Abstract
FadR is a fatty acyl-CoA dependent transcription factor that regulates genes encoding proteins involved in fatty-acid degradation and synthesis pathways. In this study, the crystal structures of Bacillus halodurans FadR, which belong to the TetR family, have been determined in three different forms: ligand-bound, ligand-free and DNA-bound at resolutions of 1.75, 2.05 and 2.80 Å, respectively. Structural and functional data showed that B. halodurans FadR was bound to its operator site without fatty acyl-CoAs. Structural comparisons among the three different forms of B. halodurans FadR revealed that the movement of DNA binding domains toward the operator DNA was blocked upon binding of ligand molecules. These findings suggest that the TetR family FadR negatively regulates the genes involved in fatty acid metabolism by binding cooperatively to the operator DNA as a dimer of dimers.
INTRODUCTION
Fatty acids are vital building blocks in lipid membranes and are carbon sources for metabolism in all living organisms. The fatty acid decomposition and biosynthesis pathways are strictly regulated. FadR is the main transcriptional regulator controlling the expression of multiple genes involved in the fatty acid degradation and biosynthesis pathways in Escherichia coli and Bacillus subtilis (1–3). The functions of FadR in E. coli and B. subtilis appear similar, however, they belong to very different structural families.
Escherichia coli FadR is a member of the GntR family of transcription factors (4,5), and a bifunctional acyl-CoA responsive transcription factor. In the absence of long-chain fatty acids, E. coli FadR binds directly to multiple fad regulons (fadE, fadF, fadG, fadH, fadBA, fadL and fadD) and acts as a repressor of the β-oxidation cycle that breaks down fatty acids into acetyl-CoAs (2,5) and transporter of fatty acids to the inner membrane (6,7). E. coli FadR also negatively regulates the transcription of universal stress genes, uspA (8) and yfcX-yfcY genes that produce FadB/A homologous proteins required for anaerobic growth conditions (9). Unsaturated fatty acid biosynthesis is activated simultaneously by FadR via transcription of biosynthetic genes (fabA and fabB) in E. coli. The products of fabA/B genes, 3-hydroxydecanoyl-acyl carrier protein (ACP) dehydratase and 3-ketoacyl-ACP synthase I, play a crucial role in dehydration, isomerization and elongation of fatty acids for the biosynthesis of unsaturated long-chain fatty acids (10,11). In addition, FadR induces expression of the iclR gene that encodes the repressor of the E. coli glycoxylate bypass operon (aceBAK) (12,13). The structures of E. coli FadR have been reported in three different forms, such as ligand-free, acyl-CoA-bound (ligand-bound) and DNA-bound (14–16). Structural studies showed that E. coli FadR is a functional homodimer and composed of two domains (15); the N-terminal DNA binding domain (N-DBD) contains a helix-turn-helix (HTH) motif, and the C-terminal domain has an acyl-CoA binding domain consisting of a seven-helix bundle, which contains a four-helix dimerization motif (14,17,18).
In B. subtilis, fatty acid degradation and biosynthesis are controlled by the FadR and FapR genes, respectively. B. subtilis FadR negatively regulates fatty acid degradation, whereas FapR negatively controls biosynthesis of fatty acids and phospholipids (19). A structural analysis revealed that B. subtilis FadR belongs to the TetR family of transcription factors, which represses five fad operons (lcfA-fadR-fadB-etfB-etfA, fadH-fadG, fadN-fadA-fadE, lcfB and fadF-acdA-rpoE) involved in the fatty acid β-oxidation cycle (20,21). B. subtilis FadR directly binds to upstream regions of the fadR, fadH, fadN, lcfB and fadF genes, and is deactivated by binding with long-chain acyl-CoAs in a similar manner as E. coli FadR. In vivo and in vitro results showed that binding to the fad regulon is inhibited by long-chain acyl-CoA (14–20 carbon atoms) and 12-metyltetradecanoyl/13-metyltetradeconoyl CoAs (4). Unlike E. coli, B. subtilis FadR is not involved in the fatty acid biosynthesis pathway. However, highly conserved FapR negatively controls expression of the fap operon (fabHA-fabF, fapR-plsx-fabD-fabG, fabI, fabHB, yhfC and plsX) involved in fatty acid and phospholipid biosynthesis in B. subtilis (19).
The crystal structures of B. subtilis FadR were determined with lauroyl-CoA and stearoyl-CoA (20,22), and its homolog Thermus thermophilus FadR with lauroyl-CoA has been reported (23). The FadR from B. halodurans is also a homologous protein to B. subtilis FadR. It contains 195 amino acid residues with 65% sequence identity to B. subtilis FadR and 21% sequence identity to T. thermophiles FadR, but no sequence identity to E. coli FadR in the GntR family.
Several structures of TetR superfamily transcriptional regulators in complex with cognate DNA have been determined, such as Streptomyces coelicolor CprB (24), Corynebacterium glutamicum CgmR (25), Pseudomonas aeruginosa DesT (26), Mycobacetrium tuberculosis Ms6564 (27), Staphylococcus aureus QacR (18), Streptomyces antibioticus SimR (28), E. coli SlmA (29) and E. coli TetR (30). Based on structural analyses, the TetR superfamily transcriptional regulators are divided into two sub-classes depending on their DNA binding mode (24). One sub-class, including E. coli TetR, S. antibioticus SimR and P. aeruginosa DesT, binds to their cognate DNA as a dimer. The other sub-class, including S. aureus QacR, E. coli SlmA, S. coelicolor CprB, C. glutamicum CgmR and Mycobacterium smegmatis Ms6564, binds as a dimer of dimers.
Although several structures of TetR family FadR proteins with fatty acyl-CoAs (C12–C18) have been determined, there are currently no structures available for a TetR family FadR protein bound to its cognate DNA. Therefore, the structural conformation and functional requirements for DNA binding by FadR and the dissociation of FadR–DNA induced by effector molecules remain unclear. In this study, we determined the structures of the ligand-bound, ligand-free and DNA-bound forms and performed gel shift assays to characterize DNA binding by FadR. The FadR was mutated and the effects on DNA binding were assessed to test structural predictions. The results show that B. halodurans FadR bound to its cognate DNA as a dimer of dimers and a comparison of the DNA-bound and ligand-bound forms revealed the induction mechanism by fatty acyl-CoA ligands.
MATERIALS AND METHODS
Protein preparation
Gene cloning, expression and purification of B. halodurans FadR were conducted as previously described (31). Native FadR was expressed in E. coli BL21 (DE3) Star pLysS cells. The purification protocol was modified slightly. The cells were resuspended in lysis buffer (20 mM Tris–HCl at pH 8.0, 0.5 M NaCl, 10% (v/v) glycerol, 1 mM PMSF and 1 mM TCEP) and homogenized using an ultrasonic processor. A 1% NP-40 solution was added during lysis to remove lipid molecules from native FadR, and the remainder of the purification procedure was the same as that for native FadR. The insoluble fraction including cellular debris was removed by centrifugation at 31 000 g for 60 min at 277 K, and the recombinant protein in the supernatant fraction was purified using three chromatographic steps described previously in detail (31). The purified proteins were concentrated to 70 mg/ml using the Centricon YM-10 (Millipore, USA) and stored at 193 K.
Site-directed mutagenesis of the FadR proteins (G42Y, Y45A, Y45F and R117A) were performed by a two-step overlapping polymerase chain reaction (PCR) method using wild-type DNA as a template. The 5΄-region of the B. halodurans fadR gene was amplified using i-Taq DNA polymerase (iNtRON, Korea), with the FadR_F oligonucleotide (5΄-GGAATTCCATATGGGAAAGAAAAAAGGACCAAAATA-3΄) as the forward primer and G42Y_R (5΄-AGGTAAATCGTGTAATCAGCTACTC-3΄), Y45A_R (5΄-TGTTAAAATAAAGCGCAATCGTGCCATC-3΄), Y45F_R (5΄-TGTTAAAATAAAGAAAAATCGTGCCATC -3΄) and R117A_R (5΄-CTTCATTTATTTTTAACGCAAGCT CTGTA-3΄) oligonucleotides as the respective reverse primers. Similarly, the 3΄-region of the B. halodurans fadR gene was amplified using the G42Y_F (5΄-GAGTAGCTGATTACACGATTTACCT-3΄), Y45A_F (5΄-GATGGCACGATTGCGCTTTATTTTAACA-3΄), Y45F_F (5΄-GATGGCACGATTTTTCTTTATTTTAACA-3΄) and R117A_F (5΄-TACAGAGCTTGCGTTAAAAATAAATGAAG-3΄) oligonucleotides as the respective forward primers and the FadR_R oligonucleotide (5΄-CCGCTCGAGTCAACGATGGCGCAACCCACC-3΄) as the reverse primer. The initial PCR products were mixed with KAPA Hifi ExTaq DNA polymerase (Kapa Biosystems, USA) and deoxynucleotides. Then, a second PCR was conducted with the FadR_F and FadR_R primer pairs. The resulting PCR products were cloned into the pET28b(+) vector. The mutant FadR sequences were confirmed by DNA sequencing. The expression and purification procedure was the same as that used for native FadR.
Crystallization
Native FadR crystals were obtained by the sitting-drop vapor diffusion method in a reservoir containing 0.1 M Tris–HCl at pH 8.5, 0.3 M MgCl2 and 25% PEG 3350. The detailed crystallization procedure for native FadR has been described previously (31). Native FadR crystals contained fatty acids derived from E. coli during purification and verified as a mixture of three fatty acids (myristic acid, palmitic acid and stearic acid) by gas chromatography-mass spectrometry (GC-MS). We attempted to grow crystals of the ligand-free FadR obtained by NP-40 treatment during purification, but it failed. The NP-40 treated FadR was used in the electrophoretic mobility shift assay (EMSA). However, a mixture of fatty acids in the purified FadR proteins was not completely removed and detected by GC-MS. In order to obtain ligand-free FadR crystals, mutant FadR_R117A crystals were grown by the hanging-drop vapor diffusion method under the same conditions as those used for native FadR crystals. Their approximate dimensions were 0.4 × 0.4 × 0.15 mm.
To prepare the DNA complex crystals, 21 base-pair (bp) HPLC-grade oligonucleotides, 21OH_F (5΄–GATGAATGAATACTCATTCAT–3΄) and 21OH_R (5΄–CATGAATGAGTATTCATTCAT–3΄), were chosen at the fadR promoter site which was the corresponding site in B. subtilis determined as the cognate DNA sequence. Each oligonucleotide was dissolved in distilled water, annealed by placing in boiling water and incubating overnight at 277K, and mixed with native FadR proteins, which were used for growing ligand-bound crystals, at a molar ratio of 0.6:1. The oligonucleotides contained the B. halodurans fadR promoter region and were homologous to B. subtilis FadR binding boxes (WTGAATGAMTANTCATTCAN, where W, M and N stand for A or T, A or C, and any bases, respectively. FadR–DNA complex crystals were obtained using the hanging-drop method in 0.2 M HEPES at pH 7.5, 10% PEG 8000 and 8% ethanol. The DNA complex crystals were further optimized using additive screening solutions (Hampton Research, USA). The crystals grew reproducibly up to a maximum size of approximately 0.05 × 0.01 × 0.3 mm within 2 days.
Data collection
Native FadR crystals were transferred into a cryoprotectant solution containing 20% (v/v) glycerol in the reservoir solution. Native data were collected to 1.75 Å resolution at 100 K using the ADSC Quantum 315 Charge-coupled device (CCD) image-plate detector on beamline 5C SB IІ at the Pohang Accelerator Laboratory (PAL), Republic of Korea. The data were collected at a wavelength of 0.97933 Å using 1° oscillation per image with a crystal-to-detector distance of 220 mm. The crystals belonged to the primitive trigonal space group P3221 with unit-cell parameters, a = b = 56.87 Å, c = 200.9 Å, α = β = 90° and γ = 120°. There was a dimer molecule in the asymmetric unit, giving a solvent fraction of 41.42%.
The cryoprotectant solution composition and mounting method for the mutant FadR_R117A crystals were the same as those for the native crystals. Mutant FadR crystals were collected to 2.05 Å resolution at a wavelength of 0.97923 Å using the ADSC Quantum 315 CCD image-plate detector on beamline 5C SB II of the PAL. The crystals belonged to the primitive trigonal space group P3221, with unit-cell parameters, a = b = 56.40 Å and c = 199.6 Å.
FadR–DNA complex crystals were transferred into a cryoprotectant solution consisting of 0.2 M HEPES at pH 7.5, 20% PEG8000 and 15% MPD. The data set was collected to 2.8 Å resolution at a wavelength of 0.97933 Å using the ADSC Quantum 270 CCD image-plate detector on beamline 7A SB I of the PAL. The crystals were in the space group P1, with the cell dimensions, a = 46.50 Å, b = 76.94 Å, c = 87.02 Å, α = 103.8°, β = 105.5° and γ = 89.64°. Two FadR dimers were in the complex with a 21 bp cognate oligonucleotide in the asymmetric unit, giving a solvent fraction of 63.11%. All data were processed and scaled using DENZO and SCALEPACK from the HKL2000 program suite (32).
Structure determination and refinement
The B. halodurans FadR structure was solved by molecular replacement at 1.75 Å resolution using the program PHASER (33) based on the structure of B. subtilis FadR (PDB ID: 1vi0) (22). The initial model was improved by iterative manual building and refinement with the COOT (34), REFMAC (35) and PHENIX programs (36). The resulting final structure had an R-work of 20.9% and R-free of 25.2%, with good stereochemistry. The crystal structure of the FadR_R117A mutant (ligand-free) was refined to 2.05 Å resolution with an R-work of 20.4% and R-free of 26.1%.
The FadR–DNA complex crystal structure was solved by molecular replacement with the native B. halodurans FadR structure as a search model, using the program PHASER (33). The initial maps showed the density of dsDNA, which was built manually using the program COOT (34). The structure was improved with iterative cycles of model building using COOT and refinement using PHENIX (34,36). The final model contained two dimeric FadRs with a 21 bp DNA molecule in the asymmetric unit. The refined structures were validated by the program Molprobity (37). All data and refinement statistics are summarized in Table 1.
Table 1. Data collection and refinement statistics.
| Dataset | Ligand-bound | Ligand-free | DNA-bound |
|---|---|---|---|
| Data collection | |||
| Resolution range (Å) | 50–1.75 (1.78–1.75)a | 50–2.05 (2.09–2.05) | 50–2.8 (2.85–2.8) |
| Space group | P3221 | P3221 | P1 |
| a, b, c (Å) | 56.87, 56.87, 200.99 | 56.4, 56.4, 199.61 | 46.5, 76.94, 87.02 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 | 103.8, 105.5, 89.6 |
| Total/unique reflections | 291 359/38 629 | 249 225/24 026 | 55 332/26 864 |
| Multiplicity | 7.5 (5.9) | 10.4 (10.7) | 2.1 (2.0) |
| Completeness (%) | 98.6 (83.0) | 99.6 (100) | 96.3 (96.2) |
| Mean I/σ(I) | 49.67 (3.43) | 53.59 (6.96) | 13.98 (1.92) |
| Rmerge (%)b | 5.2 (44.2) | 5.2 (46.1) | 10.1 (60.8) |
| Refinement statistics | |||
| Resolution range (Å) | 27.69–1.75 (1.82–1.75) | 20–2.05 (2.12–2.05) | 20–2.8 (2.9–2.8) |
| R-work/R-free (%)c | 20.9/25.2 (26.04/28.62) | 20.4/26.1 (23.7/31.8) | 20.4/26.6 (32.4/42.4) |
| No. of residues/mean B-factors (Å2) | 380/38.6 | 375/43.1 | 802/52.0 |
| No. of waters/mean B-factors (Å2) | 275/46.0 | 91/45.6 | 17/40.1 |
| No. of ligands/mean B-factors (Å2) | 49/45.1 | 7/38.3 | |
| No. of nucleotides/mean B-factors (Å2) | 42.0/54.6 | ||
| R.M.S deviations | |||
| Bond lengths (Å) | 0.01 | 0.009 | 0.004 |
| Bond angles (°) | 1.21 | 1.11 | 0.77 |
| Ramachandran plot (%)d | |||
| Favored (%) | 99 | 99 | 96.1 |
| Allowed (%) | 1 | 1 | 3.9 |
aNumbers in parentheses indicate the statistics for the last resolution shell.
bRmerge = ΣhΣi|I(h)i−<I(h)>|/ΣhΣiI(h)i, where I(h) is the intensity of reflection h, Σh is the sum over all reflections, and Σi is the sum over i measurements of reflection h.
cR-work = Σ | |Fobs| – |Fcalc| | / Σ |Fobs|, where R-free is calculated for a randomly chosen 10% of reflections, which were not used for structure refinement and R-work is calculated for the remaining reflections.
dDetermined using Molprobity.
Electrophoretic mobility shift assay
The 5΄-biotinylated oligonucleotides and their complement oligonucleotides containing the fadR promoter and impaired fadR promoter were directly synthesized for the DNA binding assay (Macrogen, Korea). All oligonucleotide sequences are listed in Supplementary Table S2. The 5΄-biotinylated oligonucleotide and 1.3-fold of the complementary oligonucleotide were added and annealed in boiling water. The dsDNA substrates were stored at −20°C until use. The EMSA was performed using LightShift® Chemiluminescent EMSA kit following the manufacturer's instruction (Pierce Biotechnology, USA). The dsDNA substrates were incubated at room temperature for 20 min with various quantities of proteins in 20 μl of reaction buffer (10 mM Tris–HCl at pH 7.5, 5 mM MgCl2, 50 mM KCl, 50 ng/μl poly dI·dC, 1 mM DTT and 2.5% glycerol). The reaction mixtures were subjected directly to 8% native polyacrylamide gel electrophoresis with 1× Tris-glycine buffer. Electrophoresis was performed at 65 V for 1 h. After electrophoresis, the gel was transferred onto a positive nylon membrane and cross-linked with ultraviolet light at 254 nm. Images were acquired by ChemiDoc™ XRS+ (Bio-Rad, USA). All experiments were carried out at least three times and quantified using Image Lab™ software (Bio-Rad, USA).
RESULTS AND DISCUSSION
Overall structure of B. halodurans FadR
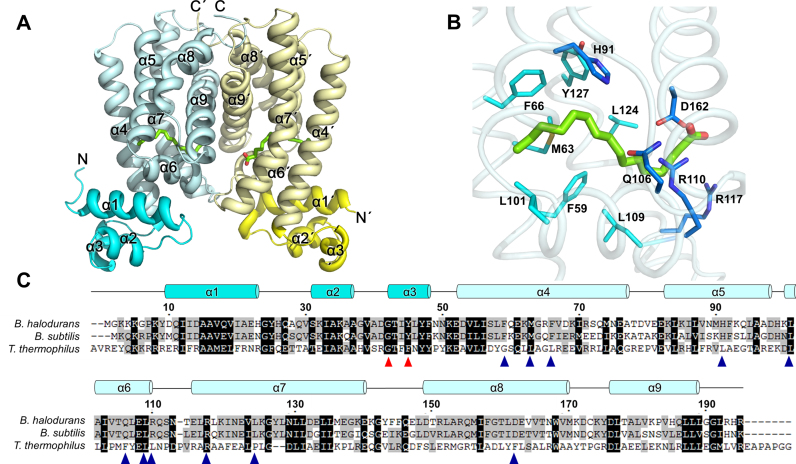
The B. halodurans FadR structure was determined by the molecular replacement method using the coordinate structure of B. subtilis FadR (PDB ID: 1vi0) as a search model and refined to 1.75 Å resolution (Figure 1A and Table 1). Among the 390 amino acid residues in the dimeric structure, two N-terminal residues in chain A, seven N-terminal residues and a C-terminal residue in chain B were not traceable. In addition, the Lys8 and Arg193 residues in chain B were assigned to alanine due to a lack of electron density. A homodimeric FadR was located in the asymmetric unit of the crystal and formed a Ω-shaped structure that was typical of the TetR family of transcription factors, composed of an N-DBD and a C-terminal effector binding domain (C-EBD). B. halodurans FadR consisted of nine helices: α1 (9–23), α2 (30–37), α3 (41–47), α4 (51–76), α5 (80–97), α6 (99–110), α7 (115–141), α8 (150–171) and α9 (177–190); it was divided into the N-DBD (helices α1–α3) and C-EBD (helices α4–α9). The N-DBD of B. halodurans FadR contained a HTH motif, which was consistent with its DNA binding activity. In the C-EBD of B. halodurans FadR, the electron density map showed that one fatty acid chain was located in each subunit, which was often observed in other FadR homologous structures as well. A fatty acyl-CoA molecule was found at each subunit in the dimeric structures of B. subtilis FadR and T. thermophilus FadR. However, we only assigned palmitic acid, which was verified as a mixture of three fatty acids (myristic acid, palmitic acid and stearic acid) using GC-MS. The two subunits formed the dimeric structure, related by the non-crystallographic 2-fold axis through octahedral coordination with a magnesium ion and six water molecules. The buried surface area in the dimer was ∼1800 Å2 (∼17% of the monomer's surface area). Dimeric B. halodurans FadR was stabilized by a substantial linkage of hydrogen bonds and hydrophobic interactions along helices α7–α9; 36 residues were involved in hydrophobic interactions and 11 residues were involved in hydrogen bonds. (PDBePISA protein–protein interaction server: http://www.ebi.ac.uk/msd-srv/prot_int/ and PDBsum generate: http://www.ebi.ac.uk/thornton-srv/databases/pdbsum/Generate.html).
Figure 1.
Overall structure of Bacillus halodurans FadR. (A) Cartoon representation of the B. halodurans FadR dimer. The monomers are colored in cyan and yellow. The DBDs are shown in dark cyan and dark yellow. Palmitic acids are drawn in green stick models. The N and C termini are labeled. (B) Close-up view of ligand binding site in B. halodurans FadR. Residues in hydrophobic core and hydrophilic patch are shown as cyan and blue sticks, respectively. (C) Sequence alignment of B. halodurans FadR and representative TetR family FadR proteins (B. subtilis and Thermus thermophilus). Every 20th residue is indicated above the sequence of B. halodurans FadR. Highly conserved residues and partially conserved residues are shaded in black and gray, respectively. The residues involved in DNA binding and ligand binding are indicated as red and blue triangles.
The hydrocarbon chain of palmitic acid was buried deeply within the ligand-binding cavity among helices α4–α8. The ligand-binding cavity was mainly surrounded by hydrophobic residues, such as Phe59, Met63, Phe66, Leu95, Leu101, Leu109, Leu124 and Tyr127; hydrophilic patches were observed, including His91, Gln106, Arg110, Arg117, Asp162 and a water molecule (Figure 1B), which were also found in the homologous structure of B. subtilis FadR. However, the amino acids involved in the ligand-binding cavity were less conserved in T. thermophilus FadR (Figure 1C).
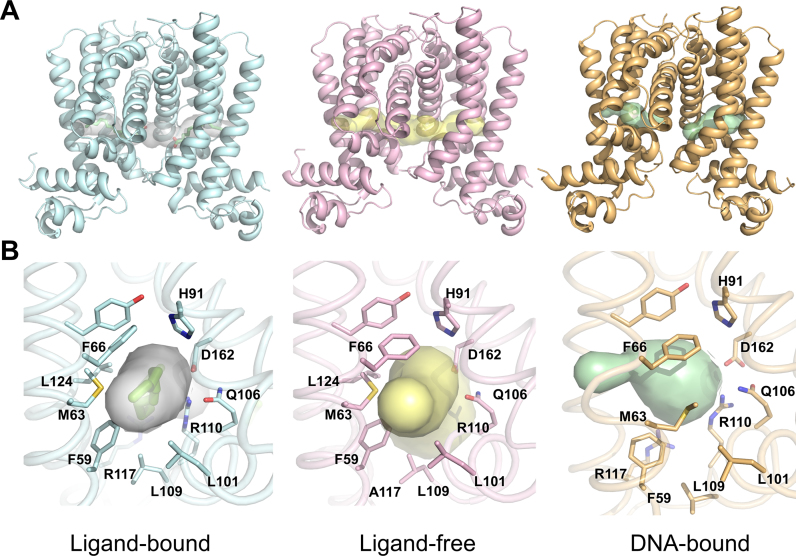
We were unsuccessful in growing the ligand-free form of B. halodurans FadR from native proteins; thus, we obtained a ligand-free structure from mutant FadR (R117A) and confirmed by an omit map and a 2fo-fc refined map around ligand binding site (Supplementary Figure S1). Arg117 is highly conserved in the TetR family FadR structures and is mainly involved in ligand binding around the hydrophobic cavity exit. The overall structure of ligand-free FadR was almost identical with that of the ligand-bound form, which had a root mean square deviation (RMSD) of 0.32 Å over 193 equivalent Cα atoms (Supplementary Figure S2). The Cα superposition plot showed that there were relatively large differences in three regions: residues 5–13, 60–80 and 110–120. Among the three regions, the 5–13 residue region showed flexibility in the N-terminus, whereas the other two regions were involved in the ligand-binding site. Residues 60–80 were main component of helix α4, which was involved in the hydrophobic cavity and directly connected with the DBD. Residues 110–115 were located in the loop between the α6 and α7 helices and were also involved in the ligand-binding site close to the exit of the cavity. When the two structures were superimposed based on the dimerization motif (helices α8 and α9), helix α4 was shifted toward the DBD in the ligand-free structure (Supplementary Figure S2). Average distance between the same positions of Cα atoms in residues 60–80 was 1.0 Å. In particular, Phe66, Met63 and Phe59 in helix α4 were shifted, and the phenyl ring of Phe66 was rotated toward the cavity (Figure 2B). The cavity volumes of ligand-bound, ligand-free and DNA-bound structures were calculated to be ∼1100 Å3, ∼750 Å3 and ∼600 Å3, respectively (GHECOM server. http://strcomp.protein.osaka-u.ac.jp/ghecom/, Figure 2A). These results indicate that binding of ligand molecules could keep the C-EBD rigid and block movement of N-DBD to bind with cognate DNA.
Figure 2.
Structural comparisons of Bacillus halodurans FadR. (A) Cartoon representations of the three B. halodurans FadR structures, ligand-bound (cyan), ligand-free (pink) and DNA-bound (orange). Each figure is drawn in same orientation. The ligand-binding cavities are illustrated as surface model. (B) The enlarged views of the ligand-binding cavities and the surrounding residues colored as in (A). The cavities are drawn using Caver.
Comparison with the other TetR family FadR proteins
The structure of B. subtilis FadR in the TetR family is well characterized and is homologous to B. halodurans FadR with ∼65% sequence identity. A recent crystallographic analysis of B. subtilis FadR showed that B. subtilis FadR is a homodimeric protein containing a stearoyl-CoA molecule in each subunit (PDB ID: 3whc) (20,22). When we superimposed the ligand-bound B. halodurans FadR on B. subtilis FadR, the RMSD for 347 Cα atoms of B. halodurans FadR at the same position as that of B. subtilis FadR was 0.8 Å. As described above, FadR is released from the cognate DNA by binding long-chain acyl-CoAs that induce conformational changes in the FadR structure (4,14,15,20). Long-chain acyl-CoA was observed at the protein surface and binding cavity in the B. subtilis FadR structure. The CoA region was anchored by several residues (Arg116, Asn120, Arg150, Arg153, Glu162΄ and Tyr174΄; prime indicates the residue in the counterpart subunit) at the surface. In particular, Arg150 and Tyr174΄ stacked the adenine moiety on both sides. The hydrocarbon chain was deeply embedded in the binding cavity, which was mainly surrounded by a hydrophobic environment and a hydrophilic patch region as seen in B. halodurans FadR. Although the CoA region was not assigned in the B. halodurans FadR structure, the binding conformation of the fatty acid hydrocarbon chain was similar. The long-chain fatty acid was bent around the C9–C11 atoms in both structures and was surrounded by the hydrophobic and hydrophilic patch (Figure 1B and Supplementary Figure S3).
T. thermophilus FadR, which is homologous to B. halodurans FadR with a sequence identity of ∼23%, was in a complex with lauroyl-CoA. The RMS distances between the Cα atoms of four T. thermophilus FadR subunits and the same positions of two B. halodurans FadR subunits were 3.6–3.9 Å for 162 Cα pairs. As shown in Figure 1, the ligand binding residues were well conserved. However, the ligand binding conformation of the T. thermophilus FadR protein was quite different from that of the Bacillus FadR protein. The structure of T. thermophilus FadR in complex with lauroyl-CoA showed a straight conformation, while that of Bacillus FadR structure showed a bent conformation around C9–C11 atoms of the hydrocarbon chain. In addition, the relatively short hydrocarbon chain was surrounded by only the hydrophobic residues in the T. thermophilus FadR structure, but long hydrocarbon chains were buried with a hydrophilic patch around the front region (C1–C9) and with hydrophobic residues in the terminal region (C10–C18) in the Bacillus FadR structures. In fact, T. thermophilus FadR is more sensitive than other FadR proteins when binding a shorter chain acyl-CoA (21,23), which could be attributed to structural differences in the ligand-binding cavity between Bacillus FadR and T. thermophilus FadR.
A regulatory protein M. tuberculosis Rv3249c (38), which is involved in regulating MmpL transporters, co-purified and co-crystallized with palmitic acid. Rv3249c represses multiple mmpL genes by binding multiple operators and is released from the promoter when bound to palmitic acid. Although the mechanism of induction by effector molecules is similar, the Rv3249c binding mode to palmitic acid was clearly different from that of B. halodurans FadR. The palmitic acid in M. tuberculosis Rv3249c was roughly parallel to the surrounding α-helices, but was almost perpendicular to the helices in B. halodurans FadR (Supplementary Figure S3B).
DNA bound structure of B. halodurans FadR
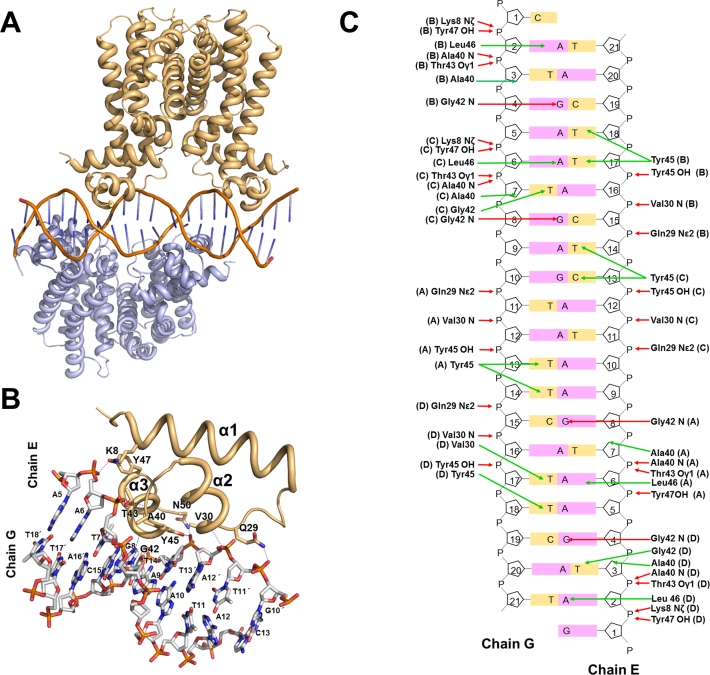
The cognate DNA sequence for B. subtilis FadR was determined by gel retardation and footprinting assays (21) and was detected near the fadR promoter region. We identified the cognate DNA of B. halodurans FadR from the corresponding fadR promoter region. The structure of the B. halodurans FadR–DNA complex was solved by molecular replacement using the native FadR dimer as a search model and refined to 2.8 Å resolution. The overall structure of the B. halodurans FadR–DNA complex is shown in Figure 3. The crystallographic data and refinement statistics are listed in (Table 1). The asymmetric unit contained two dimeric FadRs and a 21 bp fadR promoter DNA. There were no direct contacts between dimers, and each dimer bound consecutively at two inverted repeat (IR) sequences of the fadR promoter DNA (Figure 3). Overhang bases formed G-C pairs at the end of the DNA duplexes resulting in a continuous double-helical DNA filament through crystallographic symmetry. The 21 bp fadR promoter DNA substrate was pseudo-palindromic (5΄-GATGAATGAAT*ACTCATTCAT-3΄, where the asterisk represents the dyad axis). There were two consecutive 16 bp pseudo IR sequences (IR1: 5΄-GATGAATGAATACTCATTCAT-3΄ and IR2: 5΄-GATGAATGAATACTCATTCAT-3΄) separated by 4 bp in the DNA substrate. Each dimer bound to the 16 bp IR with an intervening 4 bp. The two consecutive IR sequences allowed B. halodurans FadR to bind the promoter DNA as a dimer of dimers. The two dimeric structures in the DNA complex were almost identical with an RMSD of 0.24 Å over 345 pairs of Cα atoms.
Figure 3.
Crystal structure of Bacillus halodurans FadR in complex with fadR promoter. (A) Cartoon representation of the FadR:DNA complex formed by a dimer of dimers (gold and teal) and a DNA molecule (orange). (B) Close-up view of the B. halodurans FadR–DNA interface within the monomer. The hydrogen bond interactions are drawn as blue dash lines. The DNA bases are listed, and prime indicates complementary strand of chain E. The figure has slightly different orientations from figure (A) in order to show the detailed interactions better. (C) Detailed schematic representation of FadR–DNA interactions. Residues involved in DNA interactions are shown, and each subunit is marked in parenthesis. Red and green arrows indicate hydrogen bonds and hydrophobic interactions, respectively.
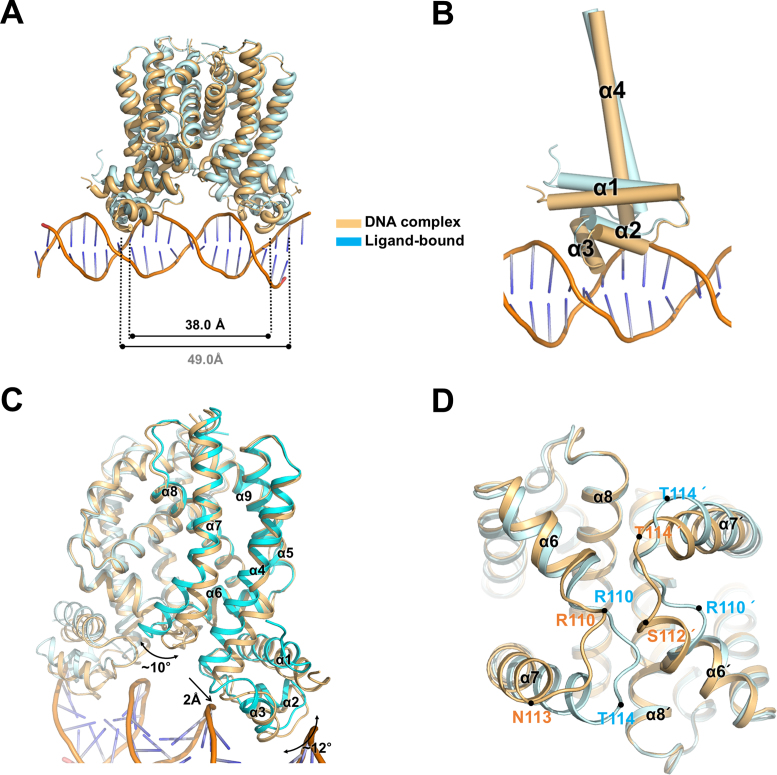
The Cα superposition between the ligand-bound and DNA complex structures indicated several conformational changes of FadR after DNA binding with an RMSD of 1.24 Å. The most noticeable structural change was movement of the N-DBD toward DNA. When B. halodurans FadR bound to DNA, the HTH motif (helices α2 and α3) in the N-DBD rotated toward the DNA major groove (Figure 4). However, the DNA showed a slight bend of ∼3.5° toward the N-DBD, which was similar to B-form DNA. The bound DNA had average roll and twist angles of 0.84° and 34.05°, while typical B-form DNA had 0.6° and 36°, respectively. The recognition helix α3 widened the major groove to ∼12.1 Å, facilitating DNA binding to the N-DBD HTH motif, while the ideal B-form DNA was 11.2 Å (Supplementary Table S1). Because each monomer interacted with the 6 bp repeated DNA sequences (5΄-ATGA(A/G)T-3΄) in the IR, the residues involved in the DNA interactions were almost identical. Most residues interacted with the DNA phosphate backbone atoms, except Gly42 and Tyr45. Gly42 made hydrophobic contacts with DNA bases and formed a hydrogen bond with N7 of the third Gua in the 6 bp repeated DNA sequence. Tyr45 also formed hydrophobic interactions with bases of the second and third Thy in the complementary strand and the hydroxyl group was hydrogen-bonded with the phosphate group of the second Thy. The Gln29, Val30, Ala40, Thr43 and Tyr47 residues interacted with the phosphate backbone (Figure 3 and Supplementary Table S2).
Figure 4.
Conformational changes of Bacillus halodurans FadR upon DNA binding. (A) Superimposition between the ligand-bound and DNA-bound FadR structures. The Cα–Cα distance (at Gly42) of recognition helices is 38.0 Å in DNA-bound, whereas the same distance in ligand-bound structure is 49.0 Å. The ligand-bound FadR is shown in cyan and the DNA-bound in orange. (B) N-DBD is bent toward the major groove of DNA upon DNA binding. Each cylinder represents α-helix and only visualizes helices α1–α4. (C) Superposition of the ligand-bound FadR and FadR–DNA complex. Conformational changes are indicated by arrows. (D) Comparison of the flexible loop regions. Flexible loops (Arg110–Asn113) in FadR–DNA complex and ligand-bound structures are indicated.
Conformational changes on DNA binding
To investigate the conformational changes upon DNA binding, we compared the structures of ligand-bound and the DNA-bound FadR based on the dimerization motif (α8–α9). Although the dimerization motif was superimposed well with an RMSD of 0.43 Å for 76 Cα atoms, several structural differences were observed. First, the N-DBDs rotated toward the DNA major groove by 12–14°. The distance between the recognition helices α3 (at residue Gly42) in the DNA-bound FadR was 38.0 Å, which was smaller than the 49.0 Å in the ligand-bound form (Figure 4A). Second, the α4 and α7 helices, which are involved in the ligand-binding cavity, were displaced. The helix α4 was shifted ∼2 Å toward the N-DBD and distorted at the center of Gly64 (Figure 4C). The movement of helix α4 decreased the size of the ligand-binding cavity, which was contributed by the combination of Phe59, Met63 and Phe66 with Gly64. The helix α7 was kinked 10° at the center of Tyr127 (Figure 4C). Third, the flexible loop region between Arg110 and Asn113 showed a significant conformational change. Residues Arg110 and Asn113 were components of the helix α6 in the FadR–DNA complex structure, while the same residues were present as a flexible loop in the ligand-bound structure, which showed that the terminal part of helix α6 was unwound upon ligand binding (Figure 4D).
When FadR bound to the operator DNA, the α4 helix was displaced toward the N-DND and the α7 helix was kinked at the center of Tyr127. These conformational changes induced the N-DBDs to rotate toward the DNA and caused the recognition helix α3 in the HTH motif to interact with the major groove in the DNA. In contrast, fatty acyl-CoA binding to FadR caused the ligand-binding cavity to be larger due to movement of the helices α4 and α7, thereby dissociating FadR from the fadR operator.
DNA binding mode of B. halodurans FadR
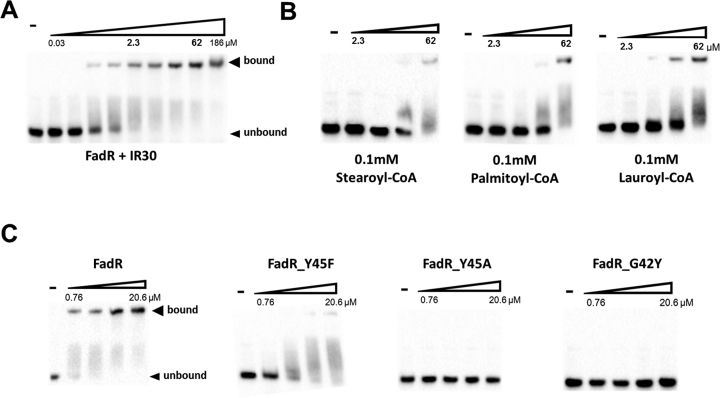
Previous studies of FadR have shown that FadR is released from the promoter by binding fatty acyl-CoA (1). Long-chain acyl-CoAs (C14–C20) detach quickly B. subtilis FadR from the operator DNA, whereas it is slowly released by lauroyl-CoA. In addition, in vitro experiments showed that unsaturated long-chain acyl-CoAs (such as palmitoleoyl(16:1)-CoA and oeloyl(18:1)-CoA) decrease DNA binding affinity more efficiently than the corresponding saturated acyl-CoAs (palmitoyl(16:0)-CoA and stearoyl(18:0)-CoA) (23). However, T. thermophilus FadR susceptibly binds shorter fatty acyl-CoAs compared with that of B. subtilis FadR (23). To identify the binding affinity of B. halodurans FadR to its operator, we performed an EMSA using 30 bp oligonucleotides containing the fadR promoter site. The EMSA was performed by mixing various amounts of B. halodurans FadR proteins with fadR promoter (IR30) either in the absence or presence of fatty acyl-CoAs. B. halodurans FadR bound to DNA tightly with a dissociation constant (Kd) of ∼100 nM in the absence of fatty acyl-CoA (Figure 5A). The DNA-binding ability of B. halodurans FadR decreased in the presence of fatty acyl-CoAs. B. halodurans FadR severely reduced DNA binding in the presence of long-chain fatty acyl-CoAs, such as stearoyl-CoAs (18:0) and palmitoyl-CoAs (16:0), yet was moderately released from the DNA by lauroyl-CoAs (12:0) (Figure 5B). In addition, palmitic acid, which was fortuitously bound in native FadR, did not affect the DNA binding (Supplementary Figure S4A). These results were consistent with homologous B. subtilis FadR (20).
Figure 5.
The EMSA result of the Bacillus halodurans FadR–fadR promoter. The DNA probe that corresponds to the promoter of B. halodurans fadR was biotinylated and mixed at 5 nM with various concentrations of the FadR proteins. FadR and its mutant proteins were diluted 3-fold in a stepwise manner. The experiments were repeated three times, and representative results are shown. The bands for the fadR promoter and FadR–moter complex (bound) indicated by arrowhead. (A) The results of native B. halodurans FadR–fadR promoter complex. (B) The results of native FadR–DNA in the presence of 0.1 mM of stearoyl-CoA, palmitoyl-CoA and lauroyl-CoA. (C) EMSA results of three FadR mutants (Y45F, Y45A and G42Y) with fadR promoter DNA.
Sequence alignment and structural analyses indicated that Gly42 and Tyr45 are highly conserved in the HTH motif of the TetR family proteins and formed multiple contacts with DNA (Supplementary Figure S5A). To probe the role of the residues, several mutations were introduced into B. halodurans FadR. G42Y (G42A was not expressed), Y45F and Y45A in the recognition helix α3 were constructed to directly perturb the FadR–DNA interaction. The mutant proteins were purified by the same method as the native FadR and subjected to EMSA experiments. Y45F showed moderate reduction of DNA binding. This is not surprising as Y45F loses a hydrogen bonding without perturbing the hydrophobic interactions with bases of the second and third Thy in the complementary strand. However, Y45A and G42Y showed significantly reduced DNA binding. The replacement of Gly42 and Y45 likely disrupted the hydrophobic interactions with DNA bases, as DNA binding was barely detectible (Figure 5C and Supplementary Table S3).
To further analyse the FadR–DNA (IR, 5΄-ATGA(A/G)T-3΄) interactions, we designed modified DNA substrates (IRm1, IRm2 and IRm3). Each DNA contained the fadR promoter sequence with 4 bp mutations in upper, middle and downstream region (Supplementary Figure S4). IRm1 would be defective in hydrophobic interactions between Tyr45 (chain D) and the third Thy in the complementary strand. IRm2 lost hydrophobic interactions between Tyr45 (chains A and C) and the second Thy. IRm3 was defective in hydrogen bonding to Gly42 (chain B) and hydrophobic interactions between Tyr45 (chain B) and third Thy. The EMSA results using modified IR substrates showed that loss of interactions between the DNA base and protein residue resulted in severe reduction of DNA binding (Supplementary Figure S4 and Supplementary Table S4). These results indicate that disrupting only a few interactions, particularly hydrophobic interactions to DNA bases, could strongly affect DNA binding ability of B. halodurans FadR.
The TetR superfamily of transcriptional regulators can be divided into two sub-classes depending on the oligomeric state upon DNA binding. One binds to DNA as a dimer and the other binds to DNA cooperatively as a dimer of dimers. Several differences exist between these two sub-classes of TetR family proteins. An analysis showed that dimeric DNA binding proteins recognize relatively short DNA (15–17bp), whereas the other binds to longer DNA (22–32bp). In addition, the dimeric TetR family proteins showed the DNA kink and induced a severe bent (15–17°), which widened the major groove. On the other hand, dimer of dimers TetR family proteins slightly widened the major groove of DNA and bent its DNA site by only 3°–3.5° (almost identical to B-form DNA). These differences are reflected to the Cα–Cα distances (at residue Gly42) in each subunit between the two sub-classes of TetR family proteins. In dimer of dimers TetR family proteins, this distance was ∼38 Å compared with ∼32 Å for dimeric TetR family proteins (Supplementary Table S5). B. halodurans FadR shared all characteristics of a dimer of the dimeric TetR superfamily proteins.
CONCLUSION
The structural mechanism of transcriptional regulation in the TetR superfamily has been elucidated (18,24,30,38). The N-DBD was translocated to the DNA major groove when the TetR transcriptional regulator binds to its operator, demonstrating a pendulum-like movement along the helix α4, which facilitated binding of the recognition helix deep into the DNA major groove. In B. halodurans FadR, hydrophobic and hydrophilic interactions with DNA bases and phosphate backbones were identified via two conserved residues (G42 and Y45). The pendulum-like rearrangement of the N-DBD decreased the distance between dimer recognition helices and resulted in a favorable state toward accepting operators by widening the major groove of DNA. When fatty acyl-CoAs bound to B. halodurans FadR, reorganization of the ligand-binding cavity lead to translocation of the helices α4 and α7, resulting in concurrent movement of the N-DBD to dissociate from DNA.
ACCESSION NUMBERS
The crystallography, atomic coordinates and structure factors reported in this paper have been deposited in the Protein Data Bank, www.pdb.org [PDB IDs: 5GP9 (B. halodurans FadR, palmitic acid bound), 5GPA (B. halodurans FadR(R117A), ligand-free) and 5GPC (B. halodurans FadR, in complex with DNA )].
Supplementary Material
ACKNOWLEDGEMENTS
We thank the staff members at Pohang Accelerator Laboratory Beamlines 7A and 5C for their help with data collection. We also thank I. Song and H. Oh on the GC-MS experiments.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Research Foundation of Korea [NRF-2014-R1A1A1A05008017]; Agriculture Research Center program of the Ministry for Food, Agriculture, Forestry and Fisheries, Korea [ARC-710003-03]. Funding for open access charge: National Research Foundation of Korea [NRF-2014-R1A1A1A05008017].
Conflict of interest statement. None declared.
REFERENCES
- 1. Fujita Y., Matsuoka H., Hirooka K.. Regulation of fatty acid metabolism in bacteria. Mol. Microbiol. 2007; 66:829–839. [DOI] [PubMed] [Google Scholar]
- 2. DiRusso C.C., Nystrom T.. The fats of Escherichia coli during infancy and old age: regulation by global regulators, alarmones and lipid intermediates. Mol. Microbiol. 1998; 27:1–8. [DOI] [PubMed] [Google Scholar]
- 3. Cronan J.E. Jr, Subrahmanyam S.. FadR, transcriptional co-ordination of metabolic expediency. Mol. Microbiol. 1998; 29:937–943. [DOI] [PubMed] [Google Scholar]
- 4. Fujita Y., Fujita T., Miwa Y., Nihashi J., Aratani Y.. Organization and transcription of the gluconate operon, gnt, of Bacillus subtilis. J. Biol. Chem. 1986; 261:13744–13753. [PubMed] [Google Scholar]
- 5. Magnuson K., Jackowski S., Rock C.O., Cronan J.E. Jr. Regulation of fatty acid biosynthesis in Escherichia coli. Microbiol. Rev. 1993; 57:522–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Black P.N., DiRusso C.C., Metzger A.K., Heimert T.L.. Cloning, sequencing, and expression of the fadD gene of Escherichia coli encoding acyl coenzyme A synthetase. J. Biol. Chem. 1992; 267:25513–25520. [PubMed] [Google Scholar]
- 7. Black P.N. Primary sequence of the Escherichia coli fadL gene encoding an outer membrane protein required for long-chain fatty acid transport. J. Bacteriol. 1991; 173:435–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nystrom T., Neidhardt F.C.. Expression and role of the universal stress protein, UspA, of Escherichia coli during growth arrest. Mol. Microbiol. 1994; 11:537–544. [DOI] [PubMed] [Google Scholar]
- 9. Campbell J.W., Morgan-Kiss R.M., Cronan J.E. Jr. A new Escherichia coli metabolic competency: growth on fatty acids by a novel anaerobic beta-oxidation pathway. Mol. Microbiol. 2003; 47:793–805. [DOI] [PubMed] [Google Scholar]
- 10. Schujman G.E., de Mendoza D.. Transcriptional control of membrane lipid synthesis in bacteria. Curr. Opin. Microbiol. 2005; 8:149–153. [DOI] [PubMed] [Google Scholar]
- 11. Lu Y.J., Zhang Y.M., Rock C.O.. Product diversity and regulation of type II fatty acid synthases. Biochem. Cell. Biol. 2004; 82:145–155. [DOI] [PubMed] [Google Scholar]
- 12. Maloy S.R., Nunn W.D.. Role of gene fadR in Escherichia coli acetate metabolism. J. Bacteriol. 1981; 148:83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gui L., Sunnarborg A., LaPorte D.C.. Regulated expression of a repressor protein: FadR activates iclR. J. Bacteriol. 1996; 178:4704–4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. van Aalten D.M., DiRusso C.C., Knudsen J.. The structural basis of acyl coenzyme A-dependent regulation of the transcription factor FadR. EMBO J. 2001; 20:2041–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. van Aalten D.M., DiRusso C.C., Knudsen J., Wierenga R.K.. Crystal structure of FadR, a fatty acid-responsive transcription factor with a novel acyl coenzyme A-binding fold. EMBO J. 2000; 19:5167–5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xu Y., Heath R.J., Li Z., Rock C.O., White S.W.. The FadR.DNA complex. Transcriptional control of fatty acid metabolism in Escherichia coli. J. Biol. Chem. 2001; 276:17373–17379. [DOI] [PubMed] [Google Scholar]
- 17. Raman N., DiRusso C.C.. Analysis of acyl coenzyme A binding to the transcription factor FadR and identification of amino acid residues in the carboxyl terminus required for ligand binding. J. Biol. Chem. 1995; 270:1092–1097. [DOI] [PubMed] [Google Scholar]
- 18. Schumacher M.A., Miller M.C., Grkovic S., Brown M.H., Skurray R.A., Brennan R.G.. Structural basis for cooperative DNA binding by two dimers of the multidrug-binding protein QacR. EMBO J. 2002; 21:1210–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schujman G.E., Paoletti L., Grossman A.D., de Mendoza D.. FapR, a bacterial transcription factor involved in global regulation of membrane lipid biosynthesis. Dev. Cell. 2003; 4:663–672. [DOI] [PubMed] [Google Scholar]
- 20. Fujihashi M., Nakatani T., Hirooka K., Matsuoka H., Fujita Y., Miki K.. Structural characterization of a ligand-bound form of Bacillus subtilis FadR involved in the regulation of fatty acid degradation. Proteins. 2014; 82:1301–1310. [DOI] [PubMed] [Google Scholar]
- 21. Matsuoka H., Hirooka K., Fujita Y.. Organization and function of the YsiA regulon of Bacillus subtilis involved in fatty acid degradation. J. Biol. Chem. 2007; 282:5180–5194. [DOI] [PubMed] [Google Scholar]
- 22. Badger J., Sauder J.M., Adams J.M., Antonysamy S., Bain K., Bergseid M.G., Buchanan S.G., Buchanan M.D., Batiyenko Y., Christopher J.A. et al. Structural analysis of a set of proteins resulting from a bacterial genomics project. Proteins. 2005; 60:787–796. [DOI] [PubMed] [Google Scholar]
- 23. Agari Y., Agari K., Sakamoto K., Kuramitsu S., Shinkai A.. TetR-family transcriptional repressor Thermus Thermophilus FadR controls fatty acid degradation. Microbiology. 2011; 157:1589–1601. [DOI] [PubMed] [Google Scholar]
- 24. Bhukya H., Bhujbalrao R., Bitra A., Anand R.. Structural and functional basis of transcriptional regulation by TetR family protein CprB from S. coelicolor A3(2). Nucleic Acids Res. 2014; 42:10122–10133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Itou H., Watanabe N., Yao M., Shirakihara Y., Tanaka I.. Crystal structures of the multidrug binding repressor Corynebacterium glutamicum CgmR in complex with inducers and with an operator. J. Mol. Biol. 2010; 403:174–184. [DOI] [PubMed] [Google Scholar]
- 26. Miller D.J., Zhang Y.M., Subramanian C., Rock C.O., White S.W.. Structural basis for the transcriptional regulation of membrane lipid homeostasis. Nat. Struct. Mol. Biol. 2010; 17:971–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yang S., Gao Z., Li T., Yang M., Zhang T., Dong Y., He Z.G.. Structural basis for interaction between Mycobacterium smegmatis Ms6564, a TetR family master regulator, and its target DNA. J. Biol. Chem. 2013; 288:23687–23695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Le T.B., Schumacher M.A., Lawson D.M., Brennan R.G., Buttner M.J.. The crystal structure of the TetR family transcriptional repressor SimR bound to DNA and the role of a flexible N-terminal extension in minor groove binding. Nucleic Acids Res. 2011; 39:9433–9447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tonthat N.K., Milam S.L., Chinnam N., Whitfill T., Margolin W., Schumacher M.A.. SlmA forms a higher-order structure on DNA that inhibits cytokinetic Z-ring formation over the nucleoid. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:10586–10591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Orth P., Schnappinger D., Hillen W., Saenger W., Hinrichs W.. Structural basis of gene regulation by the tetracycline inducible Tet repressor-operator system. Nat. Struct. Biol. 2000; 7:215–219. [DOI] [PubMed] [Google Scholar]
- 31. Park Y.W., Yeo H.K., Lee J.Y.. Crystallization and preliminary X-ray diffraction analysis of a fatty-acid metabolism regulatory protein, FadR, from Bacillus halodurans. Acta Crystallogr. F Struct. Biol. Cryst. Commun. 2012; 68:975–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Minor W., Cymborowski M., Otwinowski Z., Chruszcz M.. HKL-3000: the integration of data reduction and structure solution–from diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr. 2006; 62:859–866. [DOI] [PubMed] [Google Scholar]
- 33. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Murshudov G.N., Vagin A.A., Dodson E.J.. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997; 53:240–255. [DOI] [PubMed] [Google Scholar]
- 36. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen V.B., Arendall W.B. 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C.. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Delmar J.A., Chou T.H., Wright C.C., Licon M.H., Doh J.K., Radhakrishnan A., Kumar N., Lei H.T., Bolla J.R., Rajashankar K.R. et al. Structural basis for the regulation of the MmpL transporters of Mycobacterium tuberculosis. J. Biol. Chem. 2015; 290:28559–28574. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.