Abstract
Our understanding of translation underpins our capacity to engineer living systems. The canonical start codon (AUG) and a few near-cognates (GUG, UUG) are considered as the ‘start codons’ for translation initiation in Escherichia coli. Translation is typically not thought to initiate from the 61 remaining codons. Here, we quantified translation initiation of green fluorescent protein and nanoluciferase in E. coli from all 64 triplet codons and across a range of DNA copy number. We detected initiation of protein synthesis above measurement background for 47 codons. Translation from non-canonical start codons ranged from 0.007 to 3% relative to translation from AUG. Translation from 17 non-AUG codons exceeded the highest reported rates of non-cognate codon recognition. Translation initiation from non-canonical start codons may contribute to the synthesis of peptides in both natural and synthetic biological systems.
INTRODUCTION
The translation of messenger RNA (mRNA) to protein is one of the fundamental processes in biology. Control of translation is critical to enable precision bioengineering. One key modulator of translation is the initiation or ‘start’ codon, which is the three mRNA nucleotides that bind to N-formylmethionyl transfer RNA (tRNAfMet) (1,2). The most common start codons for known Escherichia coli genes are AUG (83% of genes), GUG (14%) and UUG (3%) (2–4). Similar percentages can be found throughout the bacterial domain (5).
The occurrence of non-canonical start codons (defined here as any codon other than AUG, GUG and UUG) in known genes is very rare. For example, only two non-canonical start codons have been confirmed in E. coli: infC (6) and pcnB (7) both begin with AUU. Approximately 0.1% of annotated start codons in eukaryotes are non-AUG (8), although recent ribosomal footprinting studies with yeast and mammalian cells suggest that non-canonical translation initiation may be more prevalent (9–12).
Translation initiation is an intricate process that has been studied in detail (e.g. 13–18). Of the mRNA sequences that modulate translation initiation, the impact of variation within the 5΄ untranslated region (5΄ UTR) (19) and the Shine-Dalgarno sequence, also known as the ribosome binding site (RBS) (20), have been systematically quantified (13,21–24). However, sequence variation within the start codon itself has not yet been systematically explored.
The number of different roles codons can adopt in translation motivated our systematic exploration of start codon variants. As examples: organisms across all domains of life naturally reassign one of the three canonical stop codons (UAA, UAG and UGA) to code for amino acids (25); in wild microbes 13 different codons have evolved to code for the proteinogenic amino acid selenocysteine (26); in engineered E. coli, 58 of the 64 codons were successfully reassigned to code for selenocysteine (27), and all instances of the UAG stop codon were recoded to UAA to allow UAG reassignment to unnatural amino acids (28).
Improvements in DNA synthesis (29), sequencing (30) and assembly (31,32), and the creation of a variety of bright fluorescent proteins (33), enabled our systematic exploration of start codon variants. These improvements have already led to systematic explorations of promoters (34–36), repressors (37,38), RBSs (34,39,40), insulators (41) and terminators (42,43). Systematic explorations of the regions immediately upstream (22,23) and downstream (44–47) of the start codon in E. coli have revealed significant impacts of the sequence surrounding the start codon on translation efficiency. However, only a few studies have explored initiation of translation via the near-cognates of AUG (48,49), and none appear to have explored initiating translation from all 64 codons. Here, we systematically quantified translation initiation of green fluorescent protein (GFP) from all 64 codons and nanoluciferase from 12 codons on plasmids designed to interrogate a range of translation initiation conditions.
MATERIALS AND METHODS
Bacterial culture
All strains were grown in lysogeny broth (LB), Rich Defined Media (RDM, Teknova) or on LB agar, supplemented with 50 μg ml−1 kanamycin, 100 μg ml−1 carbenicillin or 25 μg ml−1 chloramphenicol (Sigma) for selection. Plasmids were isolated using a QIAQuick Miniprep kit (Qiagen). Polymerase chain reaction (PCR) reaction products were purified using a GeneJet Gel extraction kit (Thermo Scientific) or NucleoSpin Gel and PCR Clean-Up kit (ClonTech). Plasmids and PCR products were sequenced using Sanger sequencing (Elim Biopharma or MCLab). All PCR and cloning reactions were performed on a S1000 Thermal Cycler (Bio-Rad). Information about the E. coli strains used in this experiment can be found in the Supplementary Data.
Construction of T7-GFP plasmids
A library of 64 plasmids was created where the start codon (AUG) in the GFP (the superfolder GFP variant (50) was used, referred to as GFP hereafter) coding sequence was replaced with each of the 64 codons. A pET20b(+) vector with a pBR322 origin of replication and an ampicillin-resistance cassette (Novagen) was used as the plasmid backbone (Supplementary Table S2). The GFP transcript had a strong RBS (AGGAGA), and the spacer between the RBS and the start codon (UAAAUAC) was designed to prevent the creation of out-of-frame canonical start codons and achieve optimal RBS-start codon spacing (51). Thirty-one variants of the 64-member library were created by one-pot Golden Gate cloning, and the remaining 33 were created in parallel reactions via plasmid amplification followed by blunt-end ligation. Bacterial cultures for plasmid construction were grown in LB supplemented with carbenicillin. Additional details about the cloning methods used can be found in the Supplementary Data.
Construction of RhaPBAD-GFP and RhaPBAD-nanoluciferase plasmids, and RhaPBAD-nanoluciferase BACs
A set of 12 codons (AUG, GUG, UUG, AUA, AUC, AUU, CUG, CAU, CGC, GGA, UAG and UGC) were selected for further exploration as potential start codons in three different expression cassettes: GFP under control of the native E. coli RhaPBAD rhamnose-inducible promoter on a p15A plasmid, nanoluciferase (Promega NanoLuc® (52), referred to as nanoluciferase hereafter) under control of RhaPBAD on a p15A plasmid, and nanoluciferase under control of RhaPBAD on a very-low-copy bacterial artificial chromosome (BAC) (53). We chose to use the BAC to better mimic physiologically-relevant expression conditions. Sequence information (Supplementary Table S2) and additional details about the cloning are available in the Supplementary Data.
Culture growth conditions for assay measurements
LB agar plates with the appropriate antibiotics were streaked from frozen glycerol stocks and incubated overnight at 37°C. Plates were stored at 4°C until ready for use. Plates were discarded after 2 weeks of storage at 4°C. Three individual colonies for each construct were used to inoculate 300 μl of LB containing the appropriate antibiotics: carbinecillin and chloramphenicol (T7-GFP plasmids), or kanamycin (all others) in a 96-well deep well culture plate (VWR). The plate was sealed with an AeraSeal gas-permeable microplate seal (E&K Scientific) and grown overnight at 37°C in a Kuhner LT-X (Lab-Therm) incubator shaking at 460 rpm with 80% relative humidity. Unless otherwise specified, all liquid cell cultures were grown under these conditions.
We used non-expressing control strains lacking a reporter gene to measure cellular autofluorescence or autoluminescence for each experiment. We used a different construct to create non-expressing control strains for each of the four reporter-backbone pairs used in this experiment. For measurements of GFP expressed from the pET20b(+) vector, we used a pET20b(+) cloning vector (Novagen) with no protein coding sequence inserted into the vector. For measurements of GFP expressed from the p15A plasmid, we used a p15A plasmid that carried a silicatein gene. For measurements of nanoluciferase expressed from the p15A plasmid, we used a p15A plasmid that carried a GFP gene. For measurements of nanoluciferase expressed from the BAC, we used a BAC that carried a GFP gene. All control vectors had the same resistance gene as the vectors expressing the reporter gene, and were transformed into the same E. coli strain used for measuring expression. The background signal measured from these control strains was used to determine the fluorescence or luminescence signal above which we considered expression to be significant.
Fluorescence measurements
For measurements of fluorescence from the T7-GFP plasmids in LB, after the overnight growth, 4 μl of each of culture was transferred into 400 μl of fresh media and grown for 2 h. A total of 4 μl of freshly prepared 100 mM IPTG was added to induce T7 RNA polymerase expression and the cells were grown for an additional 5 h. No IPTG was added to the non-expressing control cultures because we observed in previous experiments that inducing these cells inhibits cell growth. We suspect this is because the cloning vector in the non-expressing control cells contains a 300 base transcript transcribed by the T7 promoter, including a pelB leader sequence. Overexpression of this sequence appears to be toxic to the cell.
A total of 300 μl of each cell culture were centrifuged at 5000 x g for 4 min in a Beckman-Coulter Avanti J-E centrifuge with a swinging bucket rotor, and the supernatants were aspirated with a vacuum trap. The pelleted cells were re-suspended in 250 μl of 1× phosphate buffered saline (PBS) (Fisher Scientific), and 200 μl were transferred to CELLSTAR black, clear-bottom 96-well plate (Grenier, #M0562) and left overnight at 4°C to allow the GFP to mature fully. Before reading, plates were placed in the shaking incubator for 5 min to resuspend any cells that may have aggregated. Absorbance was measured at 600 nm (OD600) to estimate culture density, followed by fluorescence (excitation = 485 nm, emission = 510 nm, bandwidth = 9 nm) measured at two separate gains (high gain, sensitivity = 110; low gain, sensitivity = 65), on a BioTek Synergy H4 plate reader. We measured fluorescence at the two gain settings because the plate reader lacked sufficient dynamic range to accurately measure fluorescence from all start codons on a single gain setting (see ‘Data Analysis’ section). All raw measurements are available online (Supplementary Table S5).
For measurements of GFP expression in RDM, after the overnight growth, cultures were sub-cultured 1:100 into 300 μl of EZ RDM with the same antibiotics as the initial culture and grown for 2 h under the same conditions. Expression of GFP was then induced by supplementing the cultures with 1 mmol/l IPTG, and the cells were grown for an additional 5 h. For plate reader measurements, 50 μl of each culture were transferred to a CELLSTAR black, clear-bottom 96-well plate. A total of 150 μl of 1× PBS was added to each well. Cultures were analyzed on a BioTek Synergy H4 plate reader. Absorbance at 600 nm (OD600) was measured to estimate culture density, followed by fluorescence (excitation = 485 nm, emission = 510 nm, bandwidth = 9 nm, sensitivity = 65).
For flow cytometry, cells from the RDM expression experiment above were diluted 1/100 in PBS and measured on a BD LSRII instrument using a FITC fluorescence channel (488 nm excitation laser with a 525/50 nm emission band-pass filter). Detector voltages through forward scattering, side scattering and FITC channels were set using non-expressing control cells and pET-GFP(ATG) clones as a positive control. Measured events were triggered on a side-scattering threshold and 30 000 events were measured from each cell culture. Two measurement sets were acquired, at high- and low-gain through the FITC channel. At high gain, the mean GFP signal from the three highest expressing samples (AUG, GUG, UUG) were set off-scale, above the upper detection limit such that the remaining samples were within the range of detection. At low gain, the mean signals from highest expressing samples were within the measurable range, but means for the lowest expressing samples were off-scale below the lower detection limit. Flow cytometry data were processed in R, using the flowCore package (54) to remove non-expressing near-baseline (noise) values and log transform the data, and ggplot2 (55) to generate violin plots.
Luminescence measurements
After the overnight growth, cultures were subcultured 1:100 into 300 μl of LB with kanamycin and grown for 2 h. Expression of nanoluciferase was induced by supplementing the cultures with 4 mmol/l rhamnose, and then the cultures were incubated overnight as previously described. After the second overnight growth, 50 μl of each culture were transferred to a CELLSTAR black, clear-bottom 96-well plate. A total of 150 μl of 1× PBS was added to each well. OD600 was measured as described above.
Nanoluciferase luminescence measurements were performed using the Nano-Glo Luciferase Assay System kit (Promega, #N1110), following the manufacturer's protocol. Lysis buffer was prepared by dissolving 37.5 mg of Egg White Lysozyme (Sigma-Aldrich) in 7.5 ml of 1× Glo-Lysis Buffer (Promega). A total of 20 μl of cell culture was diluted in 180 μl of lysis buffer and incubated at room temperature for 10 min. Assay buffer was prepared by mixing 9.8 ml of nanoluciferase Assay Buffer (Promega) with 200 μl of nanoluciferase Assay Reagent (Promega). A total of 5 μl of lysed cells were mixed with 195 μl of assay buffer on a CELLSTAR white, opaque-bottom 96-well plate and incubated for 5 min at room temperature. During this part of the experiment, one of the cell solutions (of the AUC codon in the BAC) was not transferred from the lysis plate to the assay plate, so only two biological replicates measured for this codon. Luminescence was measured on a BioTek Synergy H4 plate reader for all visible wavelengths for 1 s at a gain of 100. To minimize carry-over signal from adjacent wells, codons were separated by at least one row and known high-expressing codons (AUG, GUG and UUG) were read separately.
Data analysis
Raw fluorescence and optical density measurements for each cell culture were imported into R. Wells filled only with cell culture media were used to subtract background optical density and fluorescence. For the T7-GFP measurements from LB culture, we created a calibration curve comparing the relationship between fluorescence measured at the two gain settings (Supplementary Figure S2). This curve was used to calibrate fluorescence measured at the low gain setting to an equivalent value of fluorescence measured at the high gain setting, which allowed for comparison of fluorescence measured at the two gain settings. All measurements except for the three canonical start codons were within the linear dynamic range at the high gain setting. For the three canonical start codons, we used the calibration curve (Supplementary Figure S2) to convert the low gain reading to the expected high gain reading. High gain fluorescence measurements were carried forward for downstream analysis.
Per-cell fluorescence or luminescence was calculated by dividing the fluorescence by the optical density. Mean per-cell fluorescence or luminescence for each start codon in each expression strain was calculated by averaging the per-cell measurements from three biological replicates. Per-cell expression was normalized by the per-cell expression from the AUG start codon to facilitate comparison of relative expression from different expression systems. The significance of the translation initiated from each codon was determined by comparing the expression measured from all of the codons in each experiment to the non-expressing control using Dunnett's test (56), a statistical method for comparing multiple treatments to a single control, using the R multcomp package (57).
Mass spectrometry to confirm N-terminal sequence of translated GFP proteins
GFP expression and purification
Five GFP start codon variants were selected from different expression levels (AUC, ACG, CAU, GGA and CGC). We selected these codons for five distinct reasons: first, AUC has been previously characterized for non-canonical initiation; second, ACG, despite being one base away from AUG, is not annotated as a start codon in known bacterial genes (Table 1); third, CAU expressed at a surprisingly high level despite being the reverse complement of a canonical AUG; fourth, GGA could potentially create a new RBS; and fifth, CGC did not initiate a detectable amount of translation. Genes with these five start codons were recloned into the pET20b(+) vector with a C-terminal 6x-His tag. These plasmids were transformed into BL21(DE3)pLysS. A single colony was used to inoculate 3 ml of LB supplemented with ampicillin and chloramphenicol and shaken at 37°C overnight. The overnight culture was used to inoculate 30 ml of the same media 1:100 and grown for 2 h at 37°C, followed by induction to a final concentration of 1 mmol/l IPTG. The cells were allowed to express the protein at 37°C for 5 h and harvested by centrifugation for 10 min at 4000 × g. The cells were resuspended and lysed in 1 ml BPER with 0.6 mg/ml lysozyme, vortexed and incubated for 10 min at room temperature. DNase (one unit) was added and further incubated with frequent vortexing for 10 min. The lysate was then centrifuged at 4°C for 10 min at 15 000 × g. The clarified lysate was run over 150 μl of nickel resin preequilibrated with 2 × PBS (24 mmol/l sodium phosphate buffer, 274 mmol/l NaCl, 54 mmol/l KCl) with 20 mmol/l imidazole at pH 7.5. The column was washed with 4 ml of 2 × PBS with 20 mmol/l imidazole. Green protein was eluted with 400 μl of the same buffer with 300 mmol/l imidazole.
Table 1. Annotated initiation codons in model bacterial genomes.
| Initiation codon | Number | Percentage |
|---|---|---|
| AUG | 69 447 | 81.801% |
| GUG | 11 715 | 13.799% |
| UUG | 3691 | 4.348% |
| CUG | 20 | 0.024% |
| AUU | 16 | 0.019% |
| AUC | 5 | 0.006% |
| AUA | 3 | 0.004% |
| Total | 84 897 | 100.000% |
Start codons extracted from annotated features of 69 bacterial genome and plasmid sequences.
Sample preparation for LC-MS
A total of 50 μl volume of each of the protein solution samples was aliquoted, followed by protein precipitation with the addition of 250 μl −80°C acetone. Samples were stored at −80°C for 1 h, followed by light vortex and spun at 12 500 rpm for 12 min at 4°C. The supernatant was decanted and the protein pellet was left to dry under the chemical hood for 20 min at room temperature. The protein pellet was re-suspended in 65 μl 50 mmol/l ammonium bicarbonate 0.2% protease max (Promega) followed by vortex and sonication until the pellet was fully suspended. Dithiothreitol was added to a final concentration of 5 mmol/l, incubated on a heat block at 55°C for 30 min followed by alkylation with the addition of propionamide to a final concentration of 10 mmol/l for 30 min at room temperature. A total of 2 μg of Asp N (Promega) was reconstituted in the vendor specific buffer and 0.5 μg added to each sample, followed by overnight digestion at 37°C. Formic acid was added to 1% and the acidified digest was C18 stage tip purified (Nest Group) using microspin columns and dried in a speed vac.
LC-MS
Peptide pools were reconstituted and injected onto a C18 reversed phase analytical column, 10 cm in length (New Objective). The UPLC was a Waters NanoAcquity, operated at 600 nl/min using a linear gradient from 4% mobile phase B to 35% B. Mobile phase A consisted of 0.1% formic acid, water, Mobile phase B was 0.1% formic acid, acetonitrile. The mass spectrometer was an Orbitrap Elite set to acquire data in a high/high data dependent fashion selecting and fragmenting the 10 most intense precursor ions in the HCD cell where the exclusion window was set at 60 s and multiple charge states of the same ion were allowed.
LC-MS data analysis
MS/MS data were analyzed using Preview and Byonic v2.6.49 (ProteinMetrics). Data were first analyzed in Preview to verify calibration criteria and identify likely post-translational modifications prior to Byonic analysis. All analyses used a custom .fasta file containing the target protein sequence, and were searched with a reverse-decoy strategy at a 1% false discovery rate. Byonic searches were performed using 12 ppm mass tolerances for precursor and fragment ions, allowing for semi-specific N-ragged tryptic digestion. The resulting identified peptide spectral matches were then exported for further analysis.
Bioinformatics analysis
To analyze initiation codon annotations from model bacterial species, 69 complete bacterial chromosome and plasmid sequences were collected from the National Center for Biotechnology Information databases (Supplementary Table S3). Initiation codon sequences were extracted from annotated features and compiled into a comprehensive list of 85 119 entries with Accession Number, Start Codon, Locus Tag, Gene Name and Gene Product Name extracted from GenBank annotation features ID, sequence, qualifier(locus_tag), qualifier(gene) and qualifier(product) respectively. After removal of entries due to pseudogenes and misannotations a set of 84 897 entries remained (Supplementary Table S6) for analysis of initiation codon frequencies across the replicons of model bacterial species. The BioCyc database was consulted extensively during this process (58).
RNA folding simulations of transcripts from measured plasmids was performed using both NUPACK (59) and KineFold (60). NUPACK was run using default parameters. KineFold was run using default parameters except 3 ms co-transcriptional folding parameter for pET20b(+) simulations and 20 ms co-transcriptional folding parameter for p15A and BAC simulations to account for the different RNA polymerases transcribing these vectors in vivo.
RESULTS
We were first motivated to explore non-canonical start codons when we attempted to silence translation of a dihydrofolate reductase (DHFR) gene. We changed the start codon from AUG to GUG, UUG, AUA or ACG. Surprisingly, we detected significant DHFR expression in recombinant bacterial extract (61) from all five codons (Supplementary Figure S1). We initially wondered whether the observed protein synthesis was merely an artifact of using an in vitro translation system, as similar results had been reported using rabbit reticulocyte lysate (62).
We analyzed 69 well-annotated bacterial chromosomes and endogenous plasmids (63) to determine which of the 64 codons have been annotated as start codons (Supplementary Table S3). Our approach was similar to previous efforts (5) but with a focus on well-annotated genomes. Our analysis indicated that the vast majority of annotated open reading frames (ORFs) have AUG (81.8%), GUG (13.8%) or UUG (4.35%) as the start codon, although CUG, AUU, AUC and AUA are also annotated as start codons (Table 1 and Supplementary Table S6).
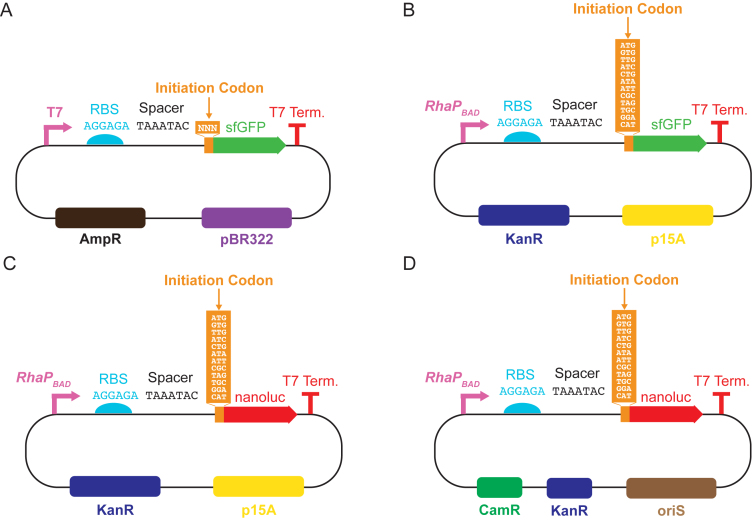
We designed a set of four plasmids with different copy numbers, promoters and reporters to experimentally quantify translation initiated from all 64 codons in E. coli (Figure 1). First, we measured the translation of GFP initiated from all 64 codons. Expression was driven by a T7 promoter and a strong RBS (AGGAGA) on a medium-copy pET20b(+) vector (Figure 1A). The spacer sequence between the RBS and the start codon (UAAAUAC) was designed to be the optimal length for promoting translation initiation (51), and also to prevent the inadvertent creation of an in-frame or out-of-frame canonical start codon.
Figure 1.
Plasmid sets used to measure translation initiation from non-canonical start codons. Plasmids varied in origin of replication (copy number), promoter and reporter gene characteristics. (A) Set of 64 pET20b(+) plasmids containing medium-copy pBR322 origin, T7 promoter and GFP reporter. (B) Set of 12 plasmids containing low-copy p15A origin, RhaPBAD rhamnose-inducible native Escherichia coli promoter and GFP reporter. (C) Set of 12 plasmids containing low-copy p15A origin, RhaPBAD rhamnose-inducible native E. coli promoter and nanoluciferase reporter. (D) Set of 12 very-low-copy bacterial artificial chromosomes (BAC) containing RhaPBAD rhamnose-inducible native E. coli promoter and nanoluciferase reporter.
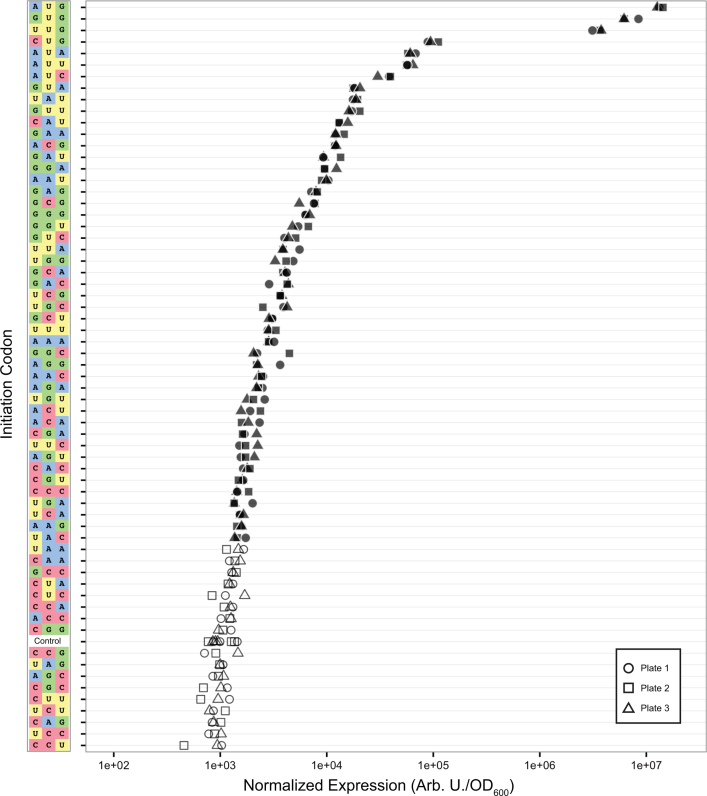
We measured fluorescence and absorbance via a plate reader from two different growth conditions. Initially, we measured expression in RDM using a single plate reader gain setting (Supplementary Figure S3). We redid our measurements after realizing that we could improve our signal-to-noise ratio by resuspending cells in PBS after growth in LB and prior to measurement, and improve our dynamic range by measuring expression at multiple plate reader gain settings (Figure 2). Measurements in the second condition had larger dynamic range than the first condition, but were otherwise similar (Supplementary Figure S4). In both cases, a strain carrying an empty cloning vector was used as a control for measuring background fluorescence. We calculated mean per-cell expression (fluorescence divided by OD600) for three biological replicates of strains expressing GFP with each of the 64 codons inserted in place of the start codon in the GFP coding sequence. The expression of GFP initiated from each start codon across the triplicate cultures was compared to the expression of the control cells using Dunnett's test, a method for comparing multiple treatments to a single control (56), assuming equal variance. Expression initiated from a codon was considered to be significant if the adjusted P-value was <0.05 (filled points, Figure 2). Of the 64 start codons tested, translation initiated from 47 at a level significantly greater than the control cells.
Figure 2.
Translation initiation from all 64 codons. Normalized per-cell fluorescence measured from three replicate cultures, grown in LB and resuspended in PBS before measurement, with each of the 64 codons as the start codon in the GFP coding sequence. Shapes represent the replicate plate number, and filled shapes represent GFP expression significantly greater (adjusted P < 0.05) than the non-expressing control (Control) as determined by Dunnett's test.
We created a codon table heat map to better visualize trends in the translation initiation strength of each of the 64 codons (Supplementary Figure S5). The seven strongest start codons have U in the second position and are the only codons that are annotated as start codons in bacterial genomes (Table 1). However, NAU and GAN also emerged as a relatively strong group of start codons. This group is not typically annotated as start codons in bacterial genomes.
We wanted to confirm that the bulk fluorescence measurements we obtained on the plate reader were not arising from a small number of highly-expressing cells. We measured the distribution of fluorescence within the population of each culture on a flow cytometer (Supplementary Figure S6). The observed distributions were unimodal for all non-canonical start codons whose expression is above the highest reported rates of non-cognate codon recognition (see ‘Discussion’ section). Additionally, the geometric means of the fluorescence of these populations were well correlated with the mean per-cell fluorescence measured on the plate reader (Supplementary Figure S7).
We examined the GFP transcript (Supplementary Table S2) for in-frame upstream and downstream start codons as a possible explanation for the observed fluorescence. We found no canonical in-frame start codons upstream of the GFP coding sequence. There is an in-frame GUG at the 16th codon in the GFP coding sequence, but any resulting protein would be truncated and non-fluorescent given that the minimal sequence needed for GFP fluorescence begins at the sixth codon (64).
We used proteolytic digestion and mass spectrometry to determine if translation began at modified start codons for five selected codons (AUC, ACG, CAU, GGA and CGC, please see ‘Materials and Methods’ section). We cloned a 6x-His tag into the C-terminus of these five genes and, following expression and purification, recovered significant amounts of protein. Little to no protein was recovered from the CGC culture, as expected. We digested proteins with AspN and analyzed the mixture via mass spectrometry. Each expressed protein released peptides of intact N-termini that included an N-terminal methionine (Supplementary Tables S7–11). ACG and AUC are one base away from AUG, while GGA and CAU would require two and three concurrent point mutations, respectively, to revert to a canonical start codon. In cultures with ACG as the start codon a small fraction of spectra (1 of 8) indicated that the N-terminal peptide might be the cognate amino acid, threonine (Mr = 119), with a mass shift of −30 Da relative to methionine (Mr = 149) (Supplementary Tables S8 and 11). Other researchers have also observed methionine in the N-terminal position of proteins whose translation initiates from GUG or UUG start codons (4,65,66).
Our initial experimental data using GFP suggested that the 64 codons could be organized into four groups: three canonical start codons (AUG, GUG, UUG), from which translation initiated at 10–100% of AUG; four near-cognates (AUA, AUC, AUU and CUG), from which translation initiated at 0.1–1% of AUG, similar to previously-reported values (48,49); 40 codons, from which translation initiated at 0.01–0.1% of AUG; and 17 codons, from which translation initiation could not be detected at a level significantly above that of the non-expressing control cells.
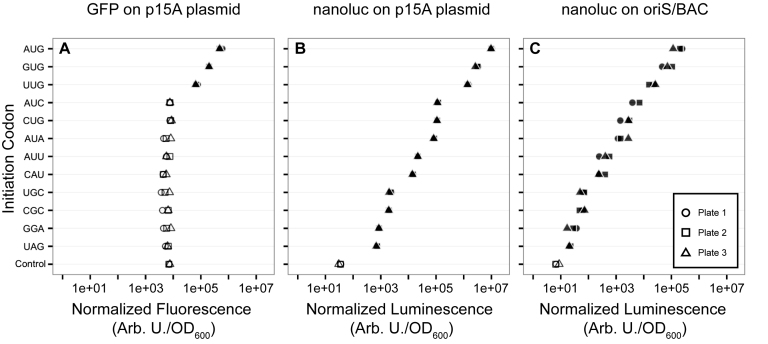
We next explored translation initiated from non-canonical start codons under more physiologically relevant conditions. We focused on 12 codons spanning the observed expression initiation range (AUG, GUG, UUG, AUA, AUC, AUU, CUG, CAU, CGC, GGA, UAG and UGC). We cloned each of the 12 codons into the first position of the coding sequence in three constructs: GFP under the control of the RhaPBAD promoter on a low-copy p15A plasmid (Figure 1B); nanoluciferase under the control of the RhaPBAD promoter on a low-copy p15A plasmid (Figure 1C); and, nanoluciferase under the control of the RhaPBAD promoter on a very-low-copy BAC (Figure 1D). The same RBS and 5΄-spacer was used in all constructs. GFP expression was quantified by measuring mean per-cell fluorescence. Nanoluciferase expression was quantified by measuring mean per-cell luminescence emitted from the nanoluciferase-catalyzed conversion of furimazine to furimamide (52). All measurements were repeated in triplicate. Measurements from serial dilutions of nanoluciferase-expressing cells indicated that, over the range of concentrations used in this work, luminescence was linear with nanoluciferase concentration (Supplementary Figure S7).
Our attempts to measure non-canonical GFP translation initiation driven by the RhaPBAD promoter on the low-copy p15A plasmid (Figure 1B) were impeded by a low signal-to-noise ratio due to significant background signal (Supplementary Figure S9). We were only able to detect significant GFP expression for the three canonical start codons AUG, GUG and UUG (Figure 3A). We therefore transitioned to an expression system with lower background signal to detect non-canonical translation initiation under more biologically relevant expression conditions. Nanoluciferase was a good reporter for this purpose because cell cultures emit negligible background luminescence and the nanoluciferase assay has a linear dynamic range greater than six orders of magnitude (52).
Figure 3.
Translation initiation from a subset of 12 codons spanning the expression range. Translation initiated from three expression cassettes, (A) GFP on a low-copy p15A plasmid, (B) nanoluciferase on a low-copy p15A plasmid and (C) nanoluciferase on a very-low-copy BAC. Transcription was driven by the RhaPBAD rhamnose-inducible native E. coli promoter. Shapes represent the replicate plate number, and filled shapes represent expression significantly greater (adjusted P < 0.05) than the non-expressing control (Control) as determined by Dunnett's test.
We measured nanoluciferase expression under transcription control of the RhaPBAD promoter on a low-copy p15A plasmid (Figure 1C). We detected significant translation initiation from all 12 codons (Figure 3B). Translation initiated from the three canonical start codons at 10–100% of AUG, from the four near-cognates at 0.1–1% of AUG and from the remaining five codons at 0.007–0.1% of AUG. Of particular note was that translation initiated from UAG (a canonical stop codon) was the lowest of the 12 codons (0.007% of AUG), but was still more than an order of magnitude higher than the signal measured from the non-expressing control (0.0003% of AUG).
We next measured nanoluciferase expression from the very-low-copy BAC. As with the p15A plasmid (Figure 3B), we measured significant expression initiated from all 12 of the start codons (Figure 3C). As expected from the lower copy number, the absolute luminescence measured from constructs on the BAC was more than an order of magnitude lower than the absolute luminescence measured from the same genes on the p15A plasmid. Translation initiated from the three canonical start codons at 10–100% of AUG, from the four near-cognates at 0.2–3% of AUG, and from the remaining five codons at 0.01–0.2% of AUG. The lowest nanoluciferase expression was again initiated from the canonical stop codon UAG (0.01% of AUG), which was still greater than the signal measured from the non-expressing control (0.004% of AUG). The relative levels of nanoluciferase translation initiated from the 12 codons on the p15A plasmid, the relative strength of nanoluciferase translation initiated from the 12 codons on the BAC, and GFP translation initiated on the pET20b(+) plasmid were well correlated (Supplementary Figure S10).
N-terminal RNA structure is known to impact translation initiation (67). We simulated RNA secondary structure around the initiation codon for the four reporter plasmids used in this work to evaluate if RNA secondary structure might contribute to translation initiation from non-canonical start codons. Both NUPACK (59) and KineFold (60) tools showed no correlation between the expected stability of the lowest energy structures and reporter expression for the nanoluciferase constructs, and a weak correlation for the pET20b(+)-GFP vector (Supplementary Figure S11 and Table S4). Additionally, there was no correlation between initiation codon GC-content and reporter expression (Supplementary Figure S10 and Table S4). These data suggest that differences in translation initiation from the start codons measured in this study were likely not caused by changes in RNA structure around the initiation codon or the GC-content of the initiation codon.
DISCUSSION
We observed translation initiation in E. coli from many non-canonical start codons, both near-cognates and non-near-cognates, at levels ranging from 0.007–3% of translation initiated from the canonical AUG start codon (Figures 2 and 3). Most of these codons have never before been identified as start codons.
We considered whether it is possible that the observed expression could be due to gene expression errors, including DNA replication errors, DNA transcription errors, tRNA misacylation or mRNA codon misreading (68). The reported rate of base misincorporation during DNA replication is ∼10−10 per base (69). The reported rate of base misincorporation during transcription ranges between 10−4 (70) and 10−5 per base (71), although in a codon that is one base away from a canonical start codon, only one out of three random mutations would result in an AUG start codon. Estimates for the rate of elongator tRNA misacylation range between 10−4 (68) and 10−6 (72), although these rates would need to be compounded with the likelihood of the mischarged amino acid being methionine (73), and discrimination by IF3 in the ribosomal P-site (74–77) for the stem-loop structure of initiator tRNAs (78,79). The rate of mRNA codon misreading (i.e. pairing a tRNA anticodon with a mismatched mRNA codon during elongation) depends on the number of mismatched bases. For single base mismatches, error rates during translation elongation have been estimated between 3.6 × 10−3 and 1 × 10−4 per codon, based on changes in activity from single amino acid substitutions in enzymatic assays (72,80,81) or radioisotope incorporation (82). For multiple base mismatches, error rates have been estimated around 3.1 × 10−4 per codon and can depend on cognate tRNA abundance (72).
Given the above, we took 3.6 × 10−3 and 3.1 × 10−4 as the highest reported error rates for single and multiple base errors in translation, respectively, noting that the experiments referenced only looked at misincorporation during translation elongation, not initiation. We calculated the expected expression due to these errors by multiplying these error rates by the expression from the AUG start codon, and compared the values to the expression level we measured for each codon (Supplementary Figure S12). The translation initiation rates we observed from non-canonical start codons were greater than the reported translation error rates for 17 non-AUG start codons from the T7-GFP plasmids. Specifically, we observed translation initiation from codons with single base mismatches from AUG at a rate ranging between 0.01 and 3% relative to AUG, and from codons with multiple base mismatches at a rate ranging between 0.007 and 0.1% relative to AUG (Figures 2 and 3). The translation initiation rates we observed were also strongly correlated to the identity of the non-canonical start codon across different genes, promoters, plasmid copy number and expression strains (Supplementary Figure S10).
Wobble base pairing occurs between mRNA codons and tRNA anticodons during translation, and may be part of the mechanistic explanation for the observations that we report in this paper. Wobble pairs occur at the third position of many codons during elongation (83). Wobble pairs also occur at the first position of GUG and UUG codons during initiation (84). We observed that G in the first position makes for a relatively stronger start codon than C in the first position (Figure 2 and Supplementary Figure S5), which could be due to the strength of the G-U wobble pair (85,86).
We observed evidence of translation initiation with N-terminal methionine from four codons (AUC, ACG, CAU and GGA), and with the N-terminal cognate amino acid in one spectrum from one codon (ACG) (Supplementary Tables S7–11). In the spectra in which we observed N-terminal methionine, it is likely that tRNAfMet is the initiating tRNA. We did not perform comprehensive mass spectrometry experiments to identify the N-terminal amino acid from the remaining codons, so we cannot be certain from which codon, with which tRNA and with which amino acid, translation is initiating.
Almost all E. coli genes with non-AUG start codons initiate with methionine as the N-terminal amino acid (4,6,7,65,66,87,88), and such events are not considered to be errors in translation initiation. By this same logic, we argue that translation initiation of genes with other non-AUG codons, in which methionine is observed as the N-terminal amino acid, should also not be considered an error. However, those wishing a strict interpretation of the central dogma could consider such events to be errors in translation initiation. All biological processes are governed by processes that imply a certain rate of unlikely events, and such unlikely events are often referred to as errors, failures or leaks. However, focusing only on the statistically likely outcome risks overlooking any advantageous aspects of rare but purposeful possibilities (89,90). Viewing non-canonical start codons without confinement to traditional dogma may reveal them as a potential feature, rather than an error, in gene expression.
For example, there may be evolutionary utility to translation initiation from non-canonical start codons. Research with yeast has shown gradual transitions of genetic sequences between genes and non-genic ORFs in related species (91). We can imagine a scenario wherein, over evolutionary time scales, point mutations could create a weak non-canonical initiation codon downstream of a RBS. The small amounts of protein produced from such an ORF, if beneficial to the organism, could select for further mutations that increased translation efficiency up to a point where the gene product more directly impacted organismal fitness. Further mutations could then be selected that tune for optimal expression dynamics in a given genetic context. Some evidence for this phenomenon exists in our start codon survey (Supplementary Table S6), in which the start codon of the pcnB (plasmid copy number B) gene alternate between AUG, GUG and UUG across the bacterial phyla Cyanobacteria, Proteobacteria, Chlamydiales and Hyperthermophiles (92).
There may also be regulatory utility to translation initiation from non-canonical start codons. The AUU start codon of infC regulates its translation (93), and a proposed mechanism for the utility of the GUG start codon is its ability to form stronger transcript secondary structures (84). Average per-cell abundances of proteins in bacteria and mammalian cells span five to seven orders of magnitude (94,95). Given that the non-canonical translation initiation shown in this paper spans about four orders of magnitude, it is possible that this level of expression could be physiologically significant and may serve as an additional mechanism for controlling protein synthesis.
Exploring changes in non-canonical translation initiation under different experimental conditions could indicate whether non-canonical translation initiation arises from error, or whether it confers an advantage through conditional regulation of gene expression. Examples of such experiments could include work with strains that have higher translational fidelity, which have been shown to increase the frequency of the programmed RF2 translational frameshift (96); initiating the translation of an essential gene, like the translation of chloramphenicol acetyltransferase from the UAG stop codon (97); measuring growth-rate-dependence of non-canonical translation initiation, similar to the growth-rate-dependent translation initiation of infC (93); and using a weaker RBS in place of the strong RBS used in this paper.
We wonder why so few genes have been annotated with non-canonical start codons in bacterial genomes. One possibility is that naturally occurring genes with non-canonical start codons are in fact exceedingly rare. Another possibility is that many naturally occurring non-canonical start codons and so-initiated proteins remain undiscovered because nobody has looked for them. In a recent E. coli whole cell shotgun proteomics experiment, approximately half of the detected spectra could not be mapped to known genes (98). E. coli ribosome profiling has also indicated that despite the extensive annotation of the E. coli genome, there may be unannotated ORFs (99). The presence of frequent but very low-level expression of proteins via non-canonical start codons would have widespread implications for genome annotation, cellular engineering and our fundamental understanding of translation initiation. We encourage reconsidering existing definitions and further exploration of what is considered a start codon.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to acknowledge Atri Choski, Geremy Clair, Steven Hallam, Sarah Munro and Ljiljana Pasa-Tolic, for helpful discussions, and Christine Chang for experimental assistance. The authors would like to thank Steve Lund for assistance with statistical data analysis. Cell sorting/flow cytometry analysis for this project was done on instruments in the Stanford Shared FACS Facility, with particularly helpful assistance from Marty Bigos and Cathy Crumpton. We would like to acknowledge Ryan Leib and Chris Adams at the Vincent Coates Foundation Mass Spectrometry Laboratory, Stanford University Mass Spectrometry (http://mass-spec.stanford.edu) for assistance in protein analysis. The authors would like to acknowledge Sara Lefort and the Friday morning Coffee Hour sponsored by the Ginzton Lab at Stanford University for providing the venue that facilitated the key conversation that inspired this project. The BAC was a generous gift from Fernan Federici of the Universidad Catolica de Chile. The authors acknowledge the financial support of the NRC/NIST Postdoctoral Research Program. Certain commercial equipment, instruments, or materials are identified in this report to specify adequately the experimental procedure. Such identification does not imply recommendation or endorsement by the National Institute of Standards and Technology, nor does it imply that the materials or equipment identified are necessarily the best available for the purpose.
SUPPLEMENTARY DATA
Supplementary data are available at NAR Online.
FUNDING
Internal NIST research and operational funding.
Conflict of interest statement. None declared.
REFERENCES
- 1. Clark B.F.C., Marcker K.A.. The role of N-Formyl-methionyl-sRNA in protein biosynthesis. J. Mol. Biol. 1966; 17:394–406. [DOI] [PubMed] [Google Scholar]
- 2. Adams J.M., Capecchi M.. N-Formylmethionyl-sRNA as the initiator of protein synthesis. Proc. Natl. Acad. Sci. U.S.A. 1966; 55:147–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nirenberg M.W., Leder P.. RNA codewords and protein synthesis. Science. 1964; 145:1399–1407. [DOI] [PubMed] [Google Scholar]
- 4. Blattner F.R., Plunkett G. III, Bloch C.A., Perna N.T., Burland V., Riley M., Collado-vides J., Glasner J.D., Rode C.K., Mayhew G.F. et al. The complete genome sequence of Escherichia coli K-12. Science. 1997; 277:1453–1462. [DOI] [PubMed] [Google Scholar]
- 5. Villegas A., Kropinski A.M.. An analysis of initiation codon utilization in the Domain Bacteria—concerns about the quality of bacterial genome annotation. Microbiology. 2008; 154:2559–2561. [DOI] [PubMed] [Google Scholar]
- 6. Sacerdot C., Fayat G., Dessen P., Springer M., Plumbridge J.A., Grunberg-Manago M., Blanquet S.. Sequence of a 1.26-kb DNA fragment containing the structural gene for E. coli initiation factor IF3: presence of an AUU initiator codon. EMBO J. 1982; 1:311–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Binns N., Masters M.. Expression of the Escherichia coli pcnB gene is translationally limited using an inefficient start codon: a second chromosomal example of translation initiated at AUU. Mol. Microbiol. 2002; 44:1287–1298. [DOI] [PubMed] [Google Scholar]
- 8. Tikole S., Sankararamakrishnan R.. A survey of mRNA sequences with a non-AUG start codon in RefSeq database. J. Biomol. Struct. Dyn. 2006; 24:33–42. [DOI] [PubMed] [Google Scholar]
- 9. Ingolia N.T., Ghaemmaghami S., Newman J.R.S., Weissman J.S.. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009; 324:218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ingolia N.T., Lareau L.F., Weissman J.S.. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011; 147:789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fritsch C., Herrmann A., Nothnagel M., Szafranski K., Huse K., Schumann F., Schreiber S., Platzer M., Krawczak M., Hampe J. et al. Genome-wide search for novel human uORFs and N-terminal protein extensions using ribosomal footprinting. Genome Res. 2012; 22:2208–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lee S., Liu B., Lee S., Huang S.-X., Shen B., Qian S.-B.. Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:E2424–E2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ringquist S., Shinedling S., Barrick D., Green L., Binkley J., Stormo G.D., Gold L.. Translation initiation in Escherichia coli: sequences within the ribosome-binding site. Mol. Microbiol. 1992; 6:1219–1229. [DOI] [PubMed] [Google Scholar]
- 14. Dreyfus M. What constitutes the signal for the initiation of protein synthesis on Escherichia coli mRNAs?. J. Mol. Biol. 1988; 204:79–94. [DOI] [PubMed] [Google Scholar]
- 15. Gold L. Posttranscriptional regulatory mechanisms in Escherichia coli. Annu. Rev. Biochem. 1988; 57:199–233. [DOI] [PubMed] [Google Scholar]
- 16. Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999; 234:187–208. [DOI] [PubMed] [Google Scholar]
- 17. Simonetti A., Marzi S., Jenner L., Myasnikov A., Romby P., Yusupova G., Klaholz B.P., Yusupov M.. A structural view of translation initiation in bacteria. Cell. Mol. Life Sci. 2009; 66:423–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gualerzi C.O., Pon C.L.. Initiation of mRNA translation in bacteria: structural and dynamic aspects. Cell. Mol. Life Sci. 2015; 72:4341–4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Billeter M.A., Dahlberg J.E., Goodman H.M., Hindley J., Weissman C.. Sequence of the first 175 nucleotides from the 5΄ terminus of Qβ RNA synthesized in vitro. Nature. 1969; 224:1083–1086. [DOI] [PubMed] [Google Scholar]
- 20. Shine J., Dalgarno L.. The 3΄-terminal sequence of Escherichia coli 16S ribosomal RNA: Complementarity to nonsense triplets and ribosome binding sites. Proc. Natl. Acad. Sci. U.S.A. 1974; 71:1342–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stormo G.D., Schneider T.D., Gold L.M.. Characterization of translation initiation sites in E. coli. Nucleic Acids Res. 1982; 10:2971–2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Matteucci M.D., Heyneker H.L.. Targeted random mutagenesis: the use of ambiguously synthesized oligonucleotides to mutagenize sequences immediately 5΄ of an ATG initiation codon. Nucleic Acids Res. 1983; 11:3113–3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hui A., Hayflick J., Dinkelspiel K., de Boer H.A.. Mutagenesis of the three bases preceding the start codon of the β-galactosidase mRNA and its effect on translation in Escherichia coli. EMBO J. 1984; 3:623–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Barrick D., Villanueba K., Childs J., Kalil R., Schneider T.D., Lawrence C.E., Gold L., Stormo G.D.. Quantitative analysis of ribosome binding sites in E. coli. Nucleic Acids Res. 1994; 22:1287–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ivanova N.N., Schwientek P., Tripp H.J., Rinke C., Pati A., Huntemann M., Visel A., Woyke T., Kyrpides N.C., Rubin E.M.. Stop codon reassignments in the wild. Science. 2014; 344:909–913. [DOI] [PubMed] [Google Scholar]
- 26. Mukai T., Englert M., Tripp H.J., Miller C., Ivanova N.N., Rubin E.M., Kyrpides N.C., Söll D.. Facile recoding of selenocysteine in nature. Angew. Chem. Int. Ed. 2016; 55:5337–5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Brocker M.J., Ho J.M.L., Church G.M., Soll D., O’Donoghue P.. Recoding the genetic code with selenocysteine. Angew. Chem. Int. Ed. 2014; 53:319–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lajoie M.J., Rovner A.J., Goodman D.B., Aerni H., Haimovich A.D., Kuznetsov G., Mercer J.A., Wang H.H., Carr P.A., Mosberg J.A. et al. Genomically recorded organisms expand biological functions. Science. 2013; 342:357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ma S., Tang N., Tian J.. DNA synthesis, assembly and applications in synthetic biology. Curr. Opin. Chem. Biol. 2012; 16:260–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shendure J., Aiden E.L., Lieberman Aiden E.. The expanding scope of DNA sequencing. Nat. Biotechnol. 2012; 30:1084–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Engler C., Gruetzner R., Kandzia R., Marillonnet S.. Golden gate shuffling: a one-pot DNA shuffling method based on Type Ils restriction enzymes. PLoS One. 2009; 4:e5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gibson D.G., Young L., Chuang R.-Y., Venter J.C., Hutchison C.A., Smith H.O.. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009; 6:343–345. [DOI] [PubMed] [Google Scholar]
- 33. Shaner N.C., Steinbach P.A., Tsien R.Y.. A guide to choosing fluorescent proteins. Nat. Methods. 2005; 2:905–909. [DOI] [PubMed] [Google Scholar]
- 34. Mutalik V.K., Guimaraes J.C., Cambray G., Lam C., Christoffersen M.J., Mai Q.-A., Tran A.B., Paull M., Keasling J.D., Arkin A.P. et al. Precise and reliable gene expression via standard transcription and translation initiation elements. Nat. Methods. 2013; 10:354–360. [DOI] [PubMed] [Google Scholar]
- 35. Alper H., Fischer C., Nevoigt E., Stephanopoulos G.. Tuning genetic control through promoter engineering. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:12678–12683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Brewster R.C., Jones D.L., Phillips R.. Tuning promoter strength through RNA polymerase binding site design in Escherichia coli. PLoS Comput. Biol. 2012; 8:e1002811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Keung A.J., Bashor C.J., Kiriakov S., Collins J.J., Khalil A.S.. Using targeted chromatin regulators to engineer combinatorial and spatial transcriptional regulation. Cell. 2014; 158:110–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stanton B.C., Nielsen A.A.K., Tamsir A., Clancy K., Peterson T., Voigt C.A.. Genomic mining of prokaryotic repressors for orthogonal logic gates. Nat. Chem. Biol. 2014; 10:99–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Salis H.M., Mirsky E.A., Voigt C.A.. Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol. 2009; 27:946–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kosuri S., Goodman D.B., Cambray G., Mutalik V.K., Gao Y., Arkin A.P., Endy D., Church G.M.. Composability of regulatory sequences controlling transcription and translation in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:14024–14029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lou C., Stanton B., Chen Y.-J., Munsky B., Voigt C.A.. Ribozyme-based insulator parts buffer synthetic circuits from genetic context. Nat. Biotechnol. 2012; 30:1137–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cambray G., Guimaraes J.C., Mutalik V.K., Lam C., Mai Q.A., Thimmaiah T., Carothers J.M., Arkin A.P., Endy D.. Measurement and modeling of intrinsic transcription terminators. Nucleic Acids Res. 2013; 41:5139–5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen Y., Liu P., Nielsen A.A.K., Brophy J.A.N., Clancy K., Peterson T., Voigt C.A.. Characterization of 582 natural and synthetic terminators and quantification of their design constraints. Nat. Methods. 2013; 10:659–664. [DOI] [PubMed] [Google Scholar]
- 44. Looman A.C., Bodlaender J., Comstock L.J., Eaton D., Jhurani P., de Boer H.A., van Knippenberg P.H.. Influence of the codon following the AUG initiation codon on the expression of a modified lacZ gene in Escherichia coli. EMBO J. 1987; 6:2489–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sprengart M.L., Fuchs E., Porter A.G.. The downstream box: an efficient and independent translation initiation signal in Escherichia coli. EMBO J. 1996; 15:665–674. [PMC free article] [PubMed] [Google Scholar]
- 46. Stenström C.M., Jin H., Major L.L., Tate W.P., Isaksson L.A.. Codon bias at the 3΄-side of the initiation codon is correlated with translation initiation efficiency in Escherichia coli. Gene. 2001; 263:273–284. [DOI] [PubMed] [Google Scholar]
- 47. Qing G., Xia B., Inouye M.. Enhancement of translation initiation by A/T-rich sequences downstream of the initiation codon in Escherichia coli. J. Mol. Microbiol. Biotechnol. 2003; 6:133–144. [DOI] [PubMed] [Google Scholar]
- 48. Sussman J.K., Simons E.L., Simons R.W.. Escherichia coli translation initiation factor 3 discriminates the initiation codon in vivo. Mol. Microbiol. 1996; 21:347–360. [DOI] [PubMed] [Google Scholar]
- 49. O’Connor M., Gregory S.T., Rajbhandary U.L., Dahlberg A.E.. Altered discrimination of start codons and initiator tRNAs by mutant initiation factor 3. RNA. 2001; 7:969–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pédelacq J.-D., Cabantous S., Tran T., Terwilliger T.C., Waldo G.S.. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 2006; 24:79–88. [DOI] [PubMed] [Google Scholar]
- 51. Chen H., Bjerknes M., Kumar R., Jay E.. Determination of the optimal aligned spacing between the Shine-Dalgarno sequence and the translation initiation codon of E. coli mRNAs. Nucleic Acids Res. 1994; 22:4953–4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hall M.P., Unch J., Binkowski B.F., Valley M.P., Butler B.L., Wood M.G., Otto P., Zimmerman K., Vidugiris G., Machleidt T. et al. Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem. Biol. 2012; 7:1848–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wild J., Hradecna Z., Szybalski W., Szygbalski W.. Conditionally amplifiable BACs: Switching from single-copy to high-copy vectors and genomic clones. Genome Res. 2002; 12:1434–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hahne F., LeMeur N., Brinkman R., Ellis B., Haaland P., Sarkar D., Spilden J., Strain E., Gentleman R.. flowCore: a Bioconductor package for high throughput flow cytometry. BMC Bioinformatics. 2009; 10:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wickham H. ggplot2: Elegant Graphics for Data Analysis. 2009; NY: Springer-Verglag. [Google Scholar]
- 56. Dunnett C.W. A multiple comparison procedure for comparing several treatments with a control. J. Am. Stat. Assoc. 1955; 50:1096–1121. [Google Scholar]
- 57. Hothorn T., Bretz F., Westfall P.. Simultaneous inference in general parametric models. Biometrical J. 2008; 50:346–363. [DOI] [PubMed] [Google Scholar]
- 58. Caspi R., Billington R., Ferrer L., Foerster H., Fulcher C.A., Keseler I.M., Kothari A., Krummenacker M., Latendresse M., Mueller L.A. et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2016; 44:D471–D480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zadeh J., Steenberg C., Bois J., Wolfe B., Pierce M., Khan A., Dirks R., Pierce N.. NUPACK: analysis and design of nucleic acid systems. J. Comput. Chem. 2010; 32:170–173. [DOI] [PubMed] [Google Scholar]
- 60. Xayaphoummine A., Bucher T., Isambert H.. Kinefold web server for RNA/DNA folding path and structure prediction including pseudoknots and knots. Nucleic Acids Res. 2005; 33:605–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Shimizu Y., Inoue A., Tomari Y., Suzuki T., Yokogawa T., Nishikawa K., Ueda T.. Cell-free translation reconstituted with purified components. Nat. Biotechnol. 2001; 19:751–755. [DOI] [PubMed] [Google Scholar]
- 62. Peabody D.S. Translation initiation at non-AUG triplets in mammalian cells. J. Biol. Chem. 1989; 264:5031–5035. [PubMed] [Google Scholar]
- 63. Hedges S.B. The origin and evolution of model organisms. Nat. Rev. Genet. 2002; 3:838–849. [DOI] [PubMed] [Google Scholar]
- 64. Li X., Zhang G., Ngo N., Zhao X., Kain S.R., Huang C.-C.. Deletions of the Aequorea victoria green fluorescent protein define the minimal domain required for fluorescence. J. Biol. Chem. 1997; 272:28545–28549. [DOI] [PubMed] [Google Scholar]
- 65. Farabaugh P.J. Sequence of the lacI gene. Nature. 1978; 274:765–769. [DOI] [PubMed] [Google Scholar]
- 66. Ishii S., Hatada E., Maekawa T., Imamoto F.. Molecular cloning and nucleotide sequencing of the nusB gene of E. coli. Nucleic Acids Res. 1984; 12:4987–4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Goodman D.B., Church G.M., Kosuri S.. Causes and effects of N-terminal codon bias in bacterial genes. Science. 2013; 342:475–479. [DOI] [PubMed] [Google Scholar]
- 68. Parker J. Errors and alternatives in reading the universal genetic code. Microbiol. Rev. 1989; 53:273–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Drake J.W. A constant rate of spontaneous mutation in DNA-based microbes. Proc. Natl. Acad. Sci. U.S.A. 1991; 88:7160–7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rosenberger R.F., Foskett G.. An estimate of the frequency of in vivo transcriptional errors at a nonsense codon in Escherichia coli. Mol. Gen. Genet. MGG. 1981; 183:561–563. [DOI] [PubMed] [Google Scholar]
- 71. Imashimizu M., Oshima T., Lubkowska L., Kashlev M.. Direct assessment of transcription fidelity by high-resolution RNA sequencing. Nucleic Acids Res. 2013; 41:9090–9104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kramer E.B., Farabaugh P.J.. The frequency of translational misreading errors in E. coli is largely determined by tRNA competition. RNA. 2007; 13:87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Blanquet S., Philippe D., Kahn D.. Properties and specificity of methionyl-tRNAfMet formyltransferase from Escherichia coli. Methods Enzymol. 1984; 106:141–152. [DOI] [PubMed] [Google Scholar]
- 74. Meinnel T., Sacerdot C., Graffe M., Blanquet S., Springer M.. Discrimination by Escherichia coli initiation factor IF3 against initiation on non-canonical codons relies on complementarity rules. J. Mol. Biol. 1999; 290:825–837. [DOI] [PubMed] [Google Scholar]
- 75. Hartz D., Binkley J., Hollingsworth T., Gold L.. Domains of initiator tRNA and initiation codon crucial for initiator tRNA selection by Escherichia coli IF3. Genes Dev. 1990; 4:1790–1800. [DOI] [PubMed] [Google Scholar]
- 76. Guillon J.M., Mechulam Y., Blanquet S., Fayat G.. Importance of formylability and anticodon stem sequence to give a tRNA(Met) an initiator identity in Escherichia coli. J. Bacteriol. 1993; 175:4507–4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Sussman J.K., Simons E.L., Simons R.W.. Escherichia coli translation initiation factor 3 discriminates the initiation codon in vivo. Mol. Microbiol. 1996; 21:347–360. [DOI] [PubMed] [Google Scholar]
- 78. Varshney U., Lee C.P., RajBhandary U.L.. From elongator tRNA to initiator tRNA. Proc. Natl. Acad. Sci. U.S.A. 1993; 90:2305–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Shetty S., Bhattacharyya S., Varshney U.. Is the cellular initiation of translation an exclusive property of the initiator tRNAs?. RNA Biol. 2015; 12:675–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Bouadloun F., Donner D., Kurland C.G.. Codon-specific missense errors in vivo. EMBO J. 1983; 2:1351–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Toth M.J., Murgola E.J., Schimmel P.. Evidence for a unique first position codon-anticodon mismatch in vivo. J. Mol. Biol. 1988; 201:451–454. [DOI] [PubMed] [Google Scholar]
- 82. Edelmann P., Gallant J.. Mistranslation in E. coli. Cell. 1977; 10:131–137. [DOI] [PubMed] [Google Scholar]
- 83. Agris P.F., Vendeix F.A.P., Graham W.D.. tRNA's wobble decoding of the genome: 40 years of modification. J. Mol. Biol. 2007; 366:1–13. [DOI] [PubMed] [Google Scholar]
- 84. Asano K. Why is start codon selection so precise in eukaryotes?. Translation. 2014; 2:e28387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Varani G., McClain W.H.. The G·U wobble base pair. EMBO Rep. 2000; 1:18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Leontis N.B., Stombaugh J., Westhof E.. The non-Watson-Crick base pairs and their associated isostericity matrices. Nucleic Acids Res. 2002; 30:3497–3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Poulis M.I., Shaw D.C., Campbell H.D., Young I.G.. In vitro synthesis of the respiratory NADH dehydrogenase of Escherichia coli. Role of UUG as initiation codon. Biochemistry. 1981; 20:4178–4185. [DOI] [PubMed] [Google Scholar]
- 88. Belin D., Hedgpeth J., Selzer G.B., Epstein R.H.. Temperature-sensitive mutation in the initiation codon of the rIIB gene of bacteriophage T4. Proc. Natl. Acad. Sci. U.S.A. 1979; 76:700–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Rutherford S.L., Lindquist S.. Hsp90 as a capacitor for morphological evolution. Nature. 1998; 396:336–342. [DOI] [PubMed] [Google Scholar]
- 90. Schwartz M.H., Waldbauer J.R., Zhang L., Pan T.. Global tRNA misacylation induced by anaerobiosis and antibiotic exposure broadly increases stress resistance in Escherichia coli. Nucleic Acids Res. 2016; 44:10292–10303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Carvunis A.-R., Rolland T., Wapinski I., Calderwood M.A., Yildirim M.A., Simonis N., Charloteaux B., Hidalgo C.A., Barbette J., Santhanam B. et al. Proto-genes and de novo gene birth. Nature. 2012; 487:370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Cao G.J., Kalapos M.P., Sarkar N.. Polyadenylated mRNA in Escherichia coli: Modulation of poly(A) RNA levels by polynucleotide phosphorylase and ribonuclease II. Biochimie. 1997; 79:211–220. [DOI] [PubMed] [Google Scholar]
- 93. Butler J.S., Springer M., Grunberg-Manago M.. AUU-to-AUG mutation in the initiator codon of the translation initiation factor IF3 abolishes translational autocontrol of its own gene (infC) in vivo. Proc. Natl. Acad. Sci. U.S.A. 1987; 84:4022–4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Taniguchi Y., Choi P.J., Li G., Chen H., Babu M., Hearn J., Emili A., Xie X.S.. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 2010; 329:533–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Vogel C., Marcotte E.M.. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 2012; 13:227–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Sipley J., Goldman E.. Increased ribosomal accuracy increases a programmed translational frameshift in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1993; 90:2315–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Varshney U., RajBhandary U.L.. Initiation of protein synthesis from a termination codon. Proc. Natl. Acad. Sci. U.S.A. 1990; 87:1586–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Schmidt A., Kochanowski K., Vedelaar S., Ahrne E., Volkmer B., Callipo L., Knoops K., Bauer M., Aebersold R., Heinemann M.. The quantitative and condition-dependent Escherichia coli proteome. Nat. Biotechnol. 2016; 34:104–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Oh E., Becker A.H., Sandikci A., Huber D., Chaba R., Gloge F., Nichols R.J., Typas A., Gross C.A., Kramer G. et al. Selective ribosome profiling reveals the cotranslational chaperone action of trigger factor in vivo. Cell. 2011; 147:1295–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.