acetylated and MED1 and P300 bind to ERα that subsequently binds a canonical ERE.
The fact that enhancers cooperate with each other from even more than a 10-kb distance suggests the establishment of a similar system between promoters and enhancers, and this effect cannot be explained without loop formation. The Mediator complex promotes these looping events and was indeed associated with SEs (17,28). Nevertheless, the precise structure of these finger-like formations and the manner in which these complexes are held together remain unknown. Another feature of these outstandingly active, open chromatin regions is the presence of P300 that acetylates histone lysine residues (53). In addition to P300, which not only establishes but also binds acetylated histones, another bromodomain-containing protein, BRD4, is also enriched in these genomic regions (54), together providing accessibility of the regulatory sites for TFs. The previously examined steady-state SEs are occupied by lineage-determining factors (e.g. PU.1, MyoD, C/EBPα), which are cell type-specific, highly expressed proteins (17). In addition to these key regulators, the results of the present study suggest that ligand-inducible TFs (NRs) also play a prominent role in the establishment of SEs. Although we have proof about several DNA–protein and protein–protein interactions in SEs, it is difficult to model how these complexes form and subsequently regulate gene expression.
Another regulatory unit, called the ‘hot spot’, where protein–protein interactions lead to the presence of several TFs at a single locus, has been described (55). Integration of the two functional units, SE and the regulatory hotspot, has revealed at least one TF-dense area in each SE region (56). This finding suggests that during adipocyte reprogramming, the high number of TFs, including C/EBP, AP-1, NRs and several other regulators, attracts more MED1. In the case of ERα-driven SEs in MCF-7 cells, we observed a similar phenomenon (Figures 2, 5, 6D); however, these regions beside their FoxA1-dependence (Supplementary Figure S4E–G) showed strong ERE-, estradiol- and ERα-dependence (Figure 3) and did not show the definitive necessity of interactions between a large number of different kinds of TFs (Figure 4D–I).
We referred to ERα-dense ‘hot spots’ as mother enhancers because these elements were detected prior to stimulation, and additional regulatory regions were referred to as daughter enhancers. Mother enhancer is special from several points of view: (i) it has typically a strong binding element specific for one single TF (Figure 3A); (ii) the predominant TF is able to recruit several other proteins such as AP2γ, FoxA1 and further co-regulators (Figures 2D and 4A); and (iii) it is located in a highly accessible regulatory region (Supplementary Figure S1C), which probably makes possible the accessibility of neighbouring regulatory regions (Figure 7). Unlike mother enhancers, TF recruitment at daughter enhancers is much less dynamic (Supplementary Figure S4D). These regions are similar to any further enhancers. They might be discriminated from typical enhancers by the proximity to mother enhancers (Figure 1A) or their higher ERα occupancy (Figure 1B–E).
Examination of the main TFs that collaborated with ERα revealed that although AP2γ was recruited at mother enhancers and permanent FoxA1-bound daughter sites upon treatment (Figure 5 and Supplementary Figure S6A–D), this recruitment appears consequential due to the presence of preoccupied mother enhancers. We had a unique observation that FoxA1 binding increased mostly at mother enhancers upon E2 treatment (Figure 4A), which, in the lack of the enrichment of its binding element, is probably due to the interaction with ERα and AP2γ. Importantly, only these sites showed increased P300 recruitment (Figure 5), and the enrichment of H3K27ac and BRD4 was also observed (Figures 5 and 6J and M).
SEs are typically formed from several enhancers. The results of the present study revealed that a large fraction of these individual ERα enhancers did not show significant FoxA1, AP2γ or P300 binding, rather, enhanced histone acetylation-dependent BRD4 enrichment (Figures 2D and 5). There are two explanations for how these elements are bound by ERα: (i) less common TFs without enriched motifs recruit ERα via protein-protein interactions or (ii) non-canonical EREs can also be occupied by an elevated number of ERα molecules. The former mechanism may also play a role similar to those of FoxA1 and AP2γ, but the latter mechanism seems to be more significant in this system.
It is well known that FoxA1 is required for and AP2γ facilitates ERα binding (10,11). However, we suggest that ERα binding is differently regulated in SEs. Our comprehensive analyses revealed that high-affinity binding sites attract ERα, even in the absence of ligand or FoxA1, however, there is no ERα-specific SE formation without this protein. In FoxA1 expressing cells estradiol treatment recruits additional ERα proteins, which occupy surrounding non-canonical EREs (Figure 7). Preformed or forming finger-like small loops facilitate the concentration of enhancers in a milieu with high ERα density that contains complexes generated from the direct and indirect interaction partners of ERα. The basic phenomenon, i.e. the presence of mother enhancers with canonical elements, governed via different signal-dependent transcription factors, was also observed in additional cellular systems, suggesting that this type of regulation generally occurs in mammalian cells. We propose that these findings may change the current view of how SEs are generated and regulated.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Lilla Ozgyin and Laszlo Nagy for the helpful discussions. The authors thank Istvan Szatmari for the critical revision of the manuscript and Nature Research Editing Service for the editorial assistance prior to submission. This study makes use of publicly available data generated by various labs, which were cited in the main text.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
University of Debrecen in the programme ‘Internal Research Grant of the Research University’ entitled ‘Dissecting the genetic and epigenetic components of gene expression regulation in the context of the 1000 genomes project’; B.L.B. is a Szodoray Fellow of the University of Debrecen, Faculty of Medicine and an alumni of the Magyary Zoltan fellowship supported by the TAMOP 4.2.4.A/2-11-1-2012-0001 grant implemented through the New Hungary Development Plan co-financed by the European Social Fund and the European Regional Development Fund; G.N. is an alumni of the Jedlik Anyos fellowship supported also by the TAMOP 4.2.4.A/2-11-1-2012-0001 grant. Funding for open access charge: University of Debrecen, Faculty of Medicine, Department of Biochemistry and Molecular Biology.
Conflict of interest statement. None declared.
REFERENCES
- 1. Davies E., Hiscox S.. New therapeutic approaches in breast cancer. Maturitas. 2011; 68:121–128. [DOI] [PubMed] [Google Scholar]
- 2. Shang Y., Hu X., DiRenzo J., Lazar M.A., Brown M.. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000; 103:843–852. [DOI] [PubMed] [Google Scholar]
- 3. Klein-Hitpass L., Schorpp M., Wagner U., Ryffel G.U.. An estrogen-responsive element derived from the 5΄ flanking region of the Xenopus vitellogenin A2 gene functions in transfected human cells. Cell. 1986; 46:1053–1061. [DOI] [PubMed] [Google Scholar]
- 4. Kumar V., Chambon P.. The estrogen receptor binds tightly to its responsive element as a ligand-induced homodimer. Cell. 1988; 55:145–156. [DOI] [PubMed] [Google Scholar]
- 5. Soule H.D., Vazguez J., Long A., Albert S., Brennan M.. A human cell line from a pleural effusion derived from a breast carcinoma. J. Natl. Cancer Inst. 1973; 51:1409–1416. [DOI] [PubMed] [Google Scholar]
- 6. Levenson A.S., Jordan V.C.. MCF-7: the first hormone-responsive breast cancer cell line. Cancer Res. 1997; 57:3071–3078. [PubMed] [Google Scholar]
- 7. Vega V.B., Lin C.-Y., Lai K.S., Kong S.L., Xie M., Su X., Teh H.F., Thomsen J.S., Yeo A.L., Sung W.K. et al. Multiplatform genome-wide identification and modeling of functional human estrogen receptor binding sites. Genome Biol. 2006; 7:R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Joseph R., Orlov Y.L., Huss M., Sun W., Kong S.L., Ukil L., Pan Y.F., Li G., Lim M., Thomsen J.S. et al. Integrative model of genomic factors for determining binding site selection by estrogen receptor-α. Mol. Syst. Biol. 2010; 6:456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vierstra J., Rynes E., Sandstrom R., Zhang M., Canfield T., Hansen R.S., Stehling-Sun S., Sabo P.J., Byron R., Humbert R. et al. Mouse regulatory DNA landscapes reveal global principles of cis-regulatory evolution. Science. 2014; 346:1007–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tan S.K., Lin Z.H., Chang C.W., Varang V., Chng K.R., Pan Y.F., Yong E.L., Sung W.K., Cheung E.. AP-2γ regulates oestrogen receptor-mediated long-range chromatin interaction and gene transcription. EMBO J. 2011; 30:2569–2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Caizzi L., Ferrero G., Cutrupi S., Cordero F., Ballaré C., Miano V., Reineri S., Ricci L., Friard O., Testori A. et al. Genome-wide activity of unliganded estrogen receptor-α in breast cancer cells. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:4892–4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Carroll J.S., Brown M.. Estrogen receptor target gene: an evolving concept. Mol. Endocrinol. 2006; 20:1707–1714. [DOI] [PubMed] [Google Scholar]
- 13. Lupien M., Eeckhoute J., Meyer C.A., Wang Q., Zhang Y., Li W., Carroll J.S., Liu X.S., Brown M.. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008; 132:958–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Robinson J.L.L., Carroll J.S.. FoxA1 is a key mediator of hormonal response in breast and prostate cancer. Front. Endocrinol. (Lausanne). 2012; 3:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Carroll J.S., Liu X.S., Brodsky A.S., Li W., Meyer C.A., Szary A.J., Eeckhoute J., Shao W., Hestermann E. V, Geistlinger T.R. et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005; 122:33–43. [DOI] [PubMed] [Google Scholar]
- 16. Lovén J., Hoke H.A., Lin C.Y., Lau A., Orlando D.A., Vakoc C.R., Bradner J.E., Lee T.I., Young R.A.. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013; 153:320–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Whyte W.A., Orlando D.A., Hnisz D., Abraham B.J., Lin C.Y., Kagey M.H., Rahl P.B., Lee T.I., Young R.A.. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013; 153:307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tolani B., Gopalakrishnan R., Punj V., Matta H., Chaudhary P.M.. Targeting Myc in KSHV-associated primary effusion lymphoma with BET bromodomain inhibitors. Oncogene. 2014; 33:2928–2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mansour M.R., Abraham B.J., Anders L., Berezovskaya A., Gutierrez A., Durbin A.D., Etchin J., Lawton L., Sallan S.E., Silverman L.B. et al. Oncogene regulation. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science. 2014; 346:1373–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hnisz D., Abraham B.J., Lee T.I., Lau A., Saint-André V., Sigova A.A., Hoke H.A., Young R.A.. Super-enhancers in the control of cell identity and disease. Cell. 2013; 155:934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K.. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010; 38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brown J.D., Lin C.Y., Duan Q., Griffin G., Federation A.J., Paranal R.M., Bair S., Newton G., Lichtman A.H., Kung A.L. et al. NF-κB directs dynamic super enhancer formation in inflammation and atherogenesis. Mol. Cell. 2014; 56:219–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Di Micco R., Fontanals-Cirera B., Low V., Ntziachristos P., Yuen S.K., Lovell C.D., Dolgalev I., Yonekubo Y., Zhang G., Rusinova E. et al. Control of embryonic stem cell identity by BRD4-dependent transcriptional elongation of super-enhancer-associated pluripotency genes. Cell Rep. 2014; 9:234–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sengupta D., Kannan A., Kern M., Moreno M.A., Vural E., Stack B., Suen J.Y., Tackett A.J., Gao L.. Disruption of BRD4 at H3K27Ac-enriched enhancer region correlates with decreased c-Myc expression in Merkel cell carcinoma. Epigenetics. 2015; 10:460–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Park S.W., Li G., Lin Y.-P., Barrero M.J., Ge K., Roeder R.G., Wei L.-N.. Thyroid hormone-induced juxtaposition of regulatory elements/factors and chromatin remodeling of Crabp1 dependent on MED1/TRAP220. Mol. Cell. 2005; 19:643–653. [DOI] [PubMed] [Google Scholar]
- 26. Kagey M.H., Newman J.J., Bilodeau S., Zhan Y., Orlando D.A., van Berkum N.L., Ebmeier C.C., Goossens J., Rahl P.B., Levine S.S. et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010; 467:430–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Malik S., Roeder R.G.. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat. Rev. Genet. 2010; 11:761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yin J., Wang G.. The Mediator complex: a master coordinator of transcription and cell lineage development. Development. 2014; 141:977–987. [DOI] [PubMed] [Google Scholar]
- 29. Tsai C.-J., Nussinov R.. Gene-specific transcription activation via long-range allosteric shape-shifting. Biochem. J. 2011; 439:15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schmidt D., Schwalie P.C., Ross-Innes C.S., Hurtado A., Brown G.D., Carroll J.S., Flicek P., Odom D.T.. A CTCF-independent role for cohesin in tissue-specific transcription. Genome Res. 2010; 20:578–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Welboren W.-J., van Driel M.A., Janssen-Megens E.M., van Heeringen S.J., Sweep F.C., Span P.N., Stunnenberg H.G.. ChIP-Seq of ERalpha and RNA polymerase II defines genes differentially responding to ligands. EMBO J. 2009; 28:1418–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Takayama K., Suzuki T., Tsutsumi S., Fujimura T., Urano T., Takahashi S., Homma Y., Aburatani H., Inoue S.. RUNX1, an androgen- and EZH2-regulated gene, has differential roles in AR-dependent and -independent prostate cancer. Oncotarget. 2015; 6:2263–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ostuni R., Piccolo V., Barozzi I., Polletti S., Termanini A., Bonifacio S., Curina A., Prosperini E., Ghisletti S., Natoli G.. Latent enhancers activated by stimulation in differentiated cells. Cell. 2013; 152:157–171. [DOI] [PubMed] [Google Scholar]
- 34. Lee S.M., Riley E.M., Meyer M.B., Benkusky N.A., Plum L.A., DeLuca H.F., Pike J.W.. 1, 25-Dihydroxyvitamin D3 Controls a Cohort of Vitamin D Receptor Target Genes in the Proximal Intestine That Is Enriched for Calcium-regulating Components. J. Biol. Chem. 2015; 290:18199–18215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Edgar R., Domrachev M., Lash A.E.. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002; 30:207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fletcher M.N.C., Castro M.A.A., Wang X., de Santiago I., O’Reilly M., Chin S.-F., Rueda O.M., Caldas C., Ponder B.A.J., Markowetz F. et al. Master regulators of FGFR2 signalling and breast cancer risk. Nat. Commun. 2013; 4:2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li W., Notani D., Ma Q., Tanasa B., Nunez E., Chen A.Y., Merkurjev D., Zhang J., Ohgi K., Song X. et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013; 498:516–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Joseph R., Orlov Y.L., Huss M., Sun W., Kong S.L., Ukil L., Pan Y.F., Li G., Lim M., Thomsen J.S. et al. Integrative model of genomic factors for determining binding site selection by estrogen receptor-α. Mol. Syst. Biol. 2010; 6:456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hu M., Yu J., Taylor J.M.G., Chinnaiyan A.M., Qin Z.S.. On the detection and refinement of transcription factor binding sites using ChIP-Seq data. Nucleic Acids Res. 2010; 38:2154–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Welboren W.-J., van Driel M.A., Janssen-Megens E.M., van Heeringen S.J., Sweep F.C., Span P.N., Stunnenberg H.G.. ChIP-Seq of ERalpha and RNA polymerase II defines genes differentially responding to ligands. EMBO J. 2009; 28:1418–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Brunelle M., Nordell Markovits A., Rodrigue S., Lupien M., Jacques P.-É., Gévry N.. The histone variant H2A.Z is an important regulator of enhancer activity. Nucleic Acids Res. 2015; 43:9742–9756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. He H.H., Meyer C.A., Chen M.W., Jordan V.C., Brown M., Liu X.S.. Differential DNase I hypersensitivity reveals factor-dependent chromatin dynamics. Genome Res. 2012; 22:1015–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu Z., Merkurjev D., Yang F., Li W., Oh S., Friedman M.J., Song X., Zhang F., Ma Q., Ohgi K.A. et al. Enhancer activation requires trans-recruitment of a mega transcription factor complex. Cell. 2014; 159:358–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Franco H.L., Nagari A., Kraus W.L.. TNFα signaling exposes latent estrogen receptor binding sites to alter the breast cancer cell transcriptome. Mol. Cell. 2015; 58:21–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Swinstead E.E., Miranda T.B., Paakinaho V., Baek S., Goldstein I., Hawkins M., Karpova T.S., Ball D., Mazza D., Lavis L.D. et al. Steroid Receptors Reprogram FoxA1 Occupancy through Dynamic Chromatin Transitions. Cell. 2016; 165:593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hurtado A., Holmes K.A., Ross-Innes C.S., Schmidt D., Carroll J.S.. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat. Genet. 2011; 43:27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Guertin M.J., Zhang X., Coonrod S.A., Hager G.L.. Transient estrogen receptor binding and p300 redistribution support a squelching mechanism for estradiol-repressed genes. Mol. Endocrinol. 2014; 28:1522–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Barta E. Command line analysis of ChIP-seq results. EMBnet.journal. 2011; 17:13–17. [Google Scholar]
- 49. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dunham I., Kundaje A., Aldred S.F., Collins P.J., Davis C.A., Doyle F., Epstein C.B., Frietze S., Harrow J., Kaul R. et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Saldanha A.J. Java Treeview–extensible visualization of microarray data. Bioinformatics. 2004; 20:3246–3248. [DOI] [PubMed] [Google Scholar]
- 53. Ogryzko V.V., Schiltz R.L., Russanova V., Howard B.H., Nakatani Y.. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996; 87:953–959. [DOI] [PubMed] [Google Scholar]
- 54. Filippakopoulos P., Picaud S., Mangos M., Keates T., Lambert J.-P., Barsyte-Lovejoy D., Felletar I., Volkmer R., Müller S., Pawson T. et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012; 149:214–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Siersbæk R., Baek S., Rabiee A., Nielsen R., Traynor S., Clark N., Sandelin A., Jensen O.N., Sung M.-H., Hager G.L.. Molecular architecture of transcription factor hotspots in early adipogenesis. 2014; 7:1434–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Siersbæk R., Rabiee A., Nielsen R., Sidoli S., Traynor S., Loft A., La Cour Poulsen L., Rogowska-Wrzesinska A., Jensen O.N., Mandrup S.. Transcription factor cooperativity in early adipogenic hotspots and super-enhancers. Cell Rep. 2014; 7:1443–1455. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.