Abstract
We examine the association of the β-, α-, and σ70-subunits of Escherichia coli RNA polymerase (RNAP) and the NusA elongation factor with transcribed regions in vivo by using chromatin immunoprecipitation. RNAP preferentially associates with the promoter-proximal region of several operons, and this preference is particularly pronounced at the lexA-dinF promoter. When cells are grown in exponential phase, little or no σ70 is associated with RNAP during early elongation. However, during stationary phase, σ70 is retained in a fraction of elongating RNAP complexes throughout the melAB operon. In contrast, σ70 is not observed in elongating RNAP complexes at the lacZYA operon during stationary phase. At both operons, NusA associates with RNAP during early elongation, and this association is greatly reduced during stationary phase. These observations suggest that in vivo association of σ70 and NusA with elongating RNAP is regulated by growth conditions.
Keywords: transcription, σ70, NusA, chromatin immunoprecipitation
The molecular understanding of transcriptional regulatory mechanisms in prokaryotes is well advanced, particularly in comparison to the understanding in eukaryotes. The association of RNA polymerase (RNAP) and associated proteins with transcribed regions of DNA has been studied extensively in vitro, and these biochemical experiments have provided great insight into the mechanisms of transcriptional initiation, elongation, and termination. In addition, there has been a great deal of genetic analysis, in which the transcriptional properties of mutant proteins and promoters have been assessed under various environmental conditions. Importantly, transcription in vitro is very efficient, occurring at rates comparable to those in living cells, and it faithfully mimics many aspects of transcription in vivo as defined by genetic analysis. However, there is very little work analyzing the association of RNAP and auxiliary proteins with transcribed regions in vivo.
Escherichia coli RNA polymerase consists of five subunits: α2ββ′ω, but this core enzyme is unable to recognize promoters and accurately initiate transcription. RNAP associates with a number of accessory proteins during transcriptional initiation, elongation, and termination. During transcriptional initiation, core RNAP associates with a σ-subunit to form an RNAP holoenzyme. There are seven σ-subunits in E. coli, and each σ-subunit allows the RNAP holoenzyme to recognize a specific sequence at a subset of promoters (1). The predominant σ factor during exponential growth in rich media is σ70.
For many years, it was widely believed that the transition to transcription elongation caused all σ70 to be released from RNAP. A number of biochemical studies showed that β and σ70 could be purified from initiating but not from elongating RNAP complexes (2-5). These data are consistent with structural studies of RNAP that suggest σ would be displaced from RNAP holoenzyme by RNA products of 12-14 nt in length (6). However, more recent in vitro studies suggest that σ70 can remain associated with elongating RNAP. First, fluorescence resonance energy transfer analysis indicates that >60% of elongating RNAP complexes at position +50 contain σ70 (7). Second, an RNA-pulldown approach indicated that only a small fraction of RNAP at position +32 (7%) purified from exponentially growing cells contains σ70 (8). However, when RNAP at position +32 was purified from stationary phase cells, the proportion of elongating RNAP containing σ70 increased to 33%. Moreover, elongating RNAP complexes that contain σ70 are five times more efficient at multiround transcription from a test promoter in vitro, suggesting that a population of RNAP retains σ70 throughout transcriptional elongation. Third, σ70 recognizes pause sequences located up to 18-bp downstream of the transcription start point (9-12). However, RNAP does not recognize a σ70-dependent pause sequence located at position +37 or +462, suggesting that RNAP complexes at these positions do not contain σ70 (10). The relationship of these in vitro results to each other and to transcription in vivo is unclear.
NusA is a transcriptional elongation factor that has associates with elongating RNAP complexes in vitro. NusA and σ70 bind to overlapping regions on the surface of RNAP (13, 14), suggesting that σ70 release from RNAP during the transition from initiation to elongation is accompanied by the association of NusA with RNAP (13, 15, 16).
Here we use chromatin immunoprecipitation (ChIP) to measure the association of RNAP, σ70, and NusA with transcribed regions of DNA in vivo. Our results demonstrate that RNAP is clustered at the promoter-proximal region of many operons, suggesting that RNAP can remain bound at or near the promoter for some time during the transition to elongation. During exponential phase, elongating RNAP does not contain σ70 at all 10 operons tested. In contrast, during stationary phase, σ70 can associate with elongating RNAP at one of the two operons tested, indicating that the association of σ70 with elongating RNAP is regulated at both an operon-specific and a growth state-dependent manner. Finally, NusA associates with transcribed DNA during early elongation, and this association is significantly reduced during stationary phase. Thus, association of σ70 and NusA with elongating RNAP is regulated by growth conditions.
Materials and Methods
Strains and Media. E. coli strain MG1655 was used for all ChIP experiments. Cells were grown to mid-exponential phase (OD650 = 0.3-0.6) or stationary phase (OD650 = 1.2-1.4). For experiments involving the gltA and sdhCDAB operons, cells were grown in LB. For experiments involving the umuDC, recN, and lexA-dinF operons, cells were grown in LB. For induced expression of these operons, cells were treated with UV light 20 min before harvesting. For experiments involving the malEFG operon, cells were grown in M9 minimal media. For induced expression of this operon, 0.4% maltose was added to the media. For experiments involving the lacZYA and melAB operons, cells were grown in M9 minimal media (noninducing conditions) or LB containing 500 μM isopropyl β-d-thiogalactoside and 0.4% melibiose (inducing conditions). For experiments involving the araBAD and araE operons, cells were grown in M9 minimal media (noninducing conditions) or LB containing 0.2% arabinose (inducing conditions). Experiments involving lacZYA and melAB were performed by using the same samples of crosslinked cells.
ChIP. ChIP was based on described procedures (17-19). Cells were grown in appropriate media, and formaldehyde was added to a final concentration of 1%. After 20 min of incubation, glycine was added to a final concentration of 0.5 M, and cells were harvested by centrifugation and washed once with Tris-buffered saline (pH 7.5). Cells were resuspended in 500 μl of lysis buffer (10 mM Tris, pH 8.0/20% sucrose/50 mM NaCl/10 mM EDTA/4 mg/ml lysozyme) and incubated at 37°C for 30 min. 500 μl of immunoprecipitation buffer (50 mM Hepes-KOH, pH 7.5/150 mM NaCl/1 mM EDTA/1% Triton X-100/0.1% sodium deoxycholate/0.1% SDS) and PMSF (final concentration 1 mM) were added to cell extract, and DNA was sheared by sonication to an average size of ≈500 bp. Insoluble cellular material was removed by microcentrifugation for 10 min, and the supernatant was transferred to a fresh tube. This supernatant (50 μl) was kept for use as the “input” sample.
Proteins were immunoprecipitated by diluting a fraction of the crosslinked cell extract with IP buffer to a final volume of 800 μl. This was then incubated with 20 μl of Protein A-Sepharose beads (Amersham Pharmacia-Pharmacia) and β mouse monoclonal (Neoclone), σ70 mouse monoclonal (Neoclone), NusA mouse monoclonal (Neoclone), or MelR rabbit polyclonal (gift from S. Busby, University of Birmingham, Birmingham, U.K.) Ab for 90 min at room temperature with gentle mixing. Samples were then washed twice with IP buffer, once with IP buffer plus 500 mM NaCl, once with wash buffer (10 mM Tris·HCl, pH 8.0/250 mM LiCl/1 mM EDTA/0.5% Nonidet-P40/0.5% sodium deoxycholate), and once with 10 mM Tris and 1 mM EDTA, pH 7.5. Immunoprecipitated complexes were eluted by incubation of beads with elution buffer (50 mM Tris·HCl, pH 7.5/10 mM EDTA/1% SDS) at 65°C for 10 min.
Immunoprecipitated samples and the corresponding “input” sample were decrosslinked by incubation for 2 h at 65°C and 6 h at 42°C in 0.5× elution buffer plus 0.8 mg/ml Pronase. DNA was purified by using a PCR purification kit (Qiagen). All ChIPs were performed at least three times. Error bars representing one SD from the mean are shown for all data.
Quantitative PCR. Quantitative PCR was performed in real time by using the Applied Biosystems 7000 and 7700 sequence detectors. Primer pairs for promoter sequences were centered around the transcriptional initiation site. Primer pairs for coding sequences were located >500 bp downstream of the initiation site and >500 bp upstream from the 3′ end of the RNA. The location of primer pairs used in the mapping experiments are specified in the figures, and primer sequences are presented in Tables 1 and 2, which are published as supporting information on the PNAS web site. Occupancy values were calculated by comparison of target regions with a region of the yabN-coding sequence as a background control. The yabN-coding sequence was chosen as this showed no β association in preliminary experiments and has been shown in numerous microarray experiments to be nontranscribed. Occupancy units represent a background-subtracted value for the association of a particular protein with a target region (19).
Immunoblot Analysis. Cell extracts were prepared as described (10). Immunoblotting was performed by using Supersignal reagents (Pierce) with mouse mAbs against β, σ70, and NusA (Neoclone, Madison, WI) as well as a polyclonal Ab against MelR (gift from E. Tamai and S. Busby, University of Birmingham, Birmingham, U.K.).
Results
Using ChIP to Determine Protein-DNA Association in Vivo. ChIP is a widely used technique for studying the association of proteins with DNA in eukaryotic cells (19, 20). Formaldehyde crosslinking of living cells allows visualization of protein-DNA interactions at a fixed point in time, and multiple protein-DNA associations can be analyzed from a single sample of crosslinked cells. Here, we adapt the ChIP methodology to study the association of RNAP and associated proteins with transcribed regions of DNA in E. coli. For all proteins studied, the level of association with the promoter and transcribed regions was significantly above the background level defined by a nontranscribed region.
For some experiments, sites of protein association were mapped by using multiple PCR primer pairs across a given genomic region. In this regard, ChIP experiments define an apparent domain of protein association, whose extent is determined by the sizes of input DNA fragments and PCR products (21). A protein that binds to a specific DNA site will show peak levels of association occurring at the actual site of binding, but detectable association on either side of the actual binding site (21). Thus, the actual site of protein association is determined by the location of the peak, and the resolution of the mapping is determined by the size of the PCR products and the number of measurements throughout the region. ChIP has been used to determine the position of protein-DNA association to a resolution of ≈50 bp (22, 23).
RNAP Associates Preferentially with the Promoter-Proximal Regions of Several Operons. We have applied ChIP to study the association of the E. coli RNAP core subunit β with transcribed regions of DNA. We determined the levels of β at 10 different operons during exponential phase under conditions that give high levels of transcription (Fig. 1). Two of these operons, sdhCDAB and gltA, are constitutively expressed (24, 25). Transcription of the remaining eight operons is activated by the following environmental conditions: umuDC, recN, and lexA-dinF by DNA-damage (26-28); melAB by addition of isopropyl β-d-thiogalactoside and melibiose (29, 30); lacZYA by isopropyl β-d-thiogalactoside (30); malEFG by maltose (31); and araBAD and araE by arabinose (32). For most operons, we determined the levels of β both at the promoter and at a distal region of the coding sequence under noninducing and inducing conditions.
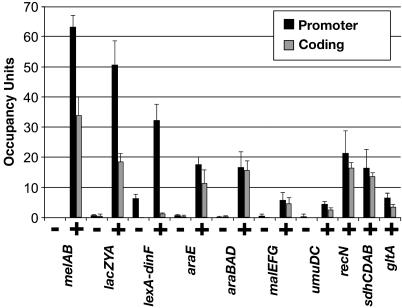
Fig. 1.
Association of β with transcribed regions. Occupancy units at the indicated promoter (black bars) and coding (gray bars) regions under noninducing (-) and inducing (+) conditions are defined as a background-subtracted ratio of binding of β to the tested region and to a control region located within the coding sequence of the predicted ORF yabN.
There is a clear increase in β association at both promoters and coding regions upon induction of all seven operons tested under both noninducing and inducing conditions (Fig. 1), demonstrating that association of β with these regions correlates with transcription. For the promoters tested, the induced level of β association varies over a 10-fold range, presumably reflecting different levels of transcriptional activity. We cannot directly compare fold-induction of β association with fold-induction at the RNA or protein level, because the level of β association under noninducing conditions is near the experimental background of the ChIP technique. In addition, RNA levels in vivo are determined both by the rate of synthesis and degradation, whereas measurements of β association are independent of the rate of mRNA decay.
In six of 10 operons tested, the association of β with coding sequences is significantly decreased as compared to association with promoter DNA (Fig. 1). More detailed mapping of the melAB and lacZYA operons (see below) confirms this preferential association of β with the promoter-proximal region. In one case, lexA-dinF, the drop in β association from the promoter to the coding sequence is particularly dramatic (>20-fold). A significant level of β association is seen at the lexA-dinF promoter even under noninducing conditions. The striking localization of RNAP at the promoter of the lexA-dinF operon (Fig. 1) suggests that RNAP is efficiently recruited to this promoter but undergoes the transition to elongation at a very slow rate. It is also possible that the pattern of β association at the lexA-dinF operon represents high level of transcription of a prematurely terminated RNA product, although such a hypothesis does not easily account for the significant level of β association during noninducing conditions. Formally, the behavior of RNAP at the lexA-dinF operon resembles the situation at the Drosophila hsp70 and Saccharomyces cerevisiae cyc1 promoters, where, under nonactivating conditions, RNAP is recruited to the promoter but does not elongate (33, 34).
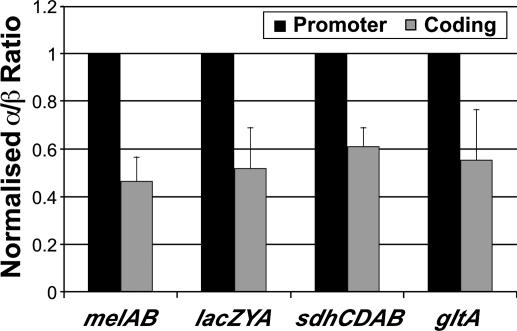
We also determined the association of the α-subunit of RNAP with the promoters and coding sequences of the melAB, lacZYA, sdhCDAB, and gltA operons under inducing conditions. Association of the α-subunit is detected at the promoter and coding regions with fold enrichments roughly comparable to those observed with the β-subunit. Interestingly, the α:β occupancy ratio is ≈2-fold higher at promoters than at coding regions at all four operons tested (Fig. 2). We speculate that this difference in α:β occupancy ratios is due to the direct association (and hence increased crosslinking) of the C-terminal domain of α with promoter DNA (35), which is not believed to occur during transcription elongation. In any event, this potential difference in crosslinking efficiency is subtle (2-fold), and it has a very small effect on the ability to detect association of the α-subunit at both promoter and coding regions.
Fig. 2.
The α:β occupancy ratio at the indicated promoter and coding regions under inducing conditions. The fold enrichments for α association at these promoters are within 20% of the fold enrichments for β association.
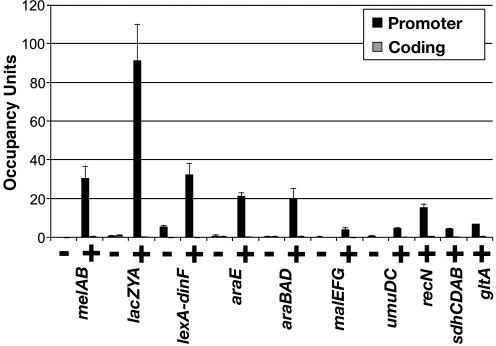
σ70 Does not Associate with Elongating RNAP During Exponential Phase. We determined the level of association of σ70 with the promoters and distal-coding sequences of the same operons in exponentially growing cells under conditions that give high levels of transcription. At all 10 operons tested, σ70 does not detectably associate with coding regions, and hence elongating RNAP, under conditions of activated transcription (Fig. 3). Analysis of seven operons, under noninducing and inducing conditions indicates that the level of σ70 association increases only at promoters (Fig. 3), whereas β association with promoters and coding sequences increases significantly upon activation of transcription (Fig. 1). This confirms that σ70 does not detectably associate with elongating RNAP at the distal regions of these operons.
Fig. 3.
Association of σ70 with transcribed regions. Occupancy units at the indicated promoter (black bars) and coding (gray bars) regions under noninducing (-) and inducing (+) conditions are defined as in Fig. 1.
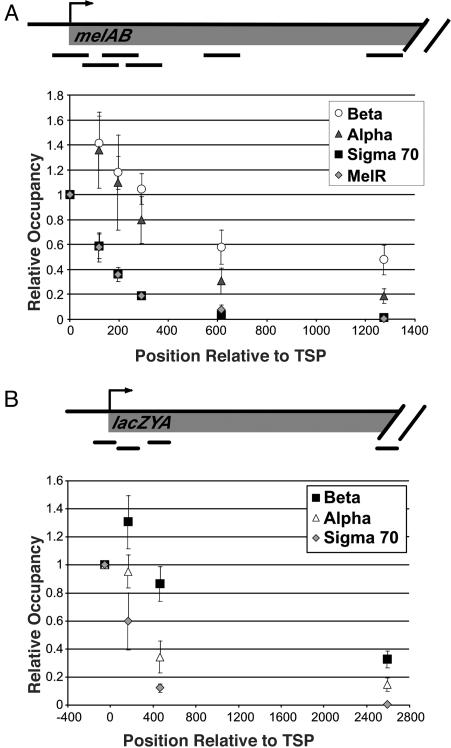
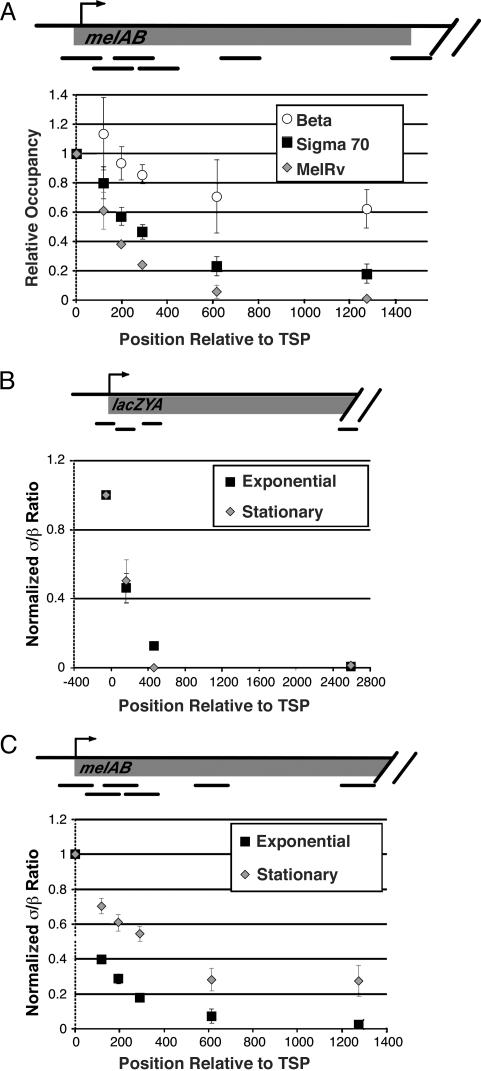
To map the location of σ70 within the melAB operon with greater precision, we compared its association profile at various points throughout the transcribed regions with the profiles of α, β, and MelR (Fig. 4A). MelR specifically recognizes four 18-bp DNA sites at the melAB promoter, the most downstream of which overlaps by 2 bp with the -35 hexamer, the most upstream binding site of σ70 (30). MelR bound at this most downstream site is believed to contact σ70 directly (36). Although MelR binds to five sites that extend 250 bp upstream of the melAB transcription start point, the downstream boundaries of MelR and σ70 binding are closely aligned. As MelR binding is almost certainly restricted to its target sequences in the promoter, a comparison of the downstream boundaries of the MelR and σ70 association will permit us to define the actual domain of σ70 association from the apparent boundary that arises from the data (21-23, 37, 38).
Fig. 4.
Binding of MelR, β, α, and σ70 across the melAB and lacZYA operons in exponential phase. Relative occupancy values at the indicated positions downstream from the RNA start site are normalized to the value at the promoter-proximal region (defined as 1).
Strikingly, the association profile of σ70 across the melAB promoter and immediate-coding region mirrors almost exactly that of MelR, indicating that very little or no σ70 is present in elongating RNAP complexes at the melAB operon (Fig. 4A). Of particular relevance to the apparent contradiction between in vitro experiments (7, 8), the σ70:β association ratio at position +59 to +189 (the second most upstream PCR product) is only 40% of the ratio observed at position -60 to +60 (the most upstream PCR product). Virtually all of the σ70 signal at +59 to +189 reflects association at the promoter, because the MelR ChIP signal at this region is 39% of the signal at -60 to +60. Mapping of σ70 association at lacZYA yields a pattern that is similar to that at melAB (Fig. 4B). In contrast, α and β are associated throughout the entire coding region, with the α:β ratio at coding sequences being ≈2-fold lower than at the promoter (see also Fig. 2). These data clearly demonstrate that, in exponentially growing cells, the vast majority of elongating RNAP complexes does not contain σ70, and that σ70 dissociates rapidly (although not necessarily immediately) after the transition from initiation to elongation in vivo.
σ70 Associates with Elongating RNAP During Stationary Phase in an Operon-Dependent Manner. RNAP isolated from cells in stationary phase have an increased proportion of σ70 at position +32 of the T7A1tR2 template as judged by an in vitro assay (8). However, it is unknown how far σ70 can travel with RNAP or whether this effect is observed on other transcribed regions. We therefore determined the level of σ70 associated with elongating RNAP in stationary phase cells at the melAB and lacZYA operons under noninducing and inducing conditions (Fig. 5A and data not shown). Under noninducing conditions, no β or σ70 was detected at the melAB or lexZYA operons (data not shown). As also observed in exponentially growing cells, significant association of σ70 with elongating RNAP is not detected at the lacZYA operon under inducing conditions (Fig. 5B).
Fig. 5.
Elongating RNAP contains σ70 at the melAB but not the lacZYA operon during stationary phase. (A) Binding of MelR, β, and σ70 to various regions across the melAB operon during stationary phase normalized to binding at the most upstream region. (B and C) The σ70:β occupancy ratio at various regions across the lacZYA (B) and melAB (C) operons. Values are normalized to those of the promoter-proximal region.
Interestingly, at the melAB operon, a significant proportion of elongating RNAP contains σ70 under inducing conditions (Fig. 5 A and C). At various positions within a 2-kb region downstream of the initiation site, there is a clear difference in the occupancy levels of MelR and σ70. Although most σ70 appears to dissociate from RNAP at or near the promoter, a significant fraction remains associated with elongating RNAP over a considerable distance. The σ70:β occupancy ratio at downstream positions is ≈25% of that observed at promoters, suggesting that this proportion of elongating RNAP contains σ70. However, if crosslinking of σ70 at downstream positions is reduced relative to that occurring at promoters, as appears to be the case with the α-subunit (Fig. 2), the proportion of RNAP containing σ70 may be even higher. In contrast to the melAB operon, the association of σ70 with the lacZYA operon is indistinguishable in stationary and exponential phase cells, indicating that under both growth conditions there is no significant association of σ70 with elongating RNAP (Fig. 5 B and C). The difference between the melAB and lacZYA operons is unlikely to reflect transcriptional activity per se, because levels of β association at these operons are roughly equivalent. Thus, although σ70 can associate with elongating RNAP in stationary phase cells, it does so in an operon-specific manner.
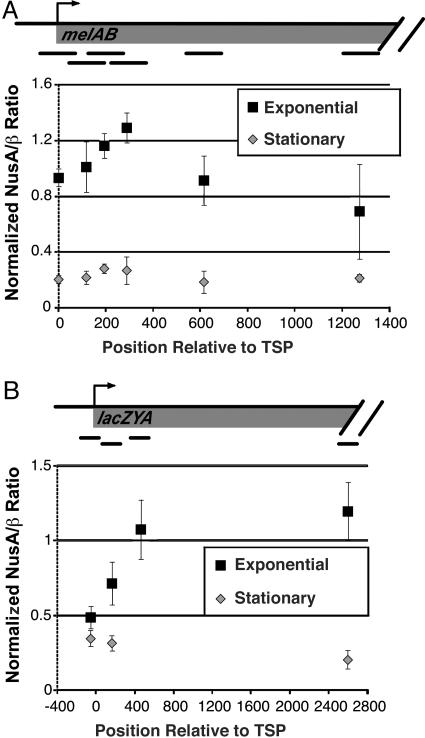
NusA Association with Elongating RNAP Decreases Significantly in Stationary Phase. NusA is a transcription elongation factor that associates directly with RNAP by contacting a region that overlaps the binding site for σ70 (13, 14). It has been proposed that NusA association occurs concommitently with dissociation of σ70 from RNAP during the transition from initiation to elongation (13, 15, 16). We determined the relative proportion of elongating RNAP complexes that contain NusA at regions spanning the melAB and lacZYA operons during both exponential phase and stationary phase (Fig. 6). At both operons tested, the peak of NusA association occurs just downstream of the peak of RNAP association, consistent with the suggestion that NusA association with RNAP occurs during early elongation after the dissociation of σ70. Unexpectedly, the level of association of NusA with elongating RNAP at both operons is significantly lower during stationary phase than during exponential phase. The cellular level of NusA is not significantly lower in stationary phase than in exponential phase (Western blotting; data not shown), consistent with previous observations (39). These observations suggest that the functional properties of either RNAP or NusA are modified when cells enter stationary phase.
Fig. 6.
NusA association with transcribed regions. Data show the NusA:β ratio at various regions across the melAB (A) and lacZYA (B) operons.
Discussion
Molecular Implications. Our work represents the first analysis of RNAP association with bacterial operons in vivo. An important conclusion from our results is that, during exponential phase, little or no elongating RNAP contains σ70 at distances of >50 bp downstream of the transcription start site. This is in contrast to the fluorescence resonance energy transfer-based in vitro studies by using reconstituted RNAP, which showed that >60% of elongating RNAP complexes located 50 bp from the promoter contain σ70 (7). Our data are more consistent with other biochemical studies showing that, for RNAP purified from exponentially growing cells, σ70 is present on only a small proportion (≈7%) of elongating complexes at position +32 on a specific DNA template (8). Our results show that this fraction is even lower at the majority of operons in vivo. This is most likely due to the presence within the cell of other proteins, such as NusA, that play a role in transcription elongation, and can therefore affect the association of σ70 with RNAP.
RNAP purified from stationary phase cells displays a significant increase (to 33%) in elongation complexes in vitro that contain σ70, and such σ70-containing complexes are 5-fold more efficient in a multiround transcription assay (8). These observations have led to the suggestion that RNAP complexes containing σ70 are immediately able to initiate another round of transcription (8). We show here that elongating RNAP can contain a significant level of σ70 in vivo, but this occurs in an operon-dependent manner. Thus, under certain conditions such as stationary phase, either the transcriptional activator protein or the promoter/operon sequence determines the degree to which σ70 associates with elongating RNAP. We suspect that the operon-specific distinction between σ70-containing or σ70-deficient forms of elongating RNAP is made at or near the promoter, because the proportion of σ70 is comparable throughout the melAB operon; i.e., σ70 does not appear to gradually dissociate as RNAP transcribes this operon. Interestingly, a recent report has shown that, in vitro, a σ70-dependent pause site 17 nt downstream of the lacUV5 promoter plays an important role in determining the proportion of elongating RNAP complex that contain σ70 (12).
It seems likely that E. coli will have other operons that, like melAB, have a significant proportion of elongating RNAP complexes that contain σ70 in stationary phase cells. Given that RNAP complexes containing σ70 appear to transcribe more efficiently in vitro (8), our results strongly suggest that transcription of specific operons in vivo can be regulated by altering the association of σ70 with RNAP during elongation. This has implications, not only for the regulation of transcription initiation, but also for transcription elongation and termination. All studies on transcriptional elongation and termination to date have focused on RNAP complexes lacking σ70. It seems likely that the elongation and termination properties of RNAP and its interaction with elongation and termination factors would be significantly altered by the presence of σ70. Therefore, we speculate that the association of σ70 with elongating RNAP may be an important factor in regulating the levels of transcriptional re-initiation, elongation, and termination. Additionally, association of σ70 with elongating RNAP could allow for σ70-dependent pausing at sequences within coding sequences that resemble a -10 hexamer (10).
We also speculate that the level of NusA association with RNAP may affect the ability of σ70 to associate with elongating RNAP. NusA and σ70 bind to overlapping sites on RNAP (13, 14), and there is evidence to suggest that NusA association with RNAP accompanies σ70 release (13, 15, 16). As the level of NusA associated with elongating RNAP is greatly reduced in stationary phase cells (Fig. 3), this may create a permissive situation to allow σ70 to associate with elongating RNAP. However, there is no direct evidence suggesting that NusA facilitates dissociation of σ70 from core RNAP, and there is evidence to suggest that NusA can associate with σ70-containing RNAP (8). We can conclude, however, that the reduced association of NusA is not sufficient for σ70 association with elongating RNAP.
ChIP for Analyzing Transcriptional Mechanisms in E. coli. ChIP is a powerful and widely used technique for elucidating transcriptional regulatory mechanisms in a wide range of eukaryotes, but it has been rarely used for analysis of transcriptional regulation in prokaryotes. Although there is a great deal of knowledge of prokaryotic transcriptional regulatory mechanism from extensive biochemical and genetic analysis, our results from ChIP provide direct and physiologically relevant information that could not be obtained by other means and that is often unexpected. First, we demonstrate that σ70 does not associate significantly with elongating RNAP in exponentially growing cells, thereby resolving an apparent discrepancy between biochemical studies (7, 8). Second, we show that RNAP is preferentially associated at some promoters in vivo, and identify an operon (lexA-dinF) where this preference is especially dramatic. Third, we make the unexpected observations of operon-specific association of σ70 and generally reduced association of NusA with elongating RNAP in stationary phase cells. We believe that ChIP will become an increasingly important approach for elucidating transcriptional regulatory mechanisms in prokaryotes.
Supplementary Material
Acknowledgments
We thank Ann Hochschild and Bryce Nickels (Harvard Medical School, Boston) for the MG1655 strain, Eiji Tamai and Steve Busby for MelR Ab, Simon Dove for assistance with preliminary experiments, and Sudanshu Dole, David Grainger, Steve Busby, Marc Schwabish, Joe Geisberg, and Zarmik Moqtaderi for helpful discussions and comments on the manuscript.
Author contributions: J.T.W. and K.S. designed research; J.T.W. performed research; J.T.W. and K.S. analyzed data; and J.T.W. and K.S. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: RNAP, RNA polymerase; ChIP, chromatin immunoprecipitation.
References
- 1.Gross, C. A., Chan, C., Dombroski, A., Gruber, T., Sharp, M., Tupy, J. & Young, B. (1998) Cold Spring Harbor Symp. Quant. Biol. 63, 141-155. [DOI] [PubMed] [Google Scholar]
- 2.Hansen, U. M. & McClure, W. R. (1980) J. Biol. Chem. 255, 9564-9570. [PubMed] [Google Scholar]
- 3.Shimamoto, N., Kamigochi, T. & Utiyama, H. (1986) J. Biol. Chem. 261, 11859-11865. [PubMed] [Google Scholar]
- 4.Stackhouse, T. M., Telesnitsky, A. P. & Meares, C. F. (1989) Biochemistry 28, 7781-7788. [DOI] [PubMed] [Google Scholar]
- 5.Krummel, B. & Chamberlin, M. J. (1989) Biochemistry 28, 7829-7842. [DOI] [PubMed] [Google Scholar]
- 6.Murakami, K. S., Masuda, S. & Darst, S. A. (2002) Science 296, 1280-1284. [DOI] [PubMed] [Google Scholar]
- 7.Mukhopadhyay, J., Kapanidis, A. N., Mekler, V., Kortkhonjia, E., Ebright, Y. W. & Ebright, R. H. (2001) Cell 106, 453-463. [DOI] [PubMed] [Google Scholar]
- 8.Bar-Nahum, G. & Nudler, E. (2001) Cell 106, 443-451. [DOI] [PubMed] [Google Scholar]
- 9.Ring, B. Z., Yarnell, W. S. & Roberts, J. W. (1996) Cell 86, 485-493. [DOI] [PubMed] [Google Scholar]
- 10.Mooney, R. A. & Landick, R. (2003) Genes Dev. 17, 2839-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brodolin, K., Zenkin, N., Mustaev, A., Mameava, D. & Heumann, H. (2004) Nat. Struct. Mol. Biol. 11, 551-557. [DOI] [PubMed] [Google Scholar]
- 12.Nickels, B. E., Mukhopadhyay, J., Garrity, S. J., Ebright, R. H. & Hochschild, A. (2004) Nat. Struct. Mol. Biol. 11, 544-550. [DOI] [PubMed] [Google Scholar]
- 13.Gill, S. C., Weitzel, S. E. & von Hippel, P. H. (1991) J. Mol. Biol. 220, 307-324. [DOI] [PubMed] [Google Scholar]
- 14.Traviglia, S. L., Datwyler, S. A., Yan, D., Ishihama, A. & Meares, C. F. (1999) Biochemistry 38, 15774-15778. [DOI] [PubMed] [Google Scholar]
- 15.Greenblatt, J. F. & Li, J. (1981) Cell 24, 421-428. [DOI] [PubMed] [Google Scholar]
- 16.Schmidt, M. C. & Chamberlin, M. J. (1984) Biochemistry 23, 197-203. [DOI] [PubMed] [Google Scholar]
- 17.Lin, D. C. & Grossman, A. D. (1998) Cell 92, 675-685. [DOI] [PubMed] [Google Scholar]
- 18.Kuras, L. & Struhl, K. (1999) Nature 399, 609-612. [DOI] [PubMed] [Google Scholar]
- 19.Aparicio, O. M., Geisberg, J. V. & Struhl, K. (2004) in Current Protocols in Molecular Biology, eds. Ausubel, F. A., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A. & Struhl, K. (Wiley, New York), pp. 21.3.1-21.3.17.
- 20.Orlando, V. (2000) Trends Biochem. Sci. 25, 99-104. [DOI] [PubMed] [Google Scholar]
- 21.Kadosh, D. & Struhl, K. (1998) Mol. Cell. Biol. 18, 5121-5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moqtaderi, Z. & Struhl, K. (2004) Mol. Cell. Biol. 24, 4118-4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wade, J. T., Hall, D. B. & Struhl, K. (2004) Nature, in press. [DOI] [PubMed]
- 24.Cunningham, L. & Guest, J. R. (1998) Microbiology 144, 2113-2123. [DOI] [PubMed] [Google Scholar]
- 25.Wilde, R. J. & Guest, J. R. (1986) J. Gen. Microbiol. 132, 3239-3251. [DOI] [PubMed] [Google Scholar]
- 26.Krueger, J. H., Elledge, S. J. & Walker, G. C. (1983) J. Bacteriol. 153, 1368-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rostas, K., Morton, S. J., Picksley, S. M. & Lloyd, R. G. (1987) Nucleic Acids Res. 15, 5041-5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kitagawa, Y., Akaboshi, H., Shinagawa, T., Horii, T., Ogawa, H. & Kato, T. (1985) Proc. Natl. Acad. Sci. USA 82, 4336-4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richmond, C. S., Glasner, J. D., Mau, R., Jin, H. & Blattner, F. R. (1999) Nucleic Acids Res. 27, 3821-3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Belyaeva, T. A., Wade, J. T., Webster, C. L., Howard, V. J., Thomas, M. S., Hyde, E. I. & Busby, S. J. (2000) Mol. Microbiol. 36, 211-222. [DOI] [PubMed] [Google Scholar]
- 31.Bedouelle, H. & Hofnung, M. (1982) Mol. Gen. Genet. 185, 82-87. [DOI] [PubMed] [Google Scholar]
- 32.Schleif, R. (2000) Trends Genet. 16, 559-565. [DOI] [PubMed] [Google Scholar]
- 33.Gilmour, D. S. & Lis, J. T. (1986) Mol. Cell. Biol. 6, 3984-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martens, C., Krett, B. & Laybourn, P. J. (2001) Mol. Microbiol. 40, 1009-1019. [DOI] [PubMed] [Google Scholar]
- 35.Ebright, R. H. & Busby, S. (1995) Curr. Opin. Genet. Dev. 5, 197-203. [DOI] [PubMed] [Google Scholar]
- 36.Grainger, D. C., Webster, C. L., Belyaeva, T. A., Hyde, E. I. & Busby, S. J. (2004) Mol. Microbiol. 51, 1297-1309. [DOI] [PubMed] [Google Scholar]
- 37.Reid, J. L., Iyer, V. R., Brown, P. O. & Struhl, K. (2000) Mol. Cell 6, 1297-1307. [DOI] [PubMed] [Google Scholar]
- 38.Ng, H. H., Robert, F., Young, R. A. & Struhl, K. (2003) Mol. Cell 11, 709-719. [DOI] [PubMed] [Google Scholar]
- 39.Ishihama, A., Honda, A., Nagasawa-Fujimori, H., Glass, R. E., Maekawa, T. & Imamoto, F. (1987) Mol. Gen. Genet. 206, 185-191. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.