Abstract
Applications that use Bacterial Artificial Chromosome (BAC) libraries often require paired-end sequences and knowledge of the physical location of each clone in plates. To facilitate obtaining this information in high-throughput, we generated pBACode vectors: a pool of BAC cloning vectors, each with a pair of random barcodes flanking its cloning site. In a pBACode BAC library, the BAC ends and their linked barcodes can be sequenced in bulk. Barcode pairs are determined by sequencing the empty pBACode vectors, which allows BAC ends to be paired according to their barcodes. For physical clone mapping, the barcodes are used as unique markers for their linked genomic sequence. After multi-dimensional pooling of BAC clones, the barcodes are sequenced and deconvoluted to locate each clone. We generated a pBACode library of 94,464 clones for the flounder Paralichthys olivaceus and obtained paired-end sequence from 95.4% of the clones. Incorporating BAC paired-ends into the genome preassembly improved its continuity by over 10-fold. Furthermore, we were able to use the barcodes to map the physical locations of each clone in just 50 pools, with up to 11 808 clones per pool. Our physical clone mapping located 90.2% of BAC clones, enabling targeted characterization of chromosomal rearrangements.
INTRODUCTION
Today, various vector-free high throughput strategies dominate de novo genome sequencing projects, such as Next Generation Sequencing (NGS) (1–4), Third Generation Sequencing (TGS) (5), Dovetail Chicago (6) and Bionano Matrix (7). However, repetitive regions are frequently missing or mis-assembled, leading to fragmented and incomplete genomes (8,9). This is particularly problematic because these poorly sequenced and assembled regions are hot spots for genetic and chromosomal variation that can cause phenotypic variation and diseases (9–12). Although expensive and cumbersome, BAC-by-BAC sequencing strategies can facilitate assembly of high-quality reference genomes, especially for genomes that are frequently used in functional studies (13–16). Furthermore, BAC libraries are essential tools for various genomic and genetic studies, including physical mapping (17), cytogenetic mapping (18), comparative genomic analysis (19, 20), targeted genome sequencing (21) and functional complementation experiments (22).
A major purpose of BAC libraries is to provide physically stable and amplifiable DNA fragments cloned in microbial hosts for further applications. In order to screen BAC libraries for a clone of interest, previous studies developed methods including probe hybridization and 4D-PCR (23). More recently, the whole genome profiling (WGP) technique provided a mean of mapping BAC clones utilizing NGS (24). In WGP, subsets of BAC clones are pooled in 2-dimensions and sequenced in parallel. Clone locations are then deconvoluted using unique genomic sequence in each clone as tags. However, in practice each pool can contain no more than 64 BAC clones because there are insufficient unique genomic sequence tags in a larger pool (25,26). Hence, a more efficient physical clone mapping method is needed.
Another rate-limiting step in BAC application is extracting sequence information from BAC clones at a large scale (27,28). To overcome a similar limitation of fosmid libraries, a high-throughput paired-end sequencing approach called Fosill (Fosmid libraries by Illumina) was recently developed (29) and has been used for the de novo assembly of several genomes (30–34). However, this strategy requires delicate DNA manipulation, and self-circularization via co-ligation of the two ends of the genomic inserts becomes much more difficult when long end sequences are desired. Furthermore, the short ends that are produced by Fosill are incompatible with emerging long-read high-throughput technology (35–37). Therefore, there is a need for a simpler and more general method for high-throughput paired-end sequencing of genomic libraries.
Oligo-based molecular indexes have been widely used in massively parallel sequencing (37–42). In addition to pre-defined indexes, random sequence barcodes are also used as tags in various functional genomics assays (43,44). For example, labeling RNA molecules with 20-bp long random barcodes before amplification reduces noise and bias in RNA-seq (45). However, high-throughput random barcoding has not been used in sequencing vector-based libraries.
To provide a high-throughput approach for BAC paired-end sequencing and physical clone mapping, we developed a random barcode based BAC library system called pBACode. The core idea of pBACode is to introduce random sequences into BAC vectors so that each BAC clone has a pair of unique barcodes flanking its cloning site. With unique barcode pairs in every BAC clone, we can obtain long and accurate sequences of BAC paired-ends and locate clones in a BAC library in high-throughput. We validated the accuracy and efficiency of pBACode by profiling a BAC library of the budding yeast S. cerevisiae. We then used pBACode to construct a BAC library for Japanese flounder Paralichthys olivaceus, whose genome had not previously been sequenced. We obtained paired-end sequences from 95.4% of the BAC clones. This data enabled a high quality assembly of the flounder genome, with a scaffold N50 length of 10.5 Mb and 90% coverage. In addition, we were able to assign 90.2% of BAC clones to unique positions in plates. This physical clone map enabled targeted BAC clone sequencing to investigate chromosome rearrangements in genes related to flatfish traits.
MATERIALS AND METHODS
PCR reaction conditions and primer sequences are listed in Supplementary Data.
Construction of pBACode-1 vectors based on pcc2FOS
PCR primers contained 20-nt of random sequence flanked by cloning sites at the 5΄ ends and sequence complementary to the pcc2FOS vector at the 3΄ ends. PCR products were NheI digested, self-ligated and transformed into Escherichia coli strain EPI300 (Epicentre) using electroporation (Bio-Rad). Transformants were cultured on LB plates containing 12.5 μg/ml chloramphenicol over night before counting. The clones were washed off plates into liquid LB, pooled together, and stored at −80°C with glycerol.
Construction of pBACode-2 vectors
PCR primers to generate the first intermediate vector contained I-SceI sites and ClaI sites. The PCR product was ClaI digested, self-ligated and propagated in E. coli DH5α.
To generate the second intermediate vector, two NsiI sites were first introduced into pcc2FOS by overlapping PCR and subcloning. Next, a kanR fragment was amplified from pHis-2 (Clontech) using primers containing a HindIII site, and then subcloned into the NsiI-pcc2FOS at the HindIII site. The LacZ-kanR-LacZ fragment of the intermediate vector was PCR amplified using primers containing 20-nt of random sequence and an NheI site or ClaI site. The PCR product was NheI and ClaI digested, subcloned into the first intermediate vector, and then transformed into E. coli strain EPI300 (Epicentre) using electroporation (Bio-Rad). Transformants were cultured on LB plates with 50 μg/ml kanamycin and 12.5 ug/ml chloramphenicol over night before counting and collecting.
BAC library construction
BAC libraries were constructed as described previously (46). Genomic DNA from yeast Saccharomyces cerevisiae strain S288C and the flounder Paralichthys olivaceus double-haploid strain 3165 was extracted from liquid culture and blood cells, respectively. Yeast protoplasts and flounder cells were evenly embedded into a gel plug of low melting temperature agarose. The gel plug was then treated with proteinase K for 48 h and partially digested by endonuclease HindIII at a concentration of 20 U/ml for 10 min at 37°C. Products were separated by pulsed-field gel electrophoresis and DNA fragments from 120 to 300 kb were purified. Either pBACode-1 or pBACode-2 vectors were digested by HindIII and dephosphorylated. The linear vectors were then ligated with the genomic fragments. The ligation product was transformed into E. coli strain DH10 (Life technologies) using electroporation and pooled and cultured on LB plates with chloramphenicol. Clones were picked into 384-well plates, cultured and stored at -80°C.
BAC end sequencing and local assembly
BAC clones were cultured in 96-well plates before mixing and DNA extraction. After digestion with NotI for pBACode-1 or I-sceI for pBACode-2, BAC DNA was sheared into 1 kb fragments by ultra-sonication (Covaris S220/E220), end-repaired (NEB end repair module), and circularized by T4 DNA ligase (NEB) at 1 ng/ul DNA. The circularized DNA was used as a template for inverse PCR amplification of BAC ends. The 3΄ ends of the PCR primers are complementary to the vector backbone while the 5΄ ends contain Illumina adapter sequences. PCR products were Illumina sequenced.
We extracted barcodes and genomic sequences from read pairs using in-house Perl scripts. Genomic sequences were then grouped according to their barcodes. Sequencing errors in barcodes were removed as described in the next section except that only one mismatch was allowed. BAC end sequences with same barcode were assembled into preliminary contigs using PHRAP (47,48). The preliminary contigs were aligned by raw reads using bowtie2 (49) to identify errors and low coverage regions, which were corrected and trimmed, respectively. Barcodes identified on more than one BAC end were detected using SEED (50). If >90% reads of the barcode belonged to one BAC end, the other BAC ends containing the barcode were discarded as contamination. Otherwise, these barcodes were considered non-unique in the BAC library and ignored in subsequent analyses. The BAC end contigs were aligned to the reference genome or genome assembly using bowtie2 (49).
Barcode pair sequencing
BAC libraries derived from pBACode-1 and pBACode-2 were digested by HindIII and NsiI, respectively, and then circularized to co-ligate barcode pairs. Barcode pairs in the circular DNA were PCR amplified with Illumina-compatible primers and sequenced on an Illumina Hiseq using 2 × 101 bp and 2 × 125 bp mode for pBACode-1-derived and pBACode-2 derived libraries, respectively. Read pairs were merged by FLASH (51) and then analyzed with custom Perl scripts. First, the sequence of vector backbone was trimmed to extract barcode pairs. Identical barcode pairs were clustered and their reads were counted. To remove sequencing errors, raw barcode pairs were compared to each other. If two barcode pairs differed by no more than two mismatches and their read counts differed by no less than 10-fold, these mismatches were considered as sequencing errors and corrected. If the read count of a hybrid read pair was no more than tenth of either of its parent pairs, it was removed while keeping parent barcode pairs. Otherwise, all three barcode pairs were considered ambiguous and discarded.
BAC library high dimensional pooling, barcode sequencing and deconvolution
For the S. cerevisiae BAC library, 1536 clones were inoculated in sixteen 96-well plates with 1.2 ml LB and 12.5 μg/ml chloramphenicol in each well. Bacteria were cultured for 48 h at 37 °C before 3D pooling (by plates, by columns, and by rows) using a liquid handling platform (Tecan Freedom EVO300).
The BAC library of the flounder P. olivaceus was cultured in 984 96-well plates and pooled in five-dimensions (by rows, by columns, by plates with same last digits, by plates with same second to last digits, and by plates with same third to last digits).
The left barcodes were PCR amplified using Illumina-compatible primers and sequenced. Every barcode was assigned to a specific location in a BAC library according to the intersection of pools in all dimensions where it occurred. Barcodes were discarded unless these criteria were satisfied: (a) A barcode must present >5% of the average sequencing depth in each pool; and (b) if a barcode occurred in multiple pools from same dimension, the reads in the most enriched pool (the putatively correct one) must represent at least two thirds of the reads from this barcode in all pools from that dimension.
Genome pre-assembly using shotgun libraries and 3-kb jumping libraries
Yeast genomic DNA was sheared to around 220 bp fragments and then sequenced using 2 × 125 bp mode on an Illumina Hiseq, yielding 8.6 M reads. A 3 kb mate-pair library was constructed and sequenced on an Illumina Hiseq, yielding 1.2 M reads. These reads were preassembled using SOAPdenovo2 (52).
Flounder genomic DNA from blood cells was sheared to around 220 bp fragments and then sequenced using 2 × 125 bp mode on an Illumina Hiseq, yielding 482.6 M reads. A 3 kb mate-pair library was constructed and sequenced by Illumina Hiseq, yielding 261.0 M reads. These reads were preassembled using ALLPATHS-LG (53).
Assembly correction
BAC-PE sequences were aligned to a draft scaffold using Bowtie2 (49). The draft can be either a pre-assembly derived from only shotgun libraries and 3-kb jumping libraries, or an intermediate assembly into which information from BAC-PEs has been incorporated to some degree. For simplicity, we use term ‘scaffold’ in its broad sense to refer to both a supercontig from a pre-assembly without BAC-PEs and a scaffold from an intermediate or final assembly with BAC-PEs. Based on the alignment result, we categorized BAC clones. Concordant BAC: those whose two ends aligned to same scaffold with opposite orientation and the inter sequence less than 300 kb. Discordant BAC: those whose two ends aligned to same scaffold with un-opposite orientation or inter sequence longer than 300 kb, or two ends aligned to different scaffolds with the sum of sequence spanned by BAC longer than 300 kb. The pair of scaffolds linked by a discordant BACs was designated as incompatible. Undefined BAC: those whose two ends aligned to different scaffolds with the sum of sequence spanned by BAC less than 300 kb. Concordant BACs were used to build tiling paths along each scaffold. If a pair of incompatible scaffolds were linked by multiple discordant BACs and at least two discordant BACs’ ends were more than 30 kb apart in both scaffolds, we looked for the tiling paths spanned by these discordant BACs and closest to these BACs’ ends. The regions between these tiling paths and the discordant BACs’ ends were considered to be mis-assembly spots and deleted.
Computational pipeline for the Japanese flounder genome assembly
de novo assembly
SSPACE (54) was used for first round of BAC-PE-based scaffolding under default parameters. BAC-PE sequences were aligned to the pre-assembly to detect and split mis-assembled scaffolds as described above. Next, we wrote a Perl script to identify BACs that unambiguously connected two scaffolds, assuming 300 kb as the maximum size of their inserts. If multiple BACs supported the same scaffold pairing and at least two BACs’ ends were more than 30 kb apart in both scaffolds, they were considered high fidelity and used by SSPACE to generate the final version of de novo assembly.
Reference-guided merging
Synteny blocks were generated by comparing the de novo assembly with the tongue sole genome using MUMmer (55) using parameter –l 10 –c 100. If two neighbor collinear blocks were connected by a BAC and the fragments spanned by the BAC were no more than 300 kb, this BAC was considered collinear. In addition, we wrote Perl scripts to identify chains of BACs that were connected by no more than three intermediate scaffolds that were not homologous to anywhere in the sole genome. If the two ends of a chain fell into two neighbor synteny blocks, they were also considered as collinear BACs. Our perl scripts merged scaffolds connected by collinear BACs to generate new synteny blocks. However, scaffolds bearing genetic markers of different flounder linkage groups were not merged. Every new synteny block was compared to the tongue sole genome using MUMmer (55). If there were contigs around the merging sites that were not collinear in the tongue sole genome, they were reordered to satisfy collinearity.
Synteny analysis
The flounder genome assembly was aligned to the stickleback genome using MUMmer (55) using parameter –l 10 –c 100 before removing multiple alignments. Alignments with the same orientations and neighbor positions in both genomes were merged into synteny blocks. Synteny blocks between the flounder and tongue sole genomes were generated in the same manner. Synteny blocks between all three of these genomes were generated by intersecting the flounder-stickleback synteny blocks with the flounder-sole ones.
RESULTS
Barcoded BAC cloning vector pools
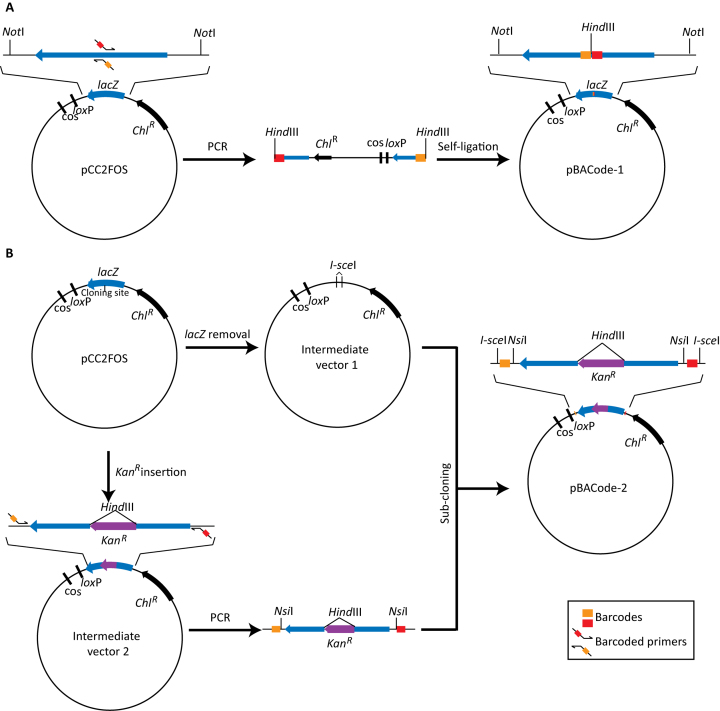
We constructed two sets of randomly barcoded BAC cloning vectors, pBACode-1 and pBACode-2 respectively, by modifying the conventional BAC vector pcc2FOS (56) so that its cloning site is flanked by a pair of 20-bp random sequences (Figure 1). In pBACode-1, we added random barcodes directly by PCR amplification using primers carrying random sequences (Figure 1A). This should generate up to 420 uniquely barcoded BAC vectors. However, one caveat of this strategy is that random barcodes can introduce stop codons into the LacZ gene. As a result, 64% clones (62 out of 97 in our pilot experiment) carrying our empty pBACode-1 pool were white on X-gal plates. This leads to false positives in the subsequent blue/white screening during BAC library construction. Indeed, about 10.9% clones were empty vectors in our yeast BAC library generated using pBACode-1.
Figure 1.
Construction of pBACode cloning vectors. (A) A pBACode-1 pool is generated by PCR amplification using pcc2FOS as a template. The 3΄ ends of primers are complementary to the vector sequences flanking the cloning sites so that the whole pcc2FOS vector except for its cloning sites is amplified. The 5΄ ends of the primers carry the cloning sites followed by 20-bp random sequences. (B) A pBACode-2 pool is generated by a combination of PCR amplification and subcloning. First, an intermediate vector is constructed based on pcc2FOS such that its lacZ sequence is replaced by an I-SceI site. The second intermediate vector is constructed by inserting the kanR selection gene marker into the cloning site of pcc2FOS, and then the lacZ-kanR-lacZ segment is amplified using primers containing random barcodes. The resulting PCR product is subcloned into the first intermediate vector. Kanamycin selection is used to eliminate no-insert vectors.
To eliminate this problem, pBACode-2 vectors were constructed by adding random barcodes outside of lacZ (Figure 1B). Generating pBACode-2 required subcloning (Figure 1B), which may limit the number of random barcodes or introduce uneven distribution of different barcodes. To determine the extent of this issue, we sequenced the barcode pairs in our pBACode-2 pool and found that it contained 6 million unique barcode pairs. in silico simulation showed that the number and distribution of random barcodes in our pool vastly surpassed the requirements of generating a large BAC library. For example, a BAC library of 100,000 clones derived from the pBACode-2 pool is expected to have fewer than 3% clones with non-unique barcodes (Supplementary Data). This prediction was supported by our pBACode-2-derived flounder BAC library, in which only 3082 out of 94 464 clones (3.3%) had non-unique barcode pairs.
Another potential caveat of the PCR-based barcoding strategy is that it could generate multiple vectors with same barcode on one side, which we call 1:N paired barcodes. Our pBACode-2 pool was generated by PCR amplification of 30 cycles using primers with random barcodes, so a barcode on one side could pair with up to 29 different barcodes on the other side. Indeed, 22.3% of our pBACode-2 vector pool of 6 × 106 clones are 1:N paired barcodes. Nevertheless, the size of a BAC library usually is much smaller than that of our pBACode-2 vector pool so that fraction of 1:N paired barcodes in the BAC library will decrease correspondingly. For example, our flounder BAC library had 94 464 clones, only 0.3% of which had 1:N paired barcodes.
BAC paired-end sequencing and physical clone mapping using the pBACode system
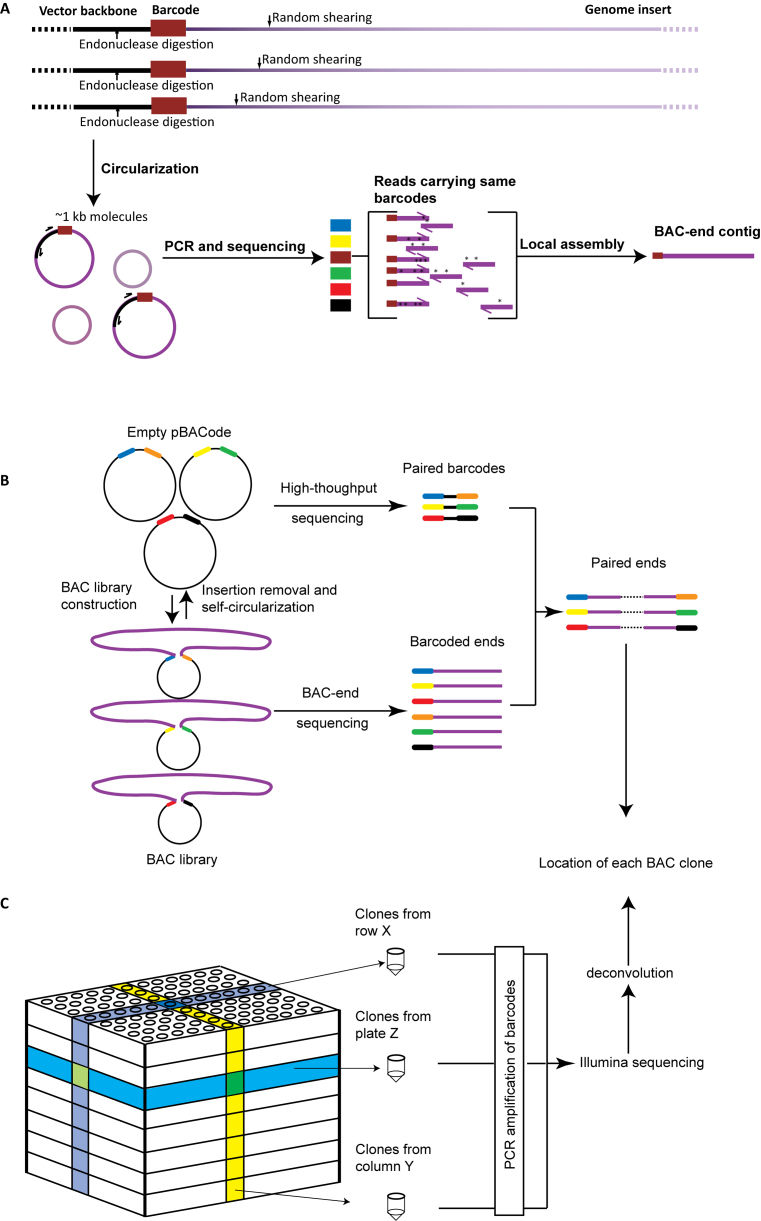
BAC ends and their linked barcodes are sequenced using an inverse PCR strategy to avoid sequencing the whole BAC plasmid (Figure 2A). Mixed BAC clones are digested with a restriction enzyme in the vector backbone outside the barcode and then randomly sheared into ∼1 kb fragments. After self-circularization, inverse PCR is used to amplify the BAC ends with their barcodes and the PCR products are sequenced in bulk (Figure 2A). As this process generates multiple reads per BAC end, reads carrying the same barcodes can be assembled to generate BAC end contigs (Figure 2A). This produces longer, more accurate sequences for genome assembly than are possible using single Illumina reads.
Figure 2.
Barcoding enables high-throughput profiling of a BAC Library. (A) The method to generate long and accurate BAC end sequences is illustrated using multiple copies of one side of a single BAC plasmid. Black thick lines represent the pBACode backbone while purple lines indicate the genomic insert. Deeper purple fragments are closer to the end of genomic insert. Different colored blocks represent different barcodes. (B) Barcode pair sequencing. Genomic inserts are removed and the empty vectors are self-circularized to co-ligate barcode pairs. This brings barcodes close enough together that an Illumina read can cover both barcodes. After the barcode, pairs are sequenced, the ends of the genomic inserts in each BAC clone are paired according to their barcoded ends. (C) Physical clone mapping. BAC clones in 96-well plates are pooled in multiple dimensions (by row, by column, and by plate, or by various combinations of plates). After BAC DNA is extracted from each pool, the barcodes are PCR amplified using primers with indexed Illumina adapters and sequenced. Each BAC clone is assigned to a specific location in the BAC library after deconvolution of their barcodes from the pooled sequences. These procedures are applicable to both pBACode-1- and pBACode-2-based genomic libraries.
One can pair BAC end contigs by sequencing barcode pairs in empty pBACode vectors (Figure 2B). To accomplish this, genomic inserts are removed and the vector backbones are circularized by self-ligation. As intramolecular ligation is far more likely than intermolecular ligation, the probability of incorrect pairing is low. To validate this approach, we sequenced the barcode pairs of the flounder BAC library after insert removal and self-circularization. 4.32 M barcode pairs were detected, much more than the number of BAC clones (94 464) (Table 1). However, 97.6% of these barcode pairs were supported by only one or two reads and differed from other barcode pairs by no more than two nucleotides, suggesting that they may be the result of sequencing errors (Supplementary Data). In addition, some low abundance pairs shared one of their barcodes with another pair (Supplementary Data), suggesting that they may be the product of intermolecular ligation. After computationally correcting or discarding these two classes of barcode pairs, we obtained 90 156 high-confidence barcode pairs (Table 1). There was no discrepancy between these barcode pairs and those identified by sequencing the pBACode-2 pool before BAC library construction, demonstrating the high accuracy of our barcode-pair sequencing strategy.
Table 1. Summary of barcode pairs and barcode-based physical clone mapping.
| Yeast S. cerevisiae | Flounder P. olivaceus | ||
|---|---|---|---|
| BAC library size | 1536 | 94 464 | |
| Barcode pair seuqencing | Clean readsa (million) | 1.5 | 30 |
| Raw barcode pairs | 52 243 | 4 315 007 | |
| Consolidated barcode pairs | 1381 (89.9%) | 90 156 (95.4%) | |
| Physical clone mapping | Pooling dimension | 3 | 5 |
| Pools | 36 | 50 | |
| Clones per pool | 96–128 | 7872–11 808 | |
| Clean readsa (million) | 0.78 | 55 | |
| Located BAC clonesb | 1324 (86.2%) | 85 173 (90.2%) | |
| Tested clones | 12 | 42 | |
| Validated clones | 12 | 42 |
aReads with QC > 20 were selected and their vector parts were trimmed.
bClones whose barcodes mapped to unique location in the BAC library by deconvolution.
After BAC-end sequencing, the relationship between barcodes and genomic sequences is established. Therefore, barcodes can be used as markers of their linked genomic sequences for physical clone mapping. In another words, instead of sequencing genomic sequences after multiple-dimensional clone pooling, one can determine the location of each clone in the BAC library by sequencing the barcodes (Figure 2C).
Profiling a yeast BAC library as a proof of principle
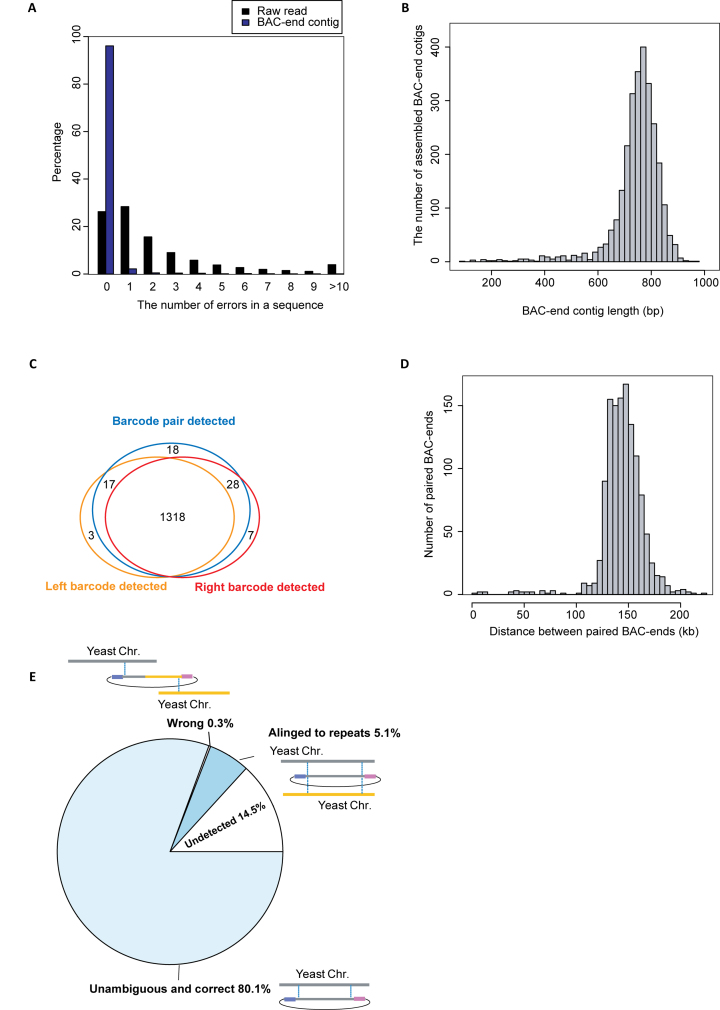
To test our method, we construct a BAC library of budding yeast S. cerevisiae using pBACode-1. The BAC library is composed of 1,536 clones (Table 1). We extracted BAC plasmids in bulk and sequenced the BAC ends using 2 × 301 bp mode on an Illumina Miseq (Figure 2A). Aligning filtered raw reads to the reference yeast genome revealed that there were 2.3 sequencing errors on average per read (Figure 3A). Next, reads were classified as coming from the left or right end according to their vector backbone sequences and then grouped according to their barcodes. After local assembly of reads with the same barcode sequence, we generated 2728 BAC end contigs that were 738 bp in length on average (Figure 3B). 96% BAC end contigs had no mismatch to the reference yeast genome (Figure 3A).
Figure 3.
Characterization of BAC-ends from the yeast BAC library. (A) The distribution of sequencing errors in filtered raw reads and BAC-end contigs, respectively. BAC-end contigs were derived from local assembly using 0.45 M reads encompassing 1373 barcodes for the left ends, and 0.45 M reads encompassing 1387 barcodes for the right ends. (B) The length distribution of the BAC-end contigs. (C) Barcode detection coverage. Left and right barcodes were sequenced with their linked BAC ends using our BAC-end sequencing protocol (Figure 2A). Barcode pairs were detected using our barcode pair sequencing protocol (Figure 2B). (D) The length distribution of genomic fragments spanned by unambiguous and correct paired BAC-end sequences. (E) Categorization of yeast BAC paired-ends. A BAC clone was considered undetected if either barcode was missing after BAC-end sequencing. A BAC clone was considered unambiguous and correct if its ends were aligned to two unique genomic loci spanning <300 kb apart in convergent orientation. An incorrect BAC clone is one whose paired-ends aligned to unique genome loci but on different chromosomes, or not in the convergent orientation on same chromosome, or more than 300 kb apart on same chromosome.
We next generated co-ligated barcode pairs by insert removal and self-circularization (Figure 2B). Barcode pairs from empty vectors were PCR amplified and Illumina sequenced, yielding 1.5M filtered reads. After correcting and discarding erroneous barcode pairs as described above (Supplementary Data), we obtained 1381 barcode pairs (Figure 3C). This represents 89.9% of BAC clones (Table 1). The barcodes of 1318 of these pairs were detected by both left and right BAC-end sequencing (Figure 3C). In 1239 (80.7%) of these BAC clones, both ends unambiguously aligned to unique loci 144.4 kb apart on average in the yeast reference genome (Figure 3D, E). For 79 clones, at least one end mapped to a repetitive region longer than the BAC end contig and could not be assigned to any unique genomic locus (Figure 3E). Just four clones mapped incorrectly to the reference genome (Figure 3E). Using these paired BAC ends, we de novo assembled a chromosome-scale draft with no errors (Supplementary Data).
The yeast BAC library was cultured in sixteen 96-well plates and pooled in three dimensions by rows (192 clones per pool), columns (128 clones per pool) and plates (96 clones per pool). In total, there were 8 row pools, 12 column pools and 16 plate pools (Table 1). We extracted BAC plasmids from each pool and amplified their barcodes in parallel using Illumina-compatible PCR primers with Illumina indexes. PCR products were sequenced in 2 × 125 bp mode on an Illumina Hiseq, yielding 0.78 M reads in total, or about 100× depth per pool on average.
Physical coordinates of each clone were deconvoluted according to which row pool, column pool, and plate pool its barcode was detected in (Figure 2B). 86.2% of BAC clones were assigned to unique locations in the library. These BAC clones covered 90.1% of the yeast reference genome. To verify the accuracy of our physical clone map, we Sanger sequenced 12 clones from the BAC library. All of them contained the expected barcodes and genomic inserts (Table 1).
We used our physical clone map to investigate the cause of the four ‘wrong’ clones, which the two ends aligned to different chromosomes or distant loci of same chromosome (Figure 3E) (Supplementary Data). Surprisingly, our physical clone map showed that these four clones were located in only two wells. For example, there were two different BAC clones in well 4I14 that represented inter-chromosomal chimeras between Chr. IX and Chr. XI (Supplementary Data). The ends of the two different BACs were <300 kb apart and in convergent orientation either on Chr. IX or on Chr. XI (Supplementary Data). A similar situation was observed in well 1N10 (Supplementary Data). A plausible explanation is that a chimeric BAC resulted from a 4-piece ligation during the BAC library construction (Supplementary Data). Supporting this explanation, a single bacteria clone from well 4I14 contained barcodes from two vectors (Supplementary Data).
de novo assembly of flounder genome
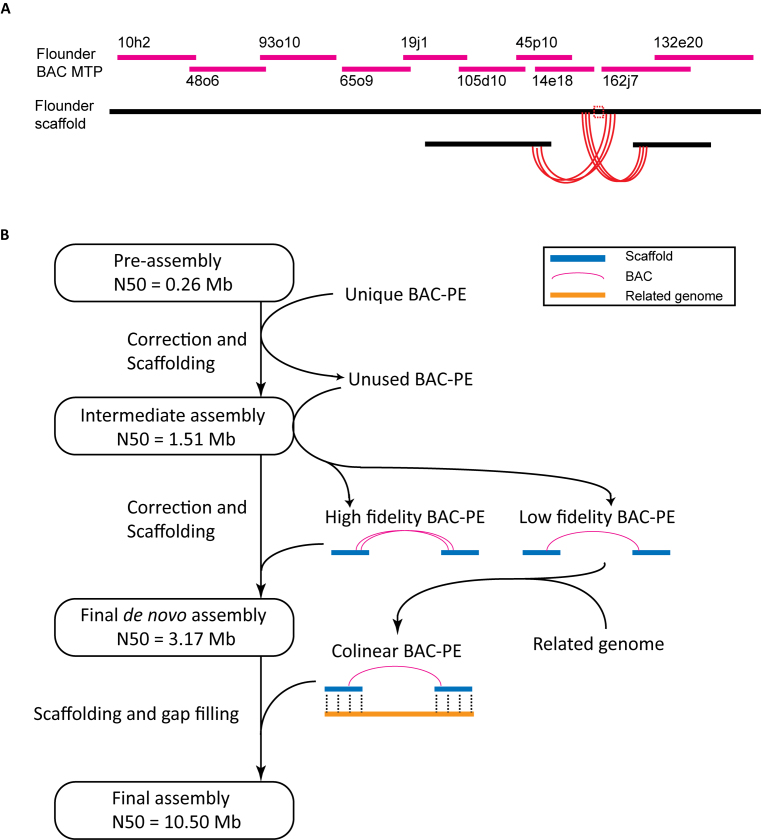
Next, we applied the pBACode-2 system to the de novo assembly of large genome that had not yet been sequenced. We constructed a BAC library of 94 464 clones using pBACode-2 (Figure 1B) of the Japanese flounder P. olivaceus, whose genome size was estimated to be around 700 Mb (57). The pre-assembly of the flounder genome, which was derived from shotgun libraries and 3-kb jumping libraries, had an N50 scaffold length of 256 kb. Paired-end sequencing of the pBACode-2 library (Figure 2) gave rise to 76 877 BAC paired-ends that aligned to unique loci in the pre-assembly, including 31 810 intra-scaffold and 45 067 inter-scaffold ones. To detect and correct pre-assembly errors, we used the intra-scaffold BAC paired-ends to generate 851 minimal tiling paths and the inter-scaffold ones to discover 71 incompatible scaffold pairs (Figure 4A). For each incompatible scaffold pair, we located and split its mis-assembly based on the positions of discordant BAC paired-ends and tiling paths (Figure 4A). Scaffolding the corrected pre-assembly by SSPACE (54) using the BAC paired-ends gave rise to an assembly composed of 10 099 scaffolds with an N50 length of 1.51 Mb. When we aligned the BAC paired-ends back to the assembly, we found that SSPACE failed to utilize 27.5% of the unique BAC paired-ends. The fraction of unused BAC paired-ends was much higher than the usual rate of chimeric clones in a BAC library (58–60), suggesting that incorporating them into the assembly could significantly improve scaffolding. To accomplish this, we developed a computation pipeline to select and utilize high fidelity BAC paired-ends (Figure 4B), resulting in an assembly with a scaffold N50 length of 3.17 Mb (genome size 618.7 Mb with 96.9 Mb Ns), more than a 10-fold improvement over the pre-assembly. Finally, using the tongue sole Cynoglossus semilaevis genome as a reference (61), we achieved an N50 length of 10.54 Mb (genome size 644.1 Mb with 122.3 Mb Ns) by reference-guided assembly (Figure 4B). The size of the final assembly is 639 Mb, covering 90% of the 700 Mb flounder genome (57).
Figure 4.
Improvement of genome assembly using BAC paired-ends (BAC-PE). Only BAC-PEs that aligned to unique loci in the pre-assembly are used. (A) BAC-PE-based mis-assembly correction. An example of a mis-assembly, defined by discordant BAC ends and tiling paths (see method) of a flounder scaffold in the pre-assembly. We deleted the mis-assembly site to split the scaffold. BAC names represent their location in plates, revealed by physical clone mapping. MTP: minimal tiling paths. Red arcs represent BAC clones whose paired ends are discordant. (B) The computation pipeline. First, the pre-assembly scaffolds were assembled using all unique BAC-PEs by SSPACE. From the unused BAC-PEs, we identified high-fidelity BAC-PEs when multiple BAC-PEs supported a scaffold pair and there were at least two BACs with ends more than certain distance (30 kb for the flounder assembly) apart in both scaffolds. After assembly using the high-fidelity BAC-PEs, low-fidelity BAC-PEs and related genome information were used for reference guided merging to generate the final assembly.
Physical clone mapping the flounder BAC library and targeted sequencing
We used the barcode-based method described above to locate the physical location of all flounder BAC clones in the library. The 94 464 clones were pooled in five dimensions so that just 50 pools were needed in total. Each pool had 7872–11 808 clones. After sequencing the barcodes and deconvolution, we assigned 90.2% of BAC clones to unique physical locations in the library (Table 1), spanning 94.9% of the assembly. The physical clone map enabled targeted characterization of genes associated with the flatfish-specific left-right asymmetry trait and sex-determination.
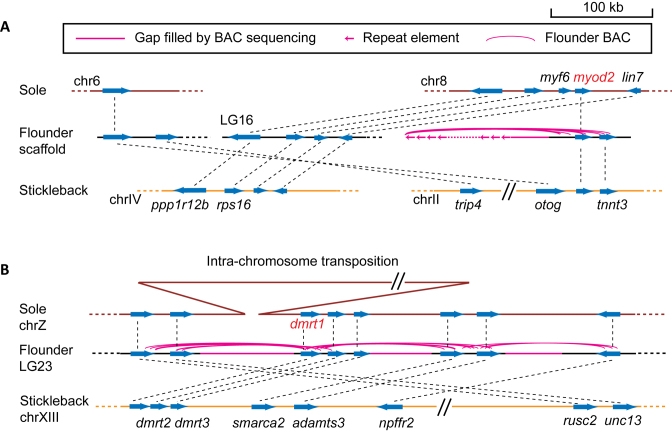
Flatfish are a monophyletic group (Order Pleuronectiformes) characterized by their left-right asymmetric adult body morphology. They share a common ancestor with the left-right symmetric three-spine stickleback (Gasterosteus aculeatus) ∼170–220 million years ago (61). myod2 has been reported to be bilaterally asymmetrically expressed during metamorphosis of flatfish (Schreiber 2013). Flounder myod2 is located on a short scaffold, and its promoter is missing in our assembly (Figure 5A). Nevertheless, myod2 was spanned by 19 BAC clones (Figure 5A), all of which were assigned unique locations in the physical clone map. We randomly selected two clones and sequenced them. The sequence of these BAC clones showed that the upstream region of flounder myod2 was different from that of the three-spine stickleback and the tongue sole. Instead, at least 80 kb upstream region is composed of tandem repeats (Figure 5A) (Supplementary Data). Most repeats have no HindIII sites, explaining why all 19 BAC clones have the same left end (Figure 5A).
Figure 5.
BAC-assisted comparative analysis. (A) myod2 upstream regions in the three fish species. The region of tandem repeats in the flounder genome was not fully assembled. Instead, its size and arrangement is based on restriction mapping. (B) Chromosomal regions around dmrt1 in the three species. The exact site of the insertion upstream of the sole dmrt1 is unknown. Genome fragments and scaffolds are drawn to scale unless specified. Genes are drawn without introns and the tapered sides represent their 3΄ ends. Dashed lines link orthologous genes.
Next, we examined a sex-determination gene. Japanese flounder and three-spine stickleback use an XY sex determination system (62, 63). On the other hand, the sex chromosomes of the tongue sole are ZW (61). dmrt1 is a sex-determination gene in the tongue sole (61), but it is not linked to sex either in the three-spine stickleback (63) or in the Japanese flounder (Supplementary Data). One possibility is that a gain-of-function mutation occurred in the tongue sole dmrt1 after it diverged from flounder. Indeed, we observed that synteny was disrupted upstream of the dmrt1 gene between the tongue sole and the Japanese flounder (Figure 5B). However, there was a gap in the flounder assembly between dmrt1 and its upstream genes (Figure 5B). To clarify the region around dmrt1, we sequenced six BAC clones surrounding dmrt1 from the library, all of which were correctly located by our physical clone map. After gap filling, we compared the syntenic region around dmrt1 between the Japanese flounder and the tongue sole and observed a 1.5 Mb extra segment in the intergenic region upstream of dmrt1 in tongue sole (Figure 5B). Future research could investigate the function of this extra segment in terms of dmrt1 expression regulation and sex determination in the tongue sole.
DISCUSSION
This paper describes pBACode, a method that incorporates random barcode pairs into BAC vectors to make BAC paired-end sequencing compatible with high-throughput sequencing. Barcodes also serve as tags for the genomic sequence of each BAC clone, enabling efficient physical clone mapping. The precision of our method was validated by a pBACode-1-based yeast BAC library. Using the pBACode-2 BAC library, we generated a high continuity genome assembly for Japanese flounder (P. olivaceus). The pBACode-2-mediated physical clone map allowed targeted sequencing to reveal complex genomic structural variations associated with important flatfish traits. The pBACode system is easy to implement, robust to sequencing errors, and compatible with all high-throughput sequencing platforms, including long read but error-prone TGS. It markedly improved the utility of BAC libraries and could also be directly adapted to fosmid libraries. One potential limitation of the pBACode-2 system is the number of barcodes in a pool of random-sequence-tagged cloning vectors. In this study, we generated a pool of six million of unique barcode pairs, sufficient complexity for a BAC library of 105 clones such that only 3.3% of clones had non-unique barcode pairs in our flounder library. Nevertheless, based on the level of complexity we observed in our library of 6 million barcode pairs, there could be more than a 10% duplication rate in a library of half a million clones. One solution is to construct a large pool of cloning vectors with hundreds of millions of unique barcode pairs. Alternatively, one could generate a large genomic library composed of multiple sub-library of 105 clones, and then perform the paired-end sequencing and physical clone mapping by sub-libraries. Finally, one could generate a large library using the pBACode-1 system, which does not require subcloning to construct the cloning vectors and allows an effectively unlimited number of unique random sequence barcodes.
Compared to another high-throughput genomic library end sequencing approach Fosill (29), pBACode enables generating long and accurate BAC end contigs. Because it is free from the constraint of co-ligating BAC ends, single end sequencing can be conducted in diverse ways to fit various purposes and sequencing platforms. In this paper, we employed inverse PCR to enrich BAC ends and add adapters for Illumina sequencing. Using Illumina mate-pair sequencing, we can sequence a BAC end that is up to 800 bp long. BAC end sequencing of pBACode libraries should be compatible with other sequencing platforms, i.e., PacBio RS II and MinION. The sample preparation for these methods may be even simpler than for Illumina, because single molecule sequencing platforms do not require enrichment and amplification of BAC ends. Long and accurate BAC end sequences will greatly improve the utility of BAC libraries for assembling large and complex genomes.
Our pBACode-based physical clone mapping approach requires a single PCR amplification for each BAC plasmid pool and is much simpler than High Information Content Fingerprinting (HICF) (64) and WGP (24). This simplicity reduces costs and cross-contamination among pools. As a result, no errors have yet been detected in our location assignments. Furthermore, identifying clones using unique barcodes enabled higher-throughput physical clone mapping. A physical clone mapping pool from our flounder library contained up to 11 808 BAC clones, two orders of magnitude greater than what is possible using WGP (25, 26). For example, mapping clones in a 384-well plate requires 16 pools using WGP methodology (26). Therefore, profiling our flounder BAC library of 94 464 clones (246 384-well plates) would have required 3936 pools. In contrast, our pBACode-based physical clone mapping required only 50 pools. However, the WGP methodology (24) does have some significant advantages over our method. WGP can assign about thirty tags to each BAC clone (25,26) while our pBACode strategy can only provide information on BAC ends. More importantly, WGP can process BAC libraries derived from any cloning vectors, while our method requires pBACode-based vectors and will not work with existing libraries. Nevertheless, pBACode adds a useful option to the genomics toolbox.
In summary, high-throughput barcoding enhanced the two most important functions of BAC libraries: paired-end sequencing and physical clone mapping. Its simplicity, high fidelity and compatibility with diverse sequencing and experimental platforms make genomic libraries of arrayed clones an appealing tool for both genomic and genetic studies, especially for those that need cloned pieces of DNA to refine the assembly or for functional experiments. We envision that a pBACode-based genomic library can greatly assist vector-free methodologies in creating high quality de novo assemblies of large and complex genomes, and facilitate the use of these reference genomes for subsequent functional studies.
ACCESSION NUMBERS
Sequencing data and genome assemblies are publicly available in the NCBI Sequence Read Archive and Genbank under project accession PRJNA344006 and PRJNA341910.
AVAILABILITY
The perl codes to process the yeast library and the flounder library are available at https://github.com/ZhichaoXu/pBACode-perl-code.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Jianhuo Fang, Jidong Lang and Lina Zhang in the Tsinghua Genomics & Synthetics Biology Core Facility for generating sequencing data and Yi Ding in the Tsinghua Drug Discovery Facility for assisting with the liquid handling platform. Tsinghua University School of Information Science and Technology provided computational facilities and assistance with the data analysis. We are very grateful to Chong-I Wu, Zhongying Zhao, Jack Chen, Cecilia Mello and Stephanie Zimmerman for helpful discussions and comments on the manuscript, and Junbiao Dai for providing S. cerevisiae S288C.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [91231109]; Modern Agro-industry Technology Research System of China [CARS-50-G02, CARS-50-Z03]; Tsinghua Qian Ren Tuan Dui funding (to M.Q.Z.). Funding for open access charge: National Natural Science Foundation of China [91231109].
Conflict of interest statement. None declared.
REFERENCES
- 1. Kang Y.J., Kim S.K., Kim M.Y., Lestari P., Kim K.H., Ha B.-K., Jun T.H., Hwang W.J., Lee T., Lee J. et al. Genome sequence of mungbean and insights into evolution within Vigna species. Nat. Commun. 2014; 5:5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhang G., Li C., Li Q., Li B., Larkin D.M., Lee C., Storz J.F., Antunes A., Greenwold M.J., Meredith R.W. et al. Comparative genomics reveals insights into avian genome evolution and adaptation. Science. 2014; 346:1311–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Koepfli K.-P., Paten B. Genome 10K Community of Scientists . Genome 10K Community of Scientists, O'Brien S.J.. The Genome 10K Project: a way forward. Annu. Rev. Anim. Biosci. 2015; 3:57–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Feuillet C., Leach J.E., Rogers J., Schnable P.S., Eversole K.. Crop genome sequencing: lessons and rationales. Trends Plant Sci. 2011; 16:77–88. [DOI] [PubMed] [Google Scholar]
- 5. VanBuren R., Bryant D., Edger P.P., Tang H., Burgess D., Challabathula D., Spittle K., Hall R., Gu J., Lyons E. et al. Single-molecule sequencing of the desiccation-tolerant grass Oropetium thomaeum. Nature. 2015; 527:508–511. [DOI] [PubMed] [Google Scholar]
- 6. Putnam N.H., O’Connell B.L., Stites J.C., Rice B.J., Blanchette M., Calef R., Troll C.J., Fields A., Hartley P.D., Sugnet C.W. et al. Chromosome-scale shotgun assembly using an in vitro method for long-range linkage. Genome Res. 2016; 26:342–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pendleton M., Sebra R., Pang A.W.C., Ummat A., Franzen O., Rausch T., Stütz A.M., Stedman W., Anantharaman T., Hastie A. et al. Assembly and diploid architecture of an individual human genome via single-molecule technologies. Nat. Methods. 2015; 12:780–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Marx V. Next-generation sequencing: the genome jigsaw. Nature. 2013; 501:263–268. [DOI] [PubMed] [Google Scholar]
- 9. Michael T.P., VanBuren R.. Progress, challenges and the future of crop genomes. Curr. Opin. Plant Biol. 2015; 24:71–81. [DOI] [PubMed] [Google Scholar]
- 10. Chan Y.F., Marks M.E., Jones F.C., Villarreal G., Shapiro M.D., Brady S.D., Southwick A.M., Absher D.M., Grimwood J., Schmutz J. et al. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science. 2010; 327:302–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li L.-B., Yu Z., Teng X., Bonini N.M.. RNA toxicity is a component of ataxin-3 degeneration in Drosophila. Nature. 2008; 453:1107–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Duyao M., Ambrose C., Myers R., Novelletto A., Persichetti F., Frontali M., Folstein S., Ross C., Franz M., Abbott M.. Trinucleotide repeat length instability and age of onset in Huntington's disease. Nat. Genet. 1993; 4:387–392. [DOI] [PubMed] [Google Scholar]
- 13. International Wheat Genome Sequencing Consortium (IWGSC) A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science. 2014; 345:1251788. [DOI] [PubMed] [Google Scholar]
- 14. Luo M.-C., Gu Y.Q., You F.M., Deal K.R., Ma Y., Hu Y., Huo N., Wang Y., Wang J., Chen S. et al. A 4-gigabase physical map unlocks the structure and evolution of the complex genome of Aegilops tauschii, the wheat D-genome progenitor. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:7940–7945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nystedt B., Street N.R., Wetterbom A., Zuccolo A., Lin Y.-C., Scofield D.G., Vezzi F., Delhomme N., Giacomello S., Alexeyenko A. et al. The Norway spruce genome sequence and conifer genome evolution. Nature. 2013; 497:579–584. [DOI] [PubMed] [Google Scholar]
- 16. Chapman J.A., Mascher M., Buluç A.N., Barry K., Georganas E., Session A., Strnadova V., Jenkins J., Sehgal S., Oliker L. et al. A whole-genome shotgun approach for assembling and anchoring the hexaploid bread wheat genome. Genome Biol. 2015; 16:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang J., Shao C., Zhang L., Liu K., Gao F., Dong Z., Xu P., Chen S.. A first generation BAC-based physical map of the half-smooth tongue sole (Cynoglossus semilaevis) genome. BMC Genomics. 2014; 15:215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hoskins R.A., Carlson J.W., Wan K.H., Park S., Mendez I., Galle S.E., Booth B.W., Pfeiffer B.D., George R.A., Svirskas R. et al. The Release 6 reference sequence of the Drosophila melanogaster genome. Genome Res. 2015; 25:445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Naville M., Chalopin D., Volff J.-N.. Interspecies insertion polymorphism analysis reveals recent activity of transposable elements in extant coelacanths. PLoS One. 2014; 9:e114382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lan H., Chen H., Chen L.-C., Wang B.-B., Sun L., Ma M.-Y., Fang S.-G., Wan Q.-H.. The first report of a Pelecaniformes defensin cluster: characterization of β-defensin genes in the crested ibis based on BAC libraries. Sci. Rep. 2014; 4:6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Smith J.J., Kuraku S., Holt C., Sauka-Spengler T., Jiang N., Campbell M.S., Yandell M.D., Manousaki T., Meyer A., Bloom O.E. et al. Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution. Nat. Genet. 2013; 45:415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bouwman P., van der Gulden H., van der Heijden I., Drost R., Klijn C.N., Prasetyanti P., Pieterse M., Wientjens E., Seibler J., Hogervorst F.B.L. et al. A high-throughput functional complementation assay for classification of BRCA1 missense variants. Cancer Discov. 2013; 3:1142–1155. [DOI] [PubMed] [Google Scholar]
- 23. Asakawa S., Abe I., Kudoh Y., Kishi N., Wang Y., Kubota R., Kudoh J., Kawasaki K., Minoshima S., Shimizu N.. Human BAC library: construction and rapid screening. Gene. 1997; 191:69–79. [DOI] [PubMed] [Google Scholar]
- 24. van Oeveren J., de Ruiter M., Jesse T., van der Poel H., Tang J., Yalcin F., Janssen A., Volpin H., Stormo K.E., Bogden R. et al. Sequence-based physical mapping of complex genomes by whole genome profiling. Genome Res. 2011; 21:618–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Poursarebani N., Nussbaumer T., Simková H., Safář J., Witsenboer H., van Oeveren J., Doležel J., Mayer K.F.X., Stein N., Schnurbusch T.. Whole-genome profiling and shotgun sequencing delivers an anchored, gene-decorated, physical map assembly of bread wheat chromosome 6A. Plant J. Cell Mol. Biol. 2014; 79:334–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sierro N., van Oeveren J., van Eijk M.J.T., Martin F., Stormo K.E., Peitsch M.C., Ivanov N.V.. Whole genome profiling physical map and ancestral annotation of tobacco Hicks Broadleaf. Plant J. Cell Mol. Biol. 2013; 75:880–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhao S., Malek J., Mahairas G., Fu L., Nierman W., Venter J.C., Adams M.D.. Human BAC ends quality assessment and sequence analyses. Genomics. 2000; 63:321–332. [DOI] [PubMed] [Google Scholar]
- 28. Zhao S., Shatsman S., Ayodeji B., Geer K., Tsegaye G., Krol M., Gebregeorgis E., Shvartsbeyn A., Russell D., Overton L. et al. Mouse BAC ends quality assessment and sequence analyses. Genome Res. 2001; 11:1736–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Williams L.J.S., Tabbaa D.G., Li N., Berlin A.M., Shea T.P., Maccallum I., Lawrence M.S., Drier Y., Getz G., Young S.K. et al. Paired-end sequencing of Fosmid libraries by Illumina. Genome Res. 2012; 22:2241–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Foote A.D., Liu Y., Thomas G.W.C., Vinař T., Alföldi J., Deng J., Dugan S., van Elk C.E., Hunter M.E., Joshi V. et al. Convergent evolution of the genomes of marine mammals. Nat. Genet. 2015; 47:272–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Keane M., Craig T., Alföldi J., Berlin A.M., Johnson J., Seluanov A., Gorbunova V., Di Palma F., Lindblad-Toh K., Church G.M. et al. The Naked Mole Rat Genome Resource: facilitating analyses of cancer and longevity-related adaptations. Bioinforma. Oxf. Engl. 2014; 30:3558–3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Neafsey D.E., Christophides G.K., Collins F.H., Emrich S.J., Fontaine M.C., Gelbart W., Hahn M.W., Howell P.I., Kafatos F.C., Lawson D. et al. The evolution of the Anopheles 16 genomes project. G3. 2013; 3:1191–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Walker B.J., Abeel T., Shea T., Priest M., Abouelliel A., Sakthikumar S., Cuomo C.A., Zeng Q., Wortman J., Young S.K. et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014; 9:e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zimin A., Stevens K.A., Crepeau M.W., Holtz-Morris A., Koriabine M., Marçais G., Puiu D., Roberts M., Wegrzyn J.L., de Jong P.J. et al. Sequencing and assembly of the 22-gb loblolly pine genome. Genetics. 2014; 196:875–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Au K.F., Underwood J.G., Lee L., Wong W.H.. Improving PacBio long read accuracy by short read alignment. PLoS One. 2012; 7:e46679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Karlsson E., Lärkeryd A., Sjödin A., Forsman M., Stenberg P.. Scaffolding of a bacterial genome using MinION nanopore sequencing. Sci. Rep. 2015; 5:11996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li R., Hsieh C.-L., Young A., Zhang Z., Ren X., Zhao Z.. Illumina synthetic long read sequencing allows recovery of missing sequences even in the ‘Finished’ C. elegans genome. Sci. Rep. 2015; 5:10814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ng P., Tan J.J.S., Ooi H.S., Lee Y.L., Chiu K.P., Fullwood M.J., Srinivasan K.G., Perbost C., Du L., Sung W.-K. et al. Multiplex sequencing of paired-end ditags (MS-PET): a strategy for the ultra-high-throughput analysis of transcriptomes and genomes. Nucleic Acids Res. 2006; 34:e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Craig D.W., Pearson J.V., Szelinger S., Sekar A., Redman M., Corneveaux J.J., Pawlowski T.L., Laub T., Nunn G., Stephan D.A. et al. Identification of genetic variants using bar-coded multiplexed sequencing. Nat. Methods. 2008; 5:887–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kozich J.J., Westcott S.L., Baxter N.T., Highlander S.K., Schloss P.D.. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 2013; 79:5112–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Adey A., Kitzman J.O., Burton J.N., Daza R., Kumar A., Christiansen L., Ronaghi M., Amini S., Gunderson K.L., Steemers F.J. et al. In vitro, long-range sequence information for de novo genome assembly via transposase contiguity. Genome Res. 2014; 24:2041–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cusanovich D.A., Daza R., Adey A., Pliner H.A., Christiansen L., Gunderson K.L., Steemers F.J., Trapnell C., Shendure J.. Epigenetics. Multiplex single-cell profiling of chromatin accessibility by combinatorial cellular indexing. Science. 2015; 348:910–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Melnikov A., Murugan A., Zhang X., Tesileanu T., Wang L., Rogov P., Feizi S., Gnirke A., Callan C.G., Kinney J.B. et al. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat. Biotechnol. 2012; 30:271–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhao W., Pollack J.L., Blagev D.P., Zaitlen N., McManus M.T., Erle D.J.. Massively parallel functional annotation of 3΄ untranslated regions. Nat. Biotechnol. 2014; 32:387–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shiroguchi K., Jia T.Z., Sims P.A., Xie X.S.. Digital RNA sequencing minimizes sequence-dependent bias and amplification noise with optimized single-molecule barcodes. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:1347–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Luo M., Wing R.A.. An improved method for plant BAC library construction. Methods Mol. Biol. 2003; 236:3–20. [DOI] [PubMed] [Google Scholar]
- 47. Ewing B., Green P.. Base-Calling of Automated Sequencer Traces Using Phred. II. Error Probabilities. Genome Res. 1998; 8:186–194. [PubMed] [Google Scholar]
- 48. Ewing B., Hillier L., Wendl M.C., Green P.. Base-Calling of Automated Sequencer Traces UsingPhred. I. Accuracy Assessment. Genome Res. 1998; 8:175–185. [DOI] [PubMed] [Google Scholar]
- 49. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bao E., Jiang T., Kaloshian I., Girke T.. SEED: efficient clustering of next-generation sequences. Bioinforma. Oxf. Engl. 2011; 27:2502–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Magoč T., Salzberg S.L.. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinforma. Oxf. Engl. 2011; 27:2957–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Luo R., Liu B., Xie Y., Li Z., Huang W., Yuan J., He G., Chen Y., Pan Q., Liu Y. et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience. 2012; 1:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gnerre S., MacCallum I., Przybylski D., Ribeiro F.J., Burton J.N., Walker B.J., Sharpe T., Hall G., Shea T.P., Sykes S. et al. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:1513–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Boetzer M., Henkel C.V., Jansen H.J., Butler D., Pirovano W.. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2011; 27:578–579. [DOI] [PubMed] [Google Scholar]
- 55. Kurtz S., Phillippy A., Delcher A.L., Smoot M., Shumway M., Antonescu C., Salzberg S.L.. Versatile and open software for comparing large genomes. Genome Biol. 2004; 5:R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wild J., Hradecna Z., Szybalski W.. Conditionally Amplifiable BACs: Switching From Single-Copy to High-Copy Vectors and Genomic Clones. Genome Res. 2002; 12:1434–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ojima Y., Yamamoto K.. Cellular DNA contents of fishes determined by flow cytometry. Kromosomo II. 1990; 57:1871–1888. [Google Scholar]
- 58. Osoegawa K., Tateno M., Woon P.Y., Frengen E., Mammoser A.G., Catanese J.J., Hayashizaki Y., de Jong P.J.. Bacterial artificial chromosome libraries for mouse sequencing and functional analysis. Genome Res. 2000; 10:116–128. [PMC free article] [PubMed] [Google Scholar]
- 59. Osoegawa K., Mammoser A.G., Wu C., Frengen E., Zeng C., Catanese J.J., de Jong P.J.. A bacterial artificial chromosome library for sequencing the complete human genome. Genome Res. 2001; 11:483–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Osoegawa K., Zhu B., Shu C.L., Ren T., Cao Q., Vessere G.M., Lutz M.M., Jensen-Seaman M.I., Zhao S., de Jong P.J.. BAC resources for the rat genome project. Genome Res. 2004; 14:780–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chen S., Zhang G., Shao C., Huang Q., Liu G., Zhang P., Song W., An N., Chalopin D., Volff J.-N. et al. Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat. Genet. 2014; 46:253–260. [DOI] [PubMed] [Google Scholar]
- 62. Yamaguchi T., Kitano T.. High temperature induces cyp26b1 mRNA expression and delays meiotic initiation of germ cells by increasing cortisol levels during gonadal sex differentiation in Japanese flounder. Biochem. Biophys. Res. Commun. 2012; 419:287–292. [DOI] [PubMed] [Google Scholar]
- 63. Kitano J., Ross J.A., Mori S., Kume M., Jones F.C., Chan Y.F., Absher D.M., Grimwood J., Schmutz J., Myers R.M. et al. A role for a neo-sex chromosome in stickleback speciation. Nature. 2009; 461:1079–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Nelson W.M., Bharti A.K., Butler E., Wei F., Fuks G., Kim H., Wing R.A., Messing J., Soderlund C.. Whole-genome validation of high-information-content fingerprinting. Plant Physiol. 2005; 139:27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.