Abstract
Transfer RNAs (tRNAs) perform essential tasks for all living cells. They are major components of the ribosomal machinery for protein synthesis and they also serve in non-ribosomal pathways for regulation and signaling metabolism. We describe the development of a genetically encoded fluorescent tRNA fusion with the potential for imaging in live Escherichia coli cells. This tRNA fusion carries a Spinach aptamer that becomes fluorescent upon binding of a cell-permeable and non-toxic fluorophore. We show that, despite having a structural framework significantly larger than any natural tRNA species, this fusion is a viable probe for monitoring tRNA stability in a cellular quality control mechanism that degrades structurally damaged tRNA. Importantly, this fusion is active in E. coli live-cell protein synthesis allowing peptidyl transfer at a rate sufficient to support cell growth, indicating that it is accommodated by translating ribosomes. Imaging analysis shows that this fusion and ribosomes are both excluded from the nucleoid, indicating that the fusion and ribosomes are in the cytosol together possibly engaged in protein synthesis. This fusion methodology has the potential for developing new tools for live-cell imaging of tRNA with the unique advantage of both stoichiometric labeling and broader application to all cells amenable to genetic engineering.
INTRODUCTION
tRNA biology is complex and has unexpected layers (1). While the primary function of a tRNA is protein synthesis on the ribosome, non-ribosomal activities involving roles in phage and viral DNA replication (2,3), nuclear import of proteins (4), pre-mRNA splicing (5) and apoptosis (6–8) are now known. To explore the potential of the diversity and complexity of tRNA biology, it is necessary to have the tools to monitor tRNA location and activity inside living cells, both spatially and temporally. However, current methodologies of live-cell imaging tRNA have limitations. Typically, a specific tRNA is isolated from the yeast bulk, labeled with a fluorophore ex vivo, and introduced by transfection into mammalian cells for analysis (9,10). First, yeast tRNA species, while readily available in relatively large quantities, often differ from their mammalian counterparts at key residues that are critical for activities unique to mammalian cells (11). Second, ex vivo labeling usually exploits specific post-transcriptional modifications (e.g. dihydrouridine and wybutosine) (12–14), which are not present in every tRNA sequence. Third, the transfection efficiency of tRNA varies substantially, depending on cell types, and it is not applicable to many types such as bacterial or yeast cells. Thus, a genetically encoded fluorescent tRNA should hold great promise for imaging its action in live cells, eliminating the need for ex vivo technologies. We show here that the fusion of a large RNA aptamer with one of the largest tRNAs, despite each having a well-defined tertiary structure, enables the tRNA to perform live-cell protein synthesis in Escherichia coli. This tRNA–aptamer fusion can be switched on with quantitative GFP (green fluorescence protein)-like fluorescence, while acting like a canonical tRNA. Most importantly, this tRNA fusion is accepted by the ribosome and supports protein synthesis at a rate sufficient for cell viability. These results suggest the potential for real-time imaging of tRNA in both ribosomal and non-ribosomal activities in live cells. The principles of developing this tRNA fusion are accessible to all cells amendable to genetic engineering.
In this work, we made tRNA fusion with a Spinach aptamer. Recent in vitro selections have isolated RNA aptamers that bind to a range of synthetic GFP-like fluorophores, generating conjugates that light up with diverse colors (15,16). One of these aptamers is known as Spinach, because its conjugate with DFHBI (3,5-difluoro-4-hydroxybenzylidene imidazolinone) emits fluorescence of a spinach color, mimicking the intrinsic chromophore of GFP (15). DFHBI is a cell-permeable and nontoxic ligand and it is selectively activated for fluorescence upon binding to Spinach. Several smaller or brighter derivatives of the original Spinach are now available (17–19), as well as other fluorescent aptamer–ligand complexes (20,21). Such aptamer-mediated fluorescence has been used as a genetically encoded sensor for real-time imaging of small molecules and metabolites in live cells (15,22–26). Further development has enabled imaging of cellular dynamics of endogenous mRNAs (27,28). In each of these examples, the Spinach–DFHBI conjugate was exploited as a sensor. None of these sensors, however, are integrated into tRNA or have the ability to image tRNA. To utilize Spinach for imaging tRNA, however, a major consideration is the large size of the aptamer (∼100 nucleotides) relative to tRNA (70–90 nucleotides) and the self-contained tertiary structure of each. The original Spinach aptamer exhibits an elongated shape of two coaxially stacked helical stems joined by a G-quadruplex (17,29), while the tRNA structure is made up of two helical arms joined by a tertiary core to form the L-shape. It is unknown if the fusion of two unrelated and distinct RNA structures can be accommodated within the complex framework of the ribosome.
Here, we show that the fusion of the original Spinach aptamer (15) with one of the largest tRNA molecules creates a hybrid that is fully active for live-cell protein synthesis. This tRNA fusion (referred hereafter as Spinach tRNA) is specifically charged with the cognate amino acid, stably brought to the ribosome by elongation factor Tu (EF-Tu), and adeptly acting in peptidyl transfer. In live-cell imaging, the fusion is found in the same sub-cellular region as the ribosome, suggesting the role as a component of the protein synthesis machinery. Given the structural complexity and intricacy of the ribosome, and the extensive and dynamic motions of the ribosome–tRNA–mRNA complex (30–33), the ability of the fusion to act as an active component of protein synthesis is unexpected. This finding provides new insight into the previously unrecognized flexibility of both the ribosome and tRNA to accommodate new motifs. Furthermore, we show that the fusion can be made with several E. coli tRNA species, each in a distinct sequence, and that the fusion can act as a probe to monitor the cellular quality control applicable to canonical tRNA. Collectively, our data suggest that fluorescent aptamers in general can be inserted into functional tRNAs for live-cell imaging of both ribosome and non-ribosome activities.
MATERIALS AND METHODS
Design of Spinach tRNA
We used E. coli tRNATyr2 (Ec tRNATyr) as the framework to introduce the Spinach aptamer (Supplementary Tables S1 and S2, Supplementary Figure S1 available online). Ec tRNATyr2 differs from Ec tRNATyr1 by only two nucleotides in the V-loop (C47:2 and A47:3 versus U47:2 and C47:3), which in crystal structure do not interfere with recognition by tyrosyl–tRNA synthetase (TyrRS) (34,35). We refer to tRNATyr2 simply as tRNATyr in this work. While Ec tRNATyr has a standard T-loop, it has a large D- and V-loop, indicating the possibility to accommodate additional motifs in these two regions. Three chimeras of Ec tRNATyr were made, in which the Spinach aptamer was inserted to the D-loop between C16 and G18 (D-Spinach), to the V-loop between C47:2 and A47:3 (V-Spinach), and to the T loop between G57 and A58 (T-Spinach). Of these, only the V-Spinach fusion was stably expressed, indicating acceptance of the new motif by enzymes such as TyrRS (34,35). Indeed, a co-crystal structure of TyrRS–tRNATyr reveals that the large V loop is well accommodated by TyrRS using a C-terminal domain that stabilizes the unique shape of the loop (35). In bacteria, tRNASer and tRNALeu also have a large V-loop, whereas in mammalian cells only tRNASer and tRNALeu have a large V-loop.
Construction of Spinach–tRNA fusions
Spinach fusions were constructed from DNA oligonucleotides (IDT, IA) and amplified in pairs (Supplementary Tables S1 and S2, Supplementary Figure S1). An example is shown for expressing V-Spinach tRNATyr from the E. coli plasmid pKK223-3 (Supplementary Figure S1). Briefly, the complementary 3΄ ends of oligo #3 and oligo #4 (each 4 μM) were hybridized in an annealing buffer (40 mM Tris–HCl, pH 7.5, 50 mM NaCl, 5 mM dithiothreitol (DTT) and 20 mM MgCl2) and extended by Sequenase (0.125 mM each dNTP) overnight at 37°C to generate double-stranded DNA segment-1, while the 3΄-ends of oligo #5 and oligo #6 (each 4 μM) were hybridized and extended by Sequenase to generate double-stranded DNA segment-2. The overnight extension time was to ensure that the reaction was complete, although shorter time was sufficient in most cases. The synthesized two segments, with mutually complementary ends, were mixed and heat-denatured to provide primers for PCR amplification of the joined segment, using the High-Fidelity DNA polymerase Phusion (NEB, MA) (15 cycles of 98°C for 30 sec, 72°C for 1 min, then 30 cycles of 98°C for 30 s, 60°C for 1 min, 72°C for 20 s). The extended joint segment was PCR amplified again with oligo #7 (containing the EcoRI restriction site) and oligo #8 (containing the PstI restriction site) and the product was cloned into the EcoRI and PstI sites of pKK223-3 for expression under the control of the pTac promoter. The gene for V-Spinach tRNALeu/CAG (CAG: the anticodon) was made similarly (Supplementary Figure S3A and B).
The gene for V-Baby-Spinach tRNAAla/UGC, V-Baby-Spinach tRNAPhe/GAA, and V-Baby-Spinach tRNAPro/UGG was each synthesized by GenScript, where the sequence of the Baby Spinach aptamer (17) was inserted into the V-loop of the respective tRNA (Supplementary Figure S3C and D). Each fusion gene was sub-cloned into the plasmid pKK223-3 at EcoRI and PstI sites and expressed in JM109 cells upon induction with 1 mM IPTG.
Northern blot analysis
E. coli CA244 cells expressing a tRNA from the pKK223-3 plasmid were grown to OD600 of 0.5–1.0 in LB with ampicillin. Cells (1.5 ml) were pelleted, resuspended in 0.3 M sodium acetate (pH 5) and extracted with 100 μl of acid-phenol (pH 5.2). The aqueous phase was precipitated with ethanol, washed with 70% (v/v) ethanol, dried, and resuspended in 15 μl TE. A sample of 2 μl was mixed with 1 μl of formamide, heated at 65°C for 5 min and electrophoresed on a denaturing 10% PAGE/7 M urea minigel. The samples were electroblotted onto Hybond N+ Nylon membrane (GE Healthcare Biosciences: Pittsburgh, PA, USA) (36). The membrane was hybridized overnight at 37°C with 75 pmol of a 32P-labeled probe (see below) in a mixture of 0.9 M NaCl, 90 mM Tris–HCl (pH 7.5), 6 mM EDTA, 0.3% SDS, 1% dry milk, and washed in 6× SSC (0.9 M NaCl, 90 mM Na-citrate) at room temperature, then in 3× SSC (0.45 M NaCl, 45 mM Na-citrate) with 0.1% SDS at 40°C. Hybridization signals were visualized by phosphorimaging and quantified using ImageQuant 5.2 software.
Probe to 5S rRNA: 5΄-TTCTGAGTTCGGCATGGGGT-3΄
Probe to the Spinach motif: 5΄-CCCGTCCTTCACCATTTCATTC-3΄
Probe to Ec tRNATyr: 5΄-TGGTGGTGGGGGAAGGATTCGAACCTTC-3΄
Suppression of lacZam
E. coli CA244 harbors an internal amber mutation in lacZ (lacZ125am) (37), which enables analysis of the suppressor activity of tRNA to synthesize the full-length β-galactosidase (β-gal). Cells expressing tRNA from the pKK223-3 plasmid were streaked on M9 agar supplemented with glucose (0.4%), CaCl2 (0.1 mM), tryptophan (40 μg/ml), thiamine (100 μg/ml), 1 mM IPTG, and 40 μg/ml X-gal and incubated at 37°C for over 24 h. Suppression was detected by the blue color on the indicator plate (34,38).
β-gal assay
The amber suppressor gene of Ec tRNATyr or V-Spinach–tRNATyr was constructed in pGFIB at EcoRI and PstI restriction sites. A control plasmid pGFIB-00 harbored no tRNA gene. Each plasmid was introduced to E. coli strain XAC-1, which has the lacZU118Am gene harboring an amber mutation at the 17th codon position for analysis of tRNA amber suppression (39,40). E. coli strain XAC-1 i−p+ containing the natural sequence of lacZ (without an amber mutation) was used as a reference. Each strain was grown at 37°C until OD550 reached 0.4–0.6, and harvested cells were resuspended in Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 50 mM β-mercaptoethanol, pH 7.0) and permeabilized by 0.005% SDS and 10% chloroform. Cell lysates were mixed with the substrate o-nitrophenyl-β-d-galactopyranoside (ONPG, 0.6 mg/ml final, in 60 mM Na2HPO4 and 40 mM NaH2PO4, pH 7.0) and incubated for up to 50 min at 30°C to develop the yellow color of the product (38). The β-gal activity was calculated from OD420 normalized by OD550 and the reaction time.
Suppression of trmDAm (Y19) to restore cell viability
The trmD gene is one of the growth-essential genes in E. coli (41,42), which encodes a tRNA methyl transferase responsible for maintaining the translational reading frame. An E. coli trmD-knockout (trmD-KO) strain was constructed by using an antibiotic marker to disrupt trmD while maintaining cell viability by expression of a plasmid-borne and arabinose (Ara)-controlled human counterpart trm5 in the presence of Ara (42,43). The trmD-KO strain was introduced with a test plasmid, derived from pET-22b, harboring the IPTG-inducible trmDWT or the variant trmDAm (Y19) gene and the lpp-controlled amber suppressor gene of Ec tRNATyr or V-Spinach tRNATyr. Each tRNA gene was isolated from pGFIB (34,39) as a PvuII fragment and was inserted into the EcoRV restriction site of pET-22b under the control of the constitutive lpp promoter. To test cell viability, individual colonies were isolated from an LB plate containing Ara at 37°C, then inoculated into fresh LB media (20–50-fold dilution) without Ara and grown overnight (at least 6 h) to deplete pre-existing human Trm5 protein. Cells were then spotted by serial dilution (from 10−1 to 10−4 fold) on LB plates with or without Ara (1% w/v) but containing IPTG (0.4 mM) to induce expression of the reporter trmD gene. Cell viability was evaluated after incubation at 37°C overnight.
Live imaging of E. coli cells expressing V-Spinach tRNA
V-Spinach tRNA was expressed from pKK223-3 in one of three E. coli strains: CA244 (Hfr (PO1) lacZ56 trpA49 relA1 spoT1) (37), Top10 (F–mcrA Δ (mrr-hsdRMS-mcrBC) Φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ (ara leu) 7697 galU galK rpsL (StrR) endA1 nupG) (Invitrogen) and JM109 (F΄ traD36 proA+B+ lacIq Δ (lacZ)M15/ Δ (lac-proAB) glnV44 e14− gyrA96 recA1 relA1 endA1 thi hsdR17) (NEB). Expression in CA244 and in Top10 was constitutive, due to the lack the lacIq repressor in these cells, whereas expression in JM109 was IPTG-inducible to switch off the lacIq repressor. Expression in all three showed no visible toxicity.
Prior to imaging analysis, optical glass slides were freshly coated with poly-d-lysine (PDL) for 1 h at room temperature and washed twice with M9 minimal media. An overnight culture of E. coli cells expressing a Spinach tRNA was grown in LB medium at 37°C supplemented with ampicillin. The overnight culture was inoculated into 3 ml fresh media (by a 100-fold dilution) and grown for 3–4 h at 37°C to OD600 of 0.3-0.4. The CA244 cells were harvested, while the JM109 cells were split into two tubes and 1 mM IPTG was added to one. At each point after induction, the culture of each tube (1.5 ml) was harvested. Cells were resuspended in 150 μl of M9 media, washed twice in M9, and plated on the PDL-treated glass slides for 20 min at 37°C. After removal of free cells by washes with M9, the plates were incubated with 200 μM DFHBI in M9 for 5 min at 37°C. This was followed by additional washes of cells in M9 to remove excess DFHBI. Live fluorescence images were obtained with a PTM camera through a 63× oil objective lens mounted on a Carl Zeiss LSM (Laser Scanning Microscopy) 510 META confocal microscope and analyzed with Metamorph software. An argon ion 488 nm laser was used for excitation and a 505–550 nm band pass filter (Carl Zeiss) was used to select the fluorescent signal. For time course analysis of fluorescence intensity, images were acquired every 30 min with 1024 × 1024 pixel resolution. The intensity of individual E. coli cells at each time point was calculated by Metamorph software and was shown as the average (n = 100 each). Images of an experiment versus its control were taken at the same excitation power, camera acquisition time and LUTs set to the same value for all pictures taken on the same channel and presented on the same figure.
To examine the physical distribution of V-Spinach tRNATyr with ribosomes, a P1 lysate was prepared from QC101 carrying the L9-mCherry fusion (44) and this lysate was used to transduce JM109 cells by selection of the kanamycin marker. The transduced cells were transformed with pKK223-3 expressing V-Spinach tRNATyr/GUA or Spinach aptamer. Overnight cultures were diluted 100-fold in LB medium supplemented with 100 μg/ml ampicillin and 50 μg/ml kanamycin and cultured at 37°C with shaking. After 1 h, 1 mM IPTG was added and growth was continued for another 2 h. Then, the cells were harvested and re-suspended in M9 medium supplemented with 100 μg/ml DFHBI reagent. After incubation in this medium for 10–20 min, cells were washed with PBS and transferred onto a 1% agarose pad (prepared in M9 media supplemented with appropriate antibiotics and IPTG) placed on glass slides. The cells were observed under a Carl Zeiss Axioplan 2 Imaging microscope equipped with a Carl Zeiss Axio-Cam CCD camera. Filters used for DFHBI and mCherry fluorescence were HQ480/40 (ex. 480/40 nm and em. 535/50) and ET-Texas Red (ex. 550 and em. 630 nm), respectively. Images were analyzed using the program Axiovision 4.7.
Total tRNA extraction
E. coli Top10 cells expressing V-Spinach tRNATyr (with the natural 5΄-GUA-3΄ anticodon) from pKK223-3 were grown overnight at 37°C in 2 l of 2 × YT, while JM109 cells expressing the wild-type tRNATyr from pKK223-3 were grown overnight in 1 × LB. During this incubation, V-Spinach tRNATyr was constitutively expressed whereas tRNATyr was expressed upon addition of 1 mM IPTG. Stationary cultures were harvested and 20 g of cell pellet were phenol extracted by vortexing at room temperature for ∼30 min with 40 ml of 50 mM sodium acetate (NaOAc, pH 5.3), 10 mM Mg (OAc)2, and 34.4 ml of liquified phenol (88%). After centrifugation the aqueous phase was collected and the phenol layer was extracted with an equal volume of 50 mM NaOAc, 10 mM Mg (OAc)2 for an additional 30 min. Total nucleic acid was pelleted from the combined aqueous phases by ethanol precipitation (addition of 1/10 volume of 20% KOAc and an equal volume of ethanol, 30 min at 25 °C) and centrifugation. DNA and low molecular weight RNAs were solubilized by vortexing the pellet with two portions of ice cold 1 M NaCl. After centrifugation the clarified supernatants were combined and two volumes of ethanol were added to precipitate the remaining nucleic acid. The resulting pellet was dissolved in 0.3 M NaOAc and DNA was selectively precipitated with 0.54 volumes of isopropanol and incubation for 10 min at room temperature, followed by centrifugation. The supernatant was reserved and the pellet was treated the same way as before. After combining the two supernatants, 0.24 volumes of isopropanol were added to precipitate total tRNA.
Total tRNA was deacylated in alkaline (0.1 M Tris–HCl, pH 9.0) for 3 h at 37°C. Each plasmid-borne tRNA was purified using a biotinylated oligo as described (11,45). The oligo for V-Spinach tRNATyr (5΄ TGG ACC CGT CCT TCA CCA TTT CAT TCA GTC GCG TC–Biotin TEG 3΄) was complementary to a major loop in the Spinach domain while the oligo for the native tRNATyr (5΄ TTT ACA GTC TGC TCC CTT TGG CCG CTC–Biotin TEG 3΄) was complementary to the sequence between the anticodon and D loops. Hybridization was in 20 mM Tris–HCl (pH 7.5), 1.8 M NaCl, 0.2 mM EDTA. The quality of purified tRNAs was checked by 12% PAGE/7M urea gel. Alternatively, V-Spinach tRNATyr was directly purified to homogeneity using a preparative 12% PAGE/7M urea gel, due to its larger size relative to tRNAs. Ec tRNATyr as purified contained a mixture of tRNATyr1 and tRNATyr2, because the oligonucleotide for affinity pull-down did not distinguish between the two. However, we estimated that the amount of Ec tRNATyr2 was >80% in the mixture (11), due to its over-expression from the pKK223-3 plasmid.
Aminoacylation of tRNA
Aminoacylation of tRNATyr and V-Spinach tRNATyr purified from cells was performed in a 30 μl reaction containing 20 mM KCl, 10 mM MgCl2, 10 mM DTT, 2 mM ATP, 50 mM HEPES, pH 7.5, and 20 μM 3H-Tyr. The tRNA concentration was corrected based on the plateau level of aminoacylation, while the enzyme concentration was corrected based on active fraction obtained from burst kinetics (46). Steady-state kinetics of aminoacylation was performed with 2 nM purified E. coli TyrRS for the wild-type tRNATyr and 5 nM enzyme for V-Spinach tRNATyr. Reactions were sampled over time and quenched on filter pads in 5% ice-cold trichloroacetic acid (TCA) (47). After extensive washes, radioactivity on air-dried filter pads was measured in a Beckman 6000S scintillation counter. Data of aminoacylation as a function of tRNA concentration were fit to the Michaelis-Menten equation. Values of Km (tRNA), kcat and kcat/Km were determined from the average of three independent experiments and were consistent with those of other Ec tRNAs (11,34,48). Errors are SDs.
Equilibrium binding of tRNA to EF-Tu-GTP
The 3΄-terminal A76 in tRNATyr (2 μM) was labeled with α-32P-ATP (0.4 μM, 6000 Ci/mmol) by Bacillus stearothermophilus CCA-adding enzyme (10 μM) (49,50) in 20 μl of 10 mM MgCl2, 1 mM DTT, and 50 mM glycine pH 9.0 (51,52). After dilution to 50 μl with TE, the reaction was extracted with phenol–chloroform–isoamyl alcohol, centrifuged through a cartridge, and then ethanol precipitated in the presence of 2 μg glycogen. Prior to charging, the labeled tRNATyr was heated at 80–85°C for 3 min in 20 μl of water, rapidly mixed with 4 μl of a 10 × AA buffer (100 mM MgCl2, 200 mM KCl, 100 mM DTT, 200 mM Tris pH 7.5), and kept at 37°C for 10–15 min. Aminoacylation (10 min at 37°C in 40 μl) was carried out in the presence of 200 μM tyrosine, 6.2 mM ATP, 0.2 mg/ml BSA and 10 μM TyrRS. After quenching with 5 μl 2.5 M NaOAc pH 5.0, the reaction was extracted with pH 5.2 phenol–chloroform–isoamyl alcohol, ethanol precipitated, dissolved in 50 μl 25 mM NaOAc pH 5.0 and stored at –20°C. The efficiency of aminoacylation was determined to be ∼44% and it was not affected by the V-Spinach fusion.
Dissociation rate constants of the ternary complex between Ec EF-Tu, Tyr-tRNATyr/GUA, and GTP were determined using a ribonuclease protection assay (53,54). EF-Tu (75 pmol) was first activated with 1.5 nmol of GTP by incubation 3 h at 37°C in 75 μl of 20 mM MgCl2, 5 mM DTT, 0.5 M NH4Cl, 1.2 mM phosphoenol pyruvate, 0.02 units/μl pyruvate kinase and 50 mM HEPES pH 7.0. The activated EF-Tu was mixed with 3 pmol of radiolabeled Tyr-tRNATyr/UGA and incubated on ice for at least 20 min to form the ternary complex. Time courses of dissociation were carried out in the presence of 0.1 mg/ml RNase A also on ice. Reaction aliquots (5 μl) were quenched in 50 μl of cold 10% TCA that contained 0.1 mg/ml bulk tRNA. Aliquots of 40 μl each were spotted on separate Whatman filter paper pads for bulk washing in cold 10% TCA three times, ethanol twice, ether once, and dried. Cerenkov counts attributable to intact 32P-labeled Tyr-tRNATyr were measured in a scintillation counter. The rate constant (koff) for dissociation of Tyr-tRNATyr from EF-Tu was determined by exponential fitting of the data. Kd values can be calculated by using a value of 1.1 × 10−4 nM−1 s−1 for the on-rate kon binding (55,56).
Programmed ribosome protein synthesis
The mRNA for translation was prepared by in vitro transcription from a synthetic double-stranded DNA template using T7 RNA polymerase (57), followed by gel purification. The sequence of the template mRNA, encoding the peptide fMYPRSKH6, is shown below, where the Shine-Dalgarno sequence is underlined and the initiation codon is in bold face.
5΄-GGGAAGGAGGUAAAAAUGUAUCCCCGUUCUAAGCACCACCACCACCACCAC-3΄
Native Ec tRNAfMet/CAU and tRNATyr/GUA were over-expressed in E. coli and purified using biotinylated oligoncleotides (11,42,45). Over-expressed V-Spinach tRNATyr/GUA was directly purified by denaturing 12% PAGE. Each tRNA was enzymatically charged with its cognate amino acid and then stored in 25 mM acetate buffer (pH 5.0) at −20°C until use. Formylation of Met-tRNAfMet/CAU was carried out during the charging reaction by including methionyl–tRNA formyltransferase with the methyl donor 10-formyltetrahydrofolate (derived from folinic acid at neutral pH) (11). The efficiency of charging was determined by doping each reaction with radiolabeled amino acid and determining both A260 and radioactive counts of product tRNA after removal of free amino acid, GTP, and protein. Tight-coupled 70S ribosomes were isolated from E. coli MRE600 cells and over-expressed His-tagged initiation and elongation factors were purified from affinity resin (11,42,58–60). These reagents were aliquoted prior to storage at −70°C.
EF-Tu-dependent hydrolysis of GTP
Reactions were carried out in 70 mM NH4Cl, 30 mM KCl, 3.5 mM MgCl2, 1 mM DTT, 0.5 mM spermidine and 50 mM Tris–HCl pH 7.5 (Buffer A) (11,61,62). A ternary complex was formed in two steps, in which EF-Tu was first incubated with limiting γ-32P-GTP (6000 Ci/mmol) for 15 min at 37°C and then with excess Tyr-tRNATyr/GUA for 15 min on ice. Free GTP was removed from the ternary complex by centrifugation through a spin cartridge (Centrispin-20; Princeton Separations). The flow-through was diluted with Buffer A to give a 2× stock of ternary complex that contained approximately 1.5 μM EF-Tu, 1.0 μM Tyr-tRNATyr and 16 nM γ-32P-GTP. In parallel, a 2× stock of 70S initiation complex was formed by incubating 0.75 μM 70S ribosome with 1.0 μM each IF1, IF2, IF3, fMet-tRNAfMet and mRNA in Buffer A supplemented with 1 mM cold GTP for 25 min at 37 °C. The 2× solutions were stored on ice until same-day use. Rapid mixing of the ternary complex and the ribosome solution was performed on a Kintek quench flow apparatus at 20°C. Aliquots of GTP hydrolysis were quenched with 40% formic acid and GTP was separated from Pi by thin layer chromatography (TLC) on PEI-cellulose in 0.5 M NaH2PO4 pH 3.5. Spots were visualized by phosphorimaging and quantified using ImageQuant 5.2.
First peptide bond formation
The first peptide bond formation was monitored for synthesis of the dipeptide fMY, catalyzed by 0.25 μM of the ribosome 70S initiation complex, which contained fMet-tRNAfMet at the P-site and Tyr-tRNATyr (or V-Spinach tRNATyr) at the A-site (62,63). A 70S initiation complex was formed by incubating 0.75 μM 70S ribosome with 0.5 μM 35S-fMet-tRNAfMet and 1.0 μM each IF1, IF2, IF3 and mRNA in Buffer A with 1 mM GTP for 25 min at 37°C. A ternary complex was formed by incubating 1.5 μM EF-Tu with 1 mM GTP in Buffer A for 15 min at 37 °C after which Tyr-tRNATyr/GUA was added to 1.0 μM and incubation continued for 15 min on ice. The 2× stocks were rapidly mixed in a Kintek quench flow apparatus at 20°C. Reaction aliquots were quenched in 0.8 M KOH. After 1–2 h at 37 °C to separate the peptide from the peptidyl-tRNA linkage, 35S-fMet was resolved from 35S-fMet-Tyr by electrophoresis on a cellulose TLC plastic sheet in PYRAC (3.48 M HOAc/62 mM pyridine; pH 2.7). Bands were visualized by phosphorimaging and quantified using ImageQuant 5.2.
RESULTS
Live-cell stability and fluorescence of Spinach–tRNA fusions
To explore the capacity of making fusions that are stable in a live cell, we began with the original Spinach, which was directly isolated from in vitro selections and was one of the largest aptamers for GFP-like fluorescence (15). We chose E. coli tRNATyr as the engineering framework, which has a naturally large D- and V-loop relative to the average tRNA, indicating the potential of these regions to accommodate a new structural motif. The total length of the fusion combining the Spinach and E. coli tRNATyr was 183 nucleotides (Supplementary Tables S1 and S2), exceeding the length of naturally existing tRNA. We expressed the fusion from a multi-copy plasmid and used Northern analysis to determine its stability in E. coli cells. While the Spinach fusion with the D- or T-loop of the tRNA was not detectable, the fusion with the V-loop was readily detected in total RNA isolated from cells (Supplementary Figures S2 and S4). The stable expression of the V-Spinach tRNA in E. coli indicates that the V-loop is a suitable site for accepting Spinach without compromising tRNA structural stability (Figure 1A). The stability of the V-Spinach tRNA was similar in two constructs: the one with the natural anticodon 5΄-GUA and the other with the amber-reading anticodon 5΄-CUA (Supplementary Figure S4).
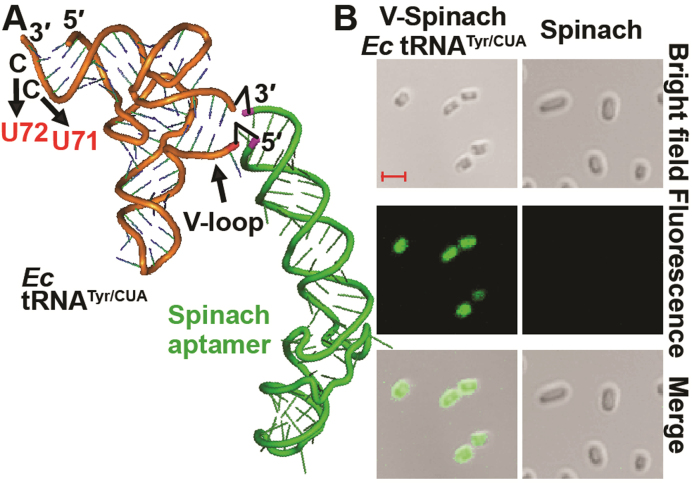
Figure 1.
Expression of a V-Spinach tRNA in E. coli. (A) The Spinach aptamer (in green) is fused to the V-loop of E. coli (Ec) tRNATyr/CUA (in orange), harboring the amber-reading anticodon 5΄-CUA-3΄. The drawing of the Spinach aptamer is based on the crystal structure of the Fab BL3-6-bound aptamer (29). The sequence of the Spinach aptamer is as described (15) and it is covalently joined to the V-loop between C47:2 and A47:3 as shown by two black up arrow icons. The U71 and U72 substitutions in Ec tRNATyr are shown by downward arrows, which replace G1-C72 and G2-C71 base pairs with G1-U72 and G2-U71, respectively. (B) Fluorescence microscope images of E. coli cells expressing the amber suppressor form of V-Spinach tRNATyr versus cells expressing the Spinach stand-alone. DIC images and scale bars (each representing 2 μm) are shown. On average, when examined in liquid cultured media, 96.4 ± 0.1% of cells expressing the V-Spinach tRNA showed the GFP-like fluorescence (n = 100).
The structural stability of the V-Spinach tRNA correlated with its stable emission of GFP-like fluorescence inside E. coli CA244 cells. Upon a brief incubation with DFHBI, 96.4 (±0.1)% of single cells that expressed the fusion showed fluorescence whereas those that expressed the stand-alone aptamer had little fluorescence (Figure 1B). The lack of fluorescence of the stand-alone Spinach was consistent with its intracellular instability in Northern analysis (Supplementary Figure S4). In cells where expression of the V-Spinach tRNA was regulated by a transcriptional repressor, fluorescence of individual cells was dependent on the inducer (Supplementary Figure S5), reaching a stable 60-fold increase over the baseline within 60 min, but a slow and small increase in fluorescence in the absence of IPTG. We attributed the latter to the IPTG-independent basal expression from the Tac promoter that drives tRNA expression from the plasmid. These results validate that the fluorescence observed in E. coli cells expressing V-Spinach tRNATyr was associated with the tRNA, rather than nonspecific effects on Spinach fluorescence.
We showed that live-cell fluorescence imaging was also applicable to the Spinach fusion with the V-loop of tRNALeu, another E. coli species with a large V-loop relative to the average (Supplementary Figure S3A and B). Furthermore, imaging was successful even with E. coli tRNAAla/UGC, tRNAPhe/GAA, and tRNAPro/UGG, each of which has the standard size of five nucleotides in the V-loop (Supplementary Figure S3C and D). In each of the latter cases, a smaller version of the original Spinach, known as the ‘Baby Spinach’, was used. The Baby Spinach retains the primary structure and fluorescence intensity of the original Spinach (17). These results support the notion that the V-loop of tRNA can accommodate aptamers of varying size.
Spinach tRNA as a probe for live-cell tRNA quality control
We determined if V-Spinach tRNATyr served as a probe to monitor the cellular quality control that degrades damaged tRNA. One of these mechanisms targets tRNA with an unstable acceptor end and attaches CCA-CCA sequence to the 3΄ end as the degradation tag (64). Structural instability in tRNA can arise from errors during transcription or post-transcriptional processing or from mutations associated with cancer development (65–67). To determine if the large size of the Spinach fusion can be recognized by this quality control, we generated a variant carrying the destabilizing U71 and U72 substitutions that created two consecutive G-U base pairs at the acceptor end (G1-U72 and G2-U71, Figure 1A and Supplementary Figure S2D). This U71-U72 variant was barely detectable in Northern analysis (Supplementary Figure S4), suggesting that it was subject to rapid degradation. The mechanism of the rapid degradation of the U71-U72 variant is likely due to the dual CCA-CCA addition at the 3΄-end (64). We also generated another variant lacking the single CCA sequence at the 3΄-end and showed that this variant was detectable, albeit with a reduced stability relative to the full-length V-Spinach tRNA (Supplementary Figure S4), indicating that the single CCA sequence can protect the tRNA from degradation.
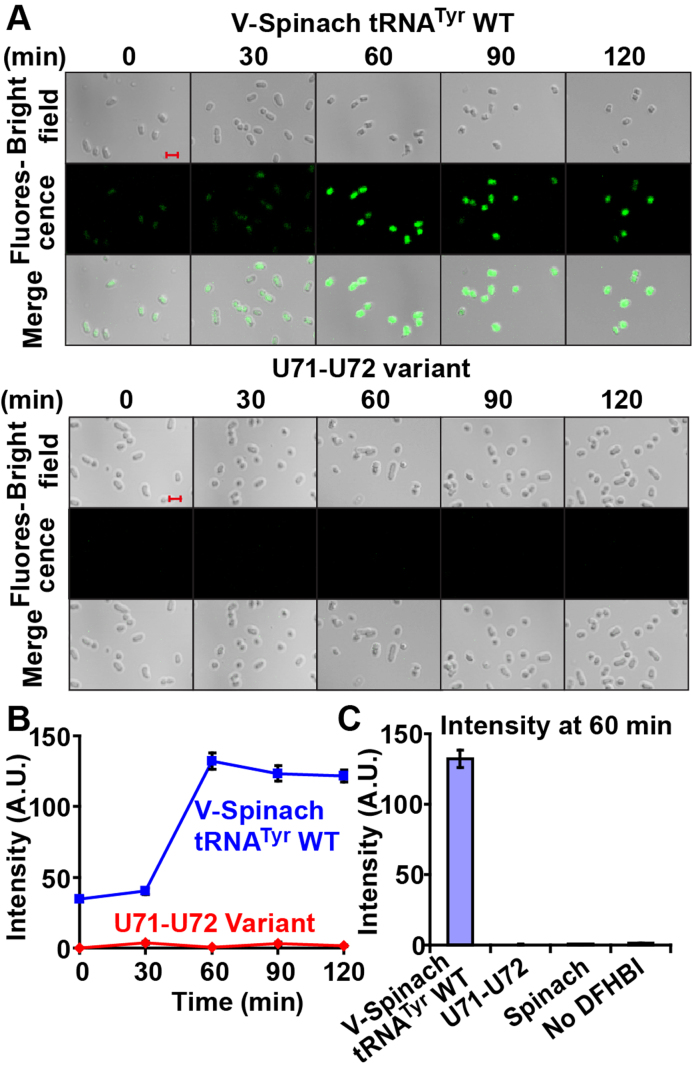
To determine the timescale of degradation, we placed an aliquot of exponentially growing cells on glass slides, incubated them with DFHBI for 5 min, washed off excess ligand, and monitored the DFHBI-dependent fluorescence over time at 22.5°C. Because cells were immobilized on glass slides, the diffusion of bound DFHBI from cells was limited. Imaging tRNA in CA244 cells represented the average level of de novo synthesis and decay. Among the more than 1000 cells examined, we observed a significant fluorescence signal above the background for V-Spinach tRNATyr with a steady increase from time 0 up to 1 hr and a stable fluorescence up to 2 h (Figure 2A). Because no maturation time is required to generate a fluorescent complex upon Spinach–DFHBI interaction (15), the progressive increase in fluorescence in the first hour might indicate the gradual transition of cells on the slide from the exponential phase to an early stationary phase. Nonetheless, the robust fluorescence indicates that de novo synthesis of the tRNA was dominant over decay within this time frame. In contrast, cells expressing the U71-U72 variant of V-Spinach tRNATyr showed no fluorescence, indicating that decay was dominant over de novo synthesis (Figure 2A and B). The lack of fluorescence from the onset was striking, indicating that degradation of the variant was rapid even with active de novo transcription. Quantification of fluorescence at 60 min confirmed high intensity from cells expressing the wild-type fusion, but virtually no intensity from cells expressing the variant, similar to cells expressing the stand-alone Spinach or cells without DFHBI (Figure 2C). Thus, the V-Spinach tRNATyr, upon the acceptor stem damage, is recognized by the appropriate cellular quality control and is degraded, indicating that it is a valid biological probe for live-cell imaging of tRNA stability.
Figure 2.
Live-cell imaging of the amber-suppressor form of V-Spinach tRNATyr /CUA in E. coli. (A) Fluorescence microscope images of E. coli cells expressing the amber suppressor form of V-Spinach tRNATyr, WT (top) or U71-U72 variant (bottom). The Spinach tRNA fusion was constitutively expressed in CA244 cells. Upon entering log-phase, an aliquot of cells was harvested, washed, suspended in M9 minimal medium and immobilized on a poly-(D)-lysine-coated glass slide. Cells were incubated with DFHBI for 5 min, and washed off the excess ligand. The diffusion of bound DFHBI from cells was limited in the immobilized condition and in minimal media. Green fluorescence was excited by 488 nm Argon ion laser and monitored under a microscope over a time course at a constant temperature of 22.5°C. DIC images and scale bars (each representing 2 μm) are shown. Few cell-to-cell variations are seen in the green channel. (B) Quantification of the time courses of fluorescence intensity. Average intensity of the pixels corresponding to the area of a single E. coli cell was quantified by image analysis software (see methods for details). Error bars are expressed as standard errors of mean (SEM), n = 100 for each of the time points. (C) The average fluorescence intensity of the native and the U71-U72 variant of V-Spinach tRNATyr at 60 min compared to negative controls, where only the Spinach stand-alone was expressed (‘Spinach’) or where V-Spinach tRNATyr/CUA was expressed in the absence of DFHBI (‘No DFHBI’). Error bars are expressed as SEM, n = 100 for each of the time points.
Spinach tRNA active in live-cell protein synthesis
We determined whether the exceptionally large size of V-Spinach tRNATyr is active in the protein synthesis machinery. We used E. coli CA244 cells, which harbored an amber mutation (5΄-UAG) in the N-terminal half of β-gal encoded by the lacZ125am locus (68). In these cells, synthesis of the full-length β-gal required suppression of the amber mutation with an amino acid donated by an amber-suppressor tRNA during live-cell protein synthesis. Importantly, despite the large and bulky structure, the fusion with the amber-reading anticodon 5΄-CUA suppressed the lacZ125am locus (Figure 3A), indicating that it was active in protein synthesis. Suppression was observed only when the fusion had the amber-reading anticodon; no suppression was observed in cells expressing Spinach stand-alone or the amber suppressor form of T- or D-Spinach tRNATyr (Figure 3A).
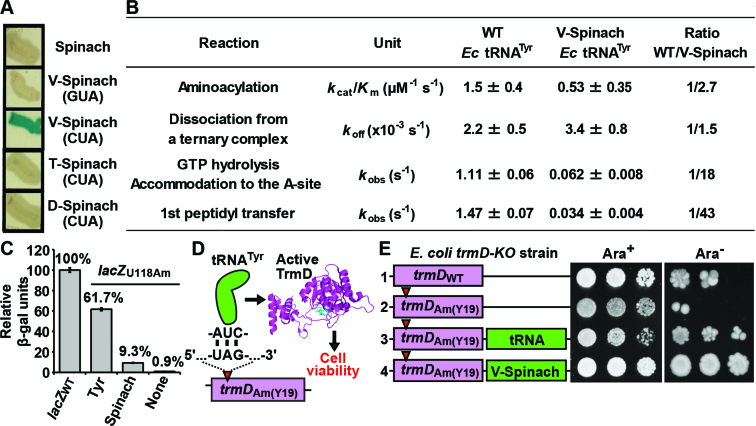
Figure 3.
Activity of V-Spinach tRNA in protein synthesis. (A) Suppression of the lacZ125am locus in E. coli CA244 cells (Yale CGSC stock center) by V-Spinach tRNATyr with the 5΄-CUA anticodon (CUA), but not by the Spinach stand-alone, V-Spinach tRNATyr with the 5΄-GUA anticodon (GUA), or T- or D-Spinach tRNATyr with the CUA anticodon (CUA). E. coli cells expressing each Spinach construct were streaked out on an LB plate with ampicillin. Three to five single colonies with each construct were picked and re-streaked to 1 cm length on a minimal M9 plate with ampicillin and X-gal. This single-colony isolation ensured the homogeneity of each clone of cells. Representative streaks on M9 plates are shown. (B) Activity of the wild-type (WT) Ec tRNATyr and V-Spinach Ec tRNATyr in major steps of protein synthesis, including aminoacylation, stability of the ternary complex, accommodation to the ribosome A-site, and formation of the first peptide bond on an mRNA template with the sequence starting with 5΄-AUG-UAU-3΄. Both V-Spinach tRNATyr and Ec tRNATyr, each containing the natural Tyr anticodon 5΄-GUA, were isolated from E. coli cells and purified to homogeneity. (C) Suppression of the amber codon 5΄-UAG at the lacZU118Am locus by the amber suppressor form of V-Spinach tRNATyr harboring the anticodon 5΄-CUA, leading to synthesis of β-gal. Measurement of β-gal activity encoded in XAC-1 cells expressing a plasmid-borne amber suppressor form of tRNATyr (Tyr), amber suppressor form of V-Spinach tRNATyr (Spinach), or none. Each activity is reported as a fraction relative to the activity encoded by the lacZWT gene (no amber mutation) in XAC-1 cells. Error bars denote SD (standard deviation), n = 3. (D) Suppression of trmDAm (Y19) by the amber suppressor form of V-Spinach tRNATyr leads to synthesis of the growth-dependent TrmD enzyme, which supports E. coli cell viability. The trmDAm (Y19) locus contains an amber codon at the Y19 position, which prevents translation of the trmD gene. This study was conducted in E. coli trmD-knockout (trmD-KO) cells where the chromosome-encoded trmD gene was disrupted with an antibiotic marker and cells were maintained viable in the presence of arabinose by a plasmid-borne and arabinose-controlled expression of the human counterpart trm5 gene (42). (E) Examination of viability of E. coli trmD-KO cells expressing a plasmid-borne trmDWT (row 1), a plasmid-borne trmDAm (Y19) (row 2), a plasmid-borne trmDAm (Y19) and the amber suppressor gene of Ec tRNATyr (row 3), and a plasmid-borne trmDAm (Y19) and the amber suppressor gene of V-Spinach tRNATyr (row 4) in the presence or absence of arabinose (Ara).
Translation of a codon involves three key steps; (i) aminoacylation of the tRNA cognate to the codon, (ii) formation of the aminoacyl-tRNA ternary complex with EF-Tu and GTP, (iii) delivery of the ternary complex to the ribosome A-site and reading of the codon by the aminoacyl-tRNA. To determine how the V-Spinach tRNATyr performed these reactions relative to the wild-type tRNATyr, we monitored the time-course of each reaction using purified enzymes and ribosomes in quantitative assays. To obtain data relevant to the biology of protein synthesis, the Spinach fusion and the wild-type tRNATyr was each isolated from cells to homogeneity, containing all natural post-transcriptional modifications and the natural Tyr anticodon 5΄-GUA. We found that the Spinach tRNA was mildly deficient relative to the wild-type tRNATyr in these key steps of protein synthesis (Figure 3B).
To assess the aminoacylation reaction, we monitored the attachment of 3H-Tyr to tRNATyr by E. coli TyrRS. The kinetic parameters of the wild-type tRNATyr (Km, kcat and kcat/Km, Supplementary Figure S6A) were closely similar to values of the transcript of E. coli tRNATyr (34), consistent with the notion that post-transcriptional modifications are not critical for TyrRS. These parameters are also similar to values of other E. coli tRNAs (11,34,48,69–71), consistent with the notion that the overall kinetics of aminoacylation is similar among different tRNA species, including the Michaelis constant and the number of catalytic turnover (72). The V-Spinach tRNATyr differed from the wild-type tRNATyr by a small increase in Km (tRNA) (from 1.7 ± 0.4 to 2.6 ± 1.2 μM) and a small decrease in kcat (from 2.5 ± 0.2 to 1.4 ± 0.3 s−1), resulting in an overall decrease of 2.7-fold in the catalytic efficiency kcat/Km (Figure 3A). Thus, while the large V-loop presents interference to aminoacylation, the effect is minor relative to the 10−6 effect observed upon the loss of a major determinant for other aminoacyl-tRNA synthetases (39,73).
To assess the stability of each charged Tyr-tRNA in a ternary complex with EF-Tu-GTP, we determined its dissociation kinetics from the ternary complex. In this assay, we preformed the ternary complex harboring each tRNA, 3΄-end labeled with 32P and charged with Tyr, and used RNase A to degrade the tRNA fraction that dissociated from the complex. The remaining labeled tRNA fraction in the ternary complex was then determined. Measurement of the dissociation constant in equilibrium conditions showed that the off rate of the fusion and of the parental tRNATyr from the respective ternary complex was similar (koff = 0.0034 ± 0.0008 versus 0.0022 ± 0.0005 s−1, Supplementary Figure S6B), indicating comparable stability of each in the respective ternary complex. These values are also similar to values of others (55), supporting the notion that EF-Tu binding focuses on the acceptor end of tRNA (74,75) and is not interfered by the Spinach insertion to the V-loop.
To assess the accommodation of each tRNA to the ribosome A-site, we assembled a ribosome post-initiation complex, where the initiator fMet-tRNAfMet was bound at the P-site next to the Tyr codon 5΄-UAU at the A-site. By analysis of GTP hydrolysis upon accommodation of each charged tRNA to the Tyr codon, we showed that the Spinach fusion induced an 18-fold slower rate relative to the wild-type tRNATyr (0.062 ± 0.008 versus 1.11 ± 0.06 s−1, Supplementary Figure S6C). In this assay, while the ribosome concentration was low (0.375 μM), it was still in excess of the ternary complex to permit one turnover. Indeed, GTP hydrolysis for Spinach tRNA and the wild-type tRNATyr both proceeded in a single exponential course over time (Supplementary Figure S6C), indicating that it was a single turnover reaction for both. The rate constant of tRNATyr was similar to values reported previously at similar ribosome concentrations (11,76). Fitting the rate data as a function of ribosome concentration showed a hyperbola curve without a lag, even at the low concentration tested here (76), indicating that substrate binding was in rapid equilibrium with the ribosome and that the reaction was not limited by binding, but by the chemistry of GTP hydrolysis (which is controlled by a slow conformational accommodation step (77)). Thus, despite the low concentration of the ribosome, binding of each ternary complex was fast relative to the kinetics of GTP hydrolysis. The 18-fold reduction in the rate of GTP hydrolysis of the V-Spinach tRNATyr relative to tRNATyr therefore reflects an impaired accommodation to the ribosome A-site prior to GTP hydrolysis.
We also monitored the first peptidyl transfer catalyzed by the ribosome, by incubating a post-initiation complex with each tRNA ternary complex and monitoring the rate of peptidyl transfer. In this assay, the limiting reagent was the post-initiation complex containing 35S-fMet-charged initiator tRNAfMet at the P-site (0.25 μM), reacting with a molar excess of each tyrosyl ternary complex. By analysis of dipeptide synthesis of 35S-fMet-Tyr, we found that the rate of each tRNA was similar to the respective rate of GTP hydrolysis (Supplementary Figure S6C and D), indicating that GTP hydrolysis remained limiting for the rate of peptide bond formation at this low ribosome concentration. These results suggest that the Spinach tRNA, despite the initial deficiency at inducing GTP hydrolysis and accommodation to the A-site, did not cause further impediment on the ribosome to slow down peptidyl transfer. However, whether Spinach tRNA was defective relative to wild-type tRNATyr in peptidyl transfer cannot be resolved by this assay. Notably, due to the rate-limiting nature of GTP hydrolysis to the first peptidyl transfer, the second peptidyl transfer was not determined. Additionally, in the time course of both GTP hydrolysis and dipeptidyl synthesis, the Spinach tRNA consistently showed a 3-4-fold reduced plateau level relative to the wild-type tRNATyr (Supplementary Figure S6C and D), further supporting the notion of a reduced accommodation prior to GTP hydrolysis.
Given that the Spinach tRNA was deficient relative to the wild-type tRNATyr in the assays above, we determined whether this deficiency reduced protein synthesis in vivo. We used the amber suppressor form of the tRNA to determine its suppression activity in E. coli XAC-1 cells, which displayed low backgrounds for reporter assays (38). XAC-1 expresses the reporter lacZU118Am, where an amber mutation is localized to position 17 of lacZ (34). Measurement of the intracellular activity of the reporter protein showed that translation of the amber codon by the wild-type tRNATyr relative to the control gene without the amber mutation (lacZWT) was 61.7%, whereas translation by the Spinach fusion was reduced to 9.3% (Figure 3C). Thus, consistent with biochemical analysis, the cell-based assay showed that the Spinach fusion performed protein synthesis with a 6-7-fold reduced activity relative to wild-type tRNATyr.
We then asked whether the reduced protein synthesis activity of the Spinach fusion compromised cell viability. Using the growth-essential trmD gene (41,42) as a reporter, which codes for a tRNA methyl transferase required for maintaining the protein synthesis reading frame (42,60,78), we showed that E. coli cells harboring an amber mutation of the Y19 codon of trmD were non-viable, unless the human counterpart gene (43) was expressed from a maintenance plasmid under the control of arabinose (Figure 3D and E, rows 1–2). This provides a cell-based assay to determine, in the absence of the human counterpart enzyme upon removal of arabinose, whether the translation of the amber codon at Y19 was sufficiently active to produce TrmD to support cell viability. Indeed, the amber-suppressor form of the wild-type tRNATyr was sufficiently active, such that cells were viable when the human gene was turned off (Figure 3E, row 3). Importantly, the amber-suppressor form of the Spinach fusion appeared to be even more active, supporting stronger cell viability in the absence of arabinose (Figure 3E, row 4). In this comparison, we suggest that the amber-suppressor form of the wild-type tRNATyr, due to its stronger suppressor activity, can interfere with the normal termination at 5΄-UAG stop codons, thus conferring a growth disadvantage to cells.
Co-exclusion of V-Spinach tRNA and ribosomes from the nucleoids
We determined if the Spinach fusion provides an imaging tool to visualize tRNA with the ribosome. In exponentially growing E. coli cells, translating ribosomes are not uniformly distributed throughout the intracellular space of cytoplasm; instead they are organized around the nucleoid lobes in segregated space (44,79). By labeling each translating ribosome with an mCherry tag fused to the ribosomal L9 protein (L9-mCherry), previous fluorescence imaging analysis showed an alternating distribution pattern of ribosomes with the DAPI-stained nucleoids (44). This specific segregation of ribosomes from the nucleoids would change when cells entered stationary phase or encountered drugs affecting transcription or translation (44). To determine the cellular distribution of V-Spinach tRNATyr relative to ribosomes, we expressed it from a multi-copy plasmid in a strain where all ribosomes carried the L9-mCherry tag. This strain, together with a control strain expressing the stand-alone Spinach, was cultured, briefly incubated with DFHBI, and imaged with appropriate filters.
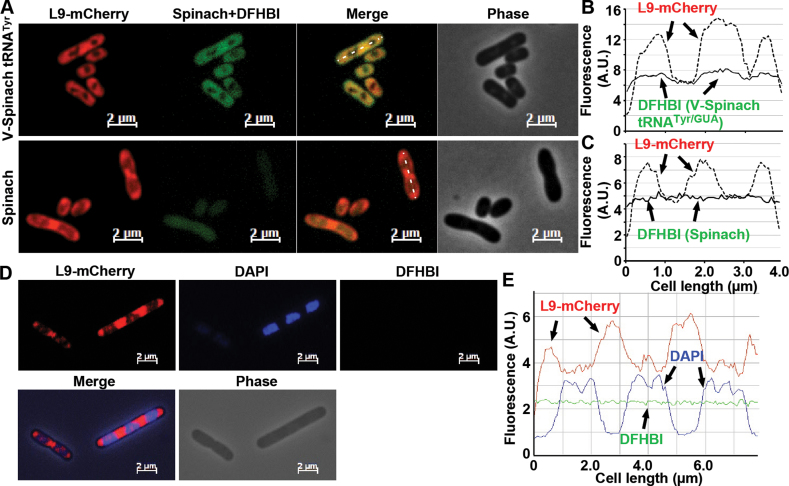
In exponentially growing E. coli cells, the L9-mCherry fluorescence was clearly visible and it exhibited the alternating pattern of ribosome distribution with the nucleoids spanning the entire length of the rod-shape (Figure 4A). Starting from one pole to the other across the average 4 μm length of single cells, the fluorescence of L9-mCherry ribosomes peaked at 1.0, 2.5 and 3.5 μm (Figure 4B), consistent with previous reports (79). Interestingly, a lengthwise intensity scan of a representative cell revealed the overlap of the L9-mCherry peaks with the GFP-like fluorescence of the V-Spinach tRNATyr peaks (Figure 4A and B), suggesting co-exclusion from the nucleoids. This overlap was not seen in cells expressing the stand-alone Spinach, which instead showed diffused and uniformly distributed fluorescence without nuclear exclusion (Figure 4A and C), or in DFHBI-stained cells without any plasmid (Figure 4D). Comparing the L9-mCherry and the GFP-like fluorescence with DAPI staining confirmed the co-exclusion of ribosomes with Spinach tRNA from the nucleoids, showing the occupancy of the nucleoids at each trough between the fluorescence peaks (Figure 4D and E). This co-exclusion was reproducibly observed in different phases of cell growth, suggesting that Spinach tRNA and ribosomes are together in the cytosol and possibly engaged in active protein synthesis.
Figure 4.
Localization of V-Spinach tRNATyr in living E. coli cells relative to L9-mCherry ribosomes. (A) E. coli JM109 cells, expressing L9-mCherry from the chromosome and V-Spinach tRNATyr/GUA (top) or the stand-alone Spinach (bottom) from pKK223-3 (induced with IPTG), were observed under microscope as described in Materials and Methods. (B and C) Lengthwise intensity scans of mCherry (dotted line) and Spinach fluorescence (solid line) of a representative cell from the two strains. (D) E. coli JM109 cells, harboring no plasmid, were imaged for L9-mCherry (left) and with DFHBI (right) and stained with DAPI (middle). (E) Lengthwise intensity scans of mCherry (red line), Spinach fluorescence (green line), and DAPI stain (blue line) of a representative cell.
DISCUSSION
This work demonstrates that the insertion of a large Spinach aptamer to the V-loop of tRNA creates a fusion that is active as an integral component of live-cell protein synthesis in E. coli. The protein synthesis activity of the fusion manifests at several levels: the suppression of separate amber mutations for synthesis of β-gal (lacZ125am, lacZu118Am), the suppression of a single mutation for synthesis of TrmD (Y19am), and the biochemical activity in individual reactions required for peptide bond formation. This range of activities in protein synthesis is unexpected, because the fusion combines two large and unrelated RNA structures in a molecular mass greatly exceeding any natural tRNA. Given that E. coli ribosomes never encountered such a fusion before and yet can readily accept it within the constraints of the protein synthesis machinery, this work reveals novel insights into the structural and dynamic flexibility of both the ribosome and the tRNA and their mutual accommodation. For the tRNA part, the V-loop is critical for assembly of the tertiary core that controls the dynamics of the two helical arms (47,58,80–82). The capacity of creating a fusion with the V-loop is high, not limited to species with a naturally large V-loop (E. coli tRNATyr and tRNALeu), but also applicable to those with a small V-loop (E. coli tRNAAla, tRNAPhe and tRNAPro). In all cases, the fluorescence intensity of the fusion is similar, indicating a similar stability among these inside the cell. For the ribosome part, the ability to use the fusion in a series of reactions leading to protein synthesis is striking, so is the efficiency with which the fusion accomplishes these reactions at a rate sufficient to support cell growth. These data emphasize that the fusion is structurally and dynamically compatible with the mechanics of protein synthesis. Because these mechanics are conserved from bacteria to humans, it is likely that the fusion, or its derivatives, will function with eukaryotic ribosomes.
Using E. coli V-Spinach tRNATyr as an example, imaging analysis reveals its versatility as a valid biological probe. First, it is a probe for the cellular quality control that degrades tRNA with structural damage at the acceptor end. By directly introducing damage to the fusion (Figure 2), we show that the quality control is active and rapid in E. coli, with little intracellular accumulation of damaged tRNA. This provides new insight into the temporal control of damage and expands the scope of the quality control from the previously established yeast and mammalian cells (64) now to bacterial cells. Second, imaging analysis identifies the fusion in overlapping positions with ribosomes in exponentially growing cells. While this overlap alone does not represent tRNA actually sitting with the ribosome, it is consistent with biochemical and cell-based data supporting the notion that the fusion is active in protein synthesis. Third, the intracellular distribution of the fusion is excluded from the nucleoids in an alternating pattern identical to that of ribosomes. This observation is intriguing, because although the fusion has a size large for a tRNA, it is small relative to the nucleoids, which are made up of millions of DNA base pairs in an irregular shape to which transcription factors bind. Nonetheless, the fusion does not freely move in and out of the space within the nucleoids but instead stays with ribosomes during the exponential growth phase with active protein synthesis. Although active protein synthesis in E. coli is coupled with transcription, this coupling is initiated inside the nucleoids to move transcription forward and to protect the nascent mRNA chain (83,84), allowing only matured mRNA to diffuse outside to the ribosome-rich region. The finding that the fusion and ribosomes are both segregated from the nucleoids suggests that they are together in the cytosol and possibly engaged in protein synthesis with minimum searching time for ribosomes to find the tRNA.
While the Spinach fusion has high potential for imaging tRNA, the current aptamer and its derivatives do not have strong enough fluorescence intensity. As shown in the localization analysis, the Spinach aptamer emits weak fluorescence relative to mCherry by more than 500-fold (Figure 4C), thus limiting single-molecular tracking of the tRNA on the ribosome. The poor fluorescence property of Spinach is also noted in imaging mRNA (85). Indeed, recent work has begun to address this issue by improving DFHBI binding and by further evolving new aptamers (18,19). For live-cell imaging of tRNA in protein synthesis, an additional option is to engineer cells to reduce the number of endogenous tRNA species competing with the Spinach fusion for the same codon. These efforts should lead to a more powerful tool for real-time imaging of tRNA in both ribosome and non-ribosome reactions.
When a stronger fluorophore is in hand, the Spinach-tRNA fusion will be an attractive tool with unique advantages of imaging tRNA not available by any existing methods. It offers the ability to monitor the trajectory of a tRNA from its biosynthesis to degradation in a live bacterial cell. One application of the tool is to determine the localization and segregation of tRNA during cell division. Specifically, when cells are in dormancy under specific conditions, while DNA replication is completely halted, low levels of translation must continue to maintain cell viability. The expression of the Spinach–tRNA fusion would identify where such low levels of translation takes place inside a cell. Second, while each cell usually expresses multiple isoacceptors for the same amino acid, one isoacceptor may dominate at a given time. For example, among the three isoacceptors for proline, the UGG isoacceptor (with the anticodon UGG) reads all four Pro codons CCN, the GGG isoacceptor reads two of the Pro codons (CC[C/U]), and the CGG isoacceptor reads the single CCG codon. As an E. coli cell progresses from the lag phase, to the exponential phase, and to the stationary phase, it changes its genetic program and codon usage for gene expression and protein synthesis. To determine which isoacceptor populates with the growth at a given time, we can create the Spinach fusion for each and identify the one that correlates with the growth trajectory. Third, the Spinach-tRNA fusion can be used to monitor the ribosome translation in real time between two specific positions in a single mRNA. By using the amber-suppressor form of the Spinach-tRNA fusion, we can monitor the translation speed between two site-specifically placed UAG codons in an mRNA and determine whether the translation proceeds at a constant speed or at a variable speed depending on the sequence context of the two UAG codons. These examples demonstrate the potential for new information on tRNA biology and protein synthesis in the context of a cell.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Samie Jaffrey, Barry Cooperman, Iris Alroy, Zeev Smilansky, Megumi Shigematsu, Jim Keen and Yolanda Covarrubias for discussion and Angela Chang, Albert Chang and Melanie Kauffman for analysis of imaging data.
Author contributions: Igarashi constructed expression plasmids, Sakaguchi, Masuda, Nitharwal, Han and Leslie performed imaging analysis, Takase, Liu and Gamper measured protein synthesis kinetics, Masuda developed cell-based assays, Ha, Sanyal and Hou contributed to data analysis and discussion, and Hou wrote the paper.
Footnotes
Present addresses:
Reiko Sakaguchi, Kyoto University, Kyoto, 606-8501, Japan.
Cuiping Liu, Huazhong Agricultural University, College of Veterinary Medicine, Wuhan, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [GM112659 to T.J.H., GM114343, GM108972 to Y.M.H.]; MDA [157681 to Y.M.H.]; National Science Foundation [PHY-1430124 to T.J.H.]. The open access publication charge for this paper has been waived by Oxford University Press - NAR.
Conflict of interest statement. None declared.
REFERENCES
- 1. Phizicky E.M., Hopper A.K.. tRNA biology charges to the front. Genes Dev. 2010; 24:1832–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhu B., Lee S.J., Tan M., Wang E.D., Richardson C.C.. Gene 5.5 protein of bacteriophage T7 in complex with Escherichia coli nucleoid protein H-NS and transfer RNA masks transfer RNA priming in T7 DNA replication. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:8050–8055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kleiman L., Jones C.P., Musier-Forsyth K.. Formation of the tRNALys packaging complex in HIV-1. FEBS Lett. 2010; 584:359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fu G., Xu T., Shi Y., Wei N., Yang X.L.. tRNA-controlled nuclear import of a human tRNA synthetase. J. Biol. Chem. 2012; 287:9330–9334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kamhi E., Raitskin O., Sperling R., Sperling J.. A potential role for initiator-tRNA in pre-mRNA splicing regulation. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:11319–11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mei Y., Yong J., Liu H., Shi Y., Meinkoth J., Dreyfuss G., Yang X.. tRNA binds to cytochrome c and inhibits caspase activation. Mol. Cell. 2010; 37:668–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mei Y., Stonestrom A., Hou Y.M., Yang X.. Apoptotic regulation and tRNA. Protein Cell. 2010; 1:795–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu C., Stonestrom A.J., Christian T., Yong J., Takase R., Hou Y.M., Yang X.. Molecular basis and consequences of the cytochrome c-tRNA interaction. J. Biol. Chem. 2016; 291:10426–10436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Barhoom S., Kaur J., Cooperman B.S., Smorodinsky N.I., Smilansky Z., Ehrlich M., Elroy-Stein O.. Quantitative single cell monitoring of protein synthesis at subcellular resolution using fluorescently labeled tRNA. Nucleic Acids Res. 2011; 39:e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Barhoom S., Farrell I., Shai B., Dahary D., Cooperman B.S., Smilansky Z., Elroy-Stein O., Ehrlich M.. Dicodon monitoring of protein synthesis (DiCoMPS) reveals levels of synthesis of a viral protein in single cells. Nucleic Acids Res. 2013; 41:e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu C., Gamper H., Liu H., Cooperman B.S., Hou Y.M.. Potential for interdependent development of tRNA determinants for aminoacylation and ribosome decoding. Nat. Commun. 2011; 2:329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Betteridge T., Liu H., Gamper H., Kirillov S., Cooperman B.S., Hou Y.M.. Fluorescent labeling of tRNAs for dynamics experiments. RNA. 2007; 13:1594–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schleich H.G., Wintermeyer W., Zachau H.G.. Replacement of wybutine by hydrazines and its effect on the active conformation of yeast tRNAPhe. Nucleic Acids Res. 1978; 5:1701–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu C., Betteridge T., Hou Y.M.. Fluorophore labeling to monitor tRNA dynamics. Methods Enzymol. 2009; 469:69–93. [DOI] [PubMed] [Google Scholar]
- 15. Paige J.S., Wu K.Y., Jaffrey S.R.. RNA mimics of green fluorescent protein. Science. 2011; 333:642–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Strack R.L., Disney M.D., Jaffrey S.R.. A superfolding Spinach2 reveals the dynamic nature of trinucleotide repeat-containing RNA. Nat. Methods. 2013; 10:1219–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Warner K.D., Chen M.C., Song W., Strack R.L., Thorn A., Jaffrey S.R., Ferre-D’Amare A.R.. Structural basis for activity of highly efficient RNA mimics of green fluorescent protein. Nat. Struct. Mol. Biol. 2014; 21:658–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ketterer S., Fuchs D., Weber W., Meier M.. Systematic reconstruction of binding and stability landscapes of the fluorogenic aptamer spinach. Nucleic Acids Res. 2015; 43:9564–9572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Autour A., Westhof E., Ryckelynck M.. iSpinach: a fluorogenic RNA aptamer optimized for in vitro applications. Nucleic Acids Res. 2016; 44:2491–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Filonov G.S., Moon J.D., Svensen N., Jaffrey S.R.. Broccoli: rapid selection of an RNA mimic of green fluorescent protein by fluorescence-based selection and directed evolution. J. Am. Chem. Soc. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dolgosheina E.V., Jeng S.C., Panchapakesan S.S., Cojocaru R., Chen P.S., Wilson P.D., Hawkins N., Wiggins P.A., Unrau P.J.. RNA mango aptamer-fluorophore: a bright, high-affinity complex for RNA labeling and tracking. ACS Chem. Biol. 2014; 9:2412–2420. [DOI] [PubMed] [Google Scholar]
- 22. Paige J.S., Nguyen-Duc T., Song W., Jaffrey S.R.. Fluorescence imaging of cellular metabolites with RNA. Science. 2012; 335:1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Strack R.L., Jaffrey S.R.. New approaches for sensing metabolites and proteins in live cells using RNA. Curr. Opin. Chem. Biol. 2013; 17:651–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Strack R.L., Song W., Jaffrey S.R.. Using Spinach-based sensors for fluorescence imaging of intracellular metabolites and proteins in living bacteria. Nat. Protoc. 2014; 9:146–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kellenberger C.A., Wilson S.C., Sales-Lee J., Hammond M.C.. RNA-based fluorescent biosensors for live cell imaging of second messengers cyclic di-GMP and cyclic AMP-GMP. J. Am. Chem. Soc. 2013; 135:4906–4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nakayama S., Luo Y., Zhou J., Dayie T.K., Sintim H.O.. Nanomolar fluorescent detection of c-di-GMP using a modular aptamer strategy. Chem. Commun. (Camb.). 2012; 48:9059–9061. [DOI] [PubMed] [Google Scholar]
- 27. Sato S., Watanabe M., Katsuda Y., Murata A., Wang D.O., Uesugi M.. Live-cell imaging of endogenous mRNAs with a small molecule. Angew. Chem. 2015; 54:1855–1858. [DOI] [PubMed] [Google Scholar]
- 28. Murata A., Sato S., Kawazoe Y., Uesugi M.. Small-molecule fluorescent probes for specific RNA targets. Chem. Commun. (Camb.). 2011; 47:4712–4714. [DOI] [PubMed] [Google Scholar]
- 29. Huang H., Suslov N.B., Li N.S., Shelke S.A., Evans M.E., Koldobskaya Y., Rice P.A., Piccirilli J.A.. A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore. Nat. Chem. Biol. 2014; 10:686–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yusupova G., Jenner L., Rees B., Moras D., Yusupov M.. Structural basis for messenger RNA movement on the ribosome. Nature. 2006; 444:391–394. [DOI] [PubMed] [Google Scholar]
- 31. Selmer M., Dunham C.M., Murphy F.V. IV, Weixlbaumer A., Petry S., Kelley A.C., Weir J.R., Ramakrishnan V.. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006; 313:1935–1942. [DOI] [PubMed] [Google Scholar]
- 32. Korostelev A., Trakhanov S., Laurberg M., Noller H.F.. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006; 126:1065–1077. [DOI] [PubMed] [Google Scholar]
- 33. Dunkle J.A., Wang L., Feldman M.B., Pulk A., Chen V.B., Kapral G.J., Noeske J., Richardson J.S., Blanchard S.C., Cate J.H.. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science. 2011; 332:981–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hou Y.M., Schimmel P.. Modeling with in vitro kinetic parameters for the elaboration of transfer RNA identity in vivo. Biochemistry. 1989; 28:4942–4947. [DOI] [PubMed] [Google Scholar]
- 35. Yaremchuk A., Kriklivyi I., Tukalo M., Cusack S.. Class I tyrosyl-tRNA synthetase has a class II mode of cognate tRNA recognition. EMBO J. 2002; 21:3829–3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gabriel K., McClain W.H.. Plasmid systems to study RNA function in Escherichia coli. J. Mol. Biol. 2001; 310:543–548. [DOI] [PubMed] [Google Scholar]
- 37. Newton W.A., Beckwith J.R., Zipser D., Brenner S.. Nonsense mutants and polarity in the lac operon of Escherichia coli. J. Mol. Biol. 1965; 14:290–296. [DOI] [PubMed] [Google Scholar]
- 38. Hou Y.M., Schimmel P.. Novel transfer RNAs that are active in Escherichia coli. Biochemistry. 1992; 31:4157–4160. [DOI] [PubMed] [Google Scholar]
- 39. Hou Y.M., Schimmel P.. A simple structural feature is a major determinant of the identity of a transfer RNA. Nature. 1988; 333:140–145. [DOI] [PubMed] [Google Scholar]
- 40. Normanly J., Masson J.M., Kleina L.G., Abelson J., Miller J.H.. Construction of two Escherichia coli amber suppressor genes: tRNAPheCUA and tRNACysCUA. Proc. Natl. Acad. Sci. U.S.A. 1986; 83:6548–6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H.. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006; 2:2006 0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gamper H.B., Masuda I., Frenkel-Morgenstern M., Hou Y.M.. Maintenance of protein synthesis reading frame by EF-P and m (1)G37-tRNA. Nat. Commun. 2015; 6:7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Christian T., Gamper H., Hou Y.M.. Conservation of structure and mechanism by Trm5 enzymes. RNA. 2013; 19:1192–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chai Q., Singh B., Peisker K., Metzendorf N., Ge X., Dasgupta S., Sanyal S.. Organization of ribosomes and nucleoids in Escherichia coli cells during growth and in quiescence. J. Biol. Chem. 2014; 289:11342–11352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yokogawa T., Kitamura Y., Nakamura D., Ohno S., Nishikawa K.. Optimization of the hybridization-based method for purification of thermostable tRNAs in the presence of tetraalkylammonium salts. Nucleic Acids Res. 2010; 38:e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fersht A.R., Ashford J.S., Bruton C.J., Jakes R., Koch G.L., Hartley B.S.. Active site titration and aminoacyl adenylate binding stoichiometry of aminoacyl-tRNA synthetases. Biochemistry. 1975; 14:1–4. [DOI] [PubMed] [Google Scholar]
- 47. Hou Y.M., Westhof E., Giege R.. An unusual RNA tertiary interaction has a role for the specific aminoacylation of a transfer RNA. Proc. Natl. Acad. Sci. U.S.A. 1993; 90:6776–6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liu C., Gamper H., Shtivelband S., Hauenstein S., Perona J.J., Hou Y.M.. Kinetic quality control of anticodon recognition by a eukaryotic aminoacyl-tRNA synthetase. J. Mol. Biol. 2007; 367:1063–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wolfson A.D., Pleiss J.A., Uhlenbeck O.C.. A new assay for tRNA aminoacylation kinetics. RNA. 1998; 4:1019–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cochella L., Brunelle J.L., Green R.. Mutational analysis reveals two independent molecular requirements during transfer RNA selection on the ribosome. Nat. Struct. Mol. Biol. 2007; 14:30–36. [DOI] [PubMed] [Google Scholar]
- 51. Zhang C.M., Liu C., Christian T., Gamper H., Rozenski J., Pan D., Randolph J.B., Wickstrom E., Cooperman B.S., Hou Y.M.. Pyrrolo-C as a molecular probe for monitoring conformations of the tRNA 3΄ end. RNA. 2008; 14:2245–2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang C.M., Liu C., Slater S., Hou Y.M.. Aminoacylation of tRNA with phosphoserine for synthesis of cysteinyl-tRNA (Cys). Nat. Struct. Mol. Biol. 2008; 15:507–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Louie A., Ribeiro N.S., Reid B.R., Jurnak F.. Relative affinities of all Escherichia coli aminoacyl-tRNAs for elongation factor Tu-GTP. J. Biol. Chem. 1984; 259:5010–5016. [PubMed] [Google Scholar]
- 54. Dale T., Sanderson L.E., Uhlenbeck O.C.. The affinity of elongation factor Tu for an aminoacyl-tRNA is modulated by the esterified amino acid. Biochemistry. 2004; 43:6159–6166. [DOI] [PubMed] [Google Scholar]
- 55. LaRiviere F.J., Wolfson A.D., Uhlenbeck O.C.. Uniform binding of aminoacyl-tRNAs to elongation factor Tu by thermodynamic compensation. Science. 2001; 294:165–168. [DOI] [PubMed] [Google Scholar]
- 56. Nazarenko I.A., Harrington K.M., Uhlenbeck O.C.. Many of the conserved nucleotides of tRNA (Phe) are not essential for ternary complex formation and peptide elongation. EMBO J. 1994; 13:2464–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dupasquier M., Kim S., Halkidis K., Gamper H., Hou Y.M.. tRNA integrity is a prerequisite for rapid CCA addition: implication for quality control. J. Mol. Biol. 2008; 379:579–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pan D., Kirillov S., Zhang C.M., Hou Y.M., Cooperman B.S.. Rapid ribosomal translocation depends on the conserved 18-55 base pair in P-site transfer RNA. Nat. Struct. Mol. Biol. 2006; 13:354–359. [DOI] [PubMed] [Google Scholar]
- 59. Pan D., Zhang C.M., Kirillov S., Hou Y.M., Cooperman B.S.. Perturbation of the tRNA tertiary core differentially affects specific steps of the elongation cycle. J. Biol. Chem. 2008; 283:18431–18440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gamper H.B., Masuda I., Frenkel-Morgenstern M., Hou Y.M.. The UGG isoacceptor of tRNAPro is naturally prone to frameshifts. Int. J. Mol. Sci. 2015; 16:14866–14883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ledoux S., Uhlenbeck O.C.. [3΄-32P]-labeling tRNA with nucleotidyltransferase for assaying aminoacylation and peptide bond formation. Methods. 2008; 44:74–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cochella L., Green R.. An active role for tRNA in decoding beyond codon:anticodon pairing. Science. 2005; 308:1178–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Youngman E.M., Brunelle J.L., Kochaniak A.B., Green R.. The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell. 2004; 117:589–599. [DOI] [PubMed] [Google Scholar]
- 64. Wilusz J.E., Whipple J.M., Phizicky E.M., Sharp P.A.. tRNAs marked with CCACCA are targeted for degradation. Science. 2011; 334:817–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wilusz J.E., Freier S.M., Spector D.L.. 3΄ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008; 135:919–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sunwoo H., Dinger M.E., Wilusz J.E., Amaral P.P., Mattick J.S., Spector D.L.. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009; 19:347–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wilusz J.E., Spector D.L.. An unexpected ending: noncanonical 3΄ end processing mechanisms. RNA. 2010; 16:259–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Bachmann B.J. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol. Rev. 1972; 36:525–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Christian T., Lipman R.S., Evilia C., Hou Y.M.. Alternative design of a tRNA core for aminoacylation. J. Mol. Biol. 2000; 303:503–514. [DOI] [PubMed] [Google Scholar]
- 70. Liu C., Sanders J.M., Pascal J.M., Hou Y.M.. Adaptation to tRNA acceptor stem structure by flexible adjustment in the catalytic domain of class I tRNA synthetases. RNA. 2012; 18:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Naganuma M., Sekine S., Chong Y.E., Guo M., Yang X.L., Gamper H., Hou Y.M., Schimmel P., Yokoyama S.. The selective tRNA aminoacylation mechanism based on a single G*U pair. Nature. 2014; 510:507–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ibba M., Francklyn C., Cusack S.. The Aminoacyl-tRNA Synthetases. 2005; Georgetown: Landes Bioscience. [Google Scholar]
- 73. Hamann C.S., Hou Y.M.. Enzymatic aminoacylation of tRNA acceptor stem helices with cysteine is dependent on a single nucleotide. Biochemistry. 1995; 34:6527–6532. [DOI] [PubMed] [Google Scholar]
- 74. Nissen P., Kjeldgaard M., Thirup S., Polekhina G., Reshetnikova L., Clark B.F., Nyborg J.. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog [see comments]. Science. 1995; 270:1464–1472. [DOI] [PubMed] [Google Scholar]
- 75. Nissen P., Thirup S., Kjeldgaard M., Nyborg J.. The crystal structure of Cys-tRNACys-EF-Tu-GDPNP reveals general and specific features in the ternary complex and in tRNA. Structure. 1999; 7:143–156. [DOI] [PubMed] [Google Scholar]
- 76. Ledoux S., Olejniczak M., Uhlenbeck O.C.. A sequence element that tunes Escherichia coli tRNA (Ala)(GGC) to ensure accurate decoding. Nat. Struct. Mol. Biol. 2009; 16:359–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Pape T., Wintermeyer W., Rodnina M.. Induced fit in initial selection and proofreading of aminoacyl-tRNA on the ribosome. EMBO J. 1999; 18:3800–3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Bystrom A.S., Bjork G.R.. Chromosomal location and cloning of the gene (trmD) responsible for the synthesis of tRNA (m1G) methyltransferase in Escherichia coli K-12. Mol. Gen. Genet. 1982; 188:440–446. [DOI] [PubMed] [Google Scholar]
- 79. Bakshi S., Siryaporn A., Goulian M., Weisshaar J.C.. Superresolution imaging of ribosomes and RNA polymerase in live Escherichia coli cells. Mol. Microbiol. 2012; 85:21–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Hou Y.M. Structural elements that contribute to an unusual tertiary interaction in a transfer RNA. Biochemistry. 1994; 33:4677–4681. [DOI] [PubMed] [Google Scholar]
- 81. Hamann C.S., Hou Y.M.. An RNA structural determinant for tRNA recognition. Biochemistry. 1997; 36:7967–7972. [DOI] [PubMed] [Google Scholar]
- 82. Hamann C.S., Hou Y.M.. Probing a tRNA core that contributes to aminoacylation. J. Mol. Biol. 2000; 295:777–789. [DOI] [PubMed] [Google Scholar]
- 83. Sanamrad A., Persson F., Lundius E.G., Fange D., Gynna A.H., Elf J.. Single-particle tracking reveals that free ribosomal subunits are not excluded from the Escherichia coli nucleoid. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:11413–11418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Bakshi S., Choi H., Weisshaar J.C.. The spatial biology of transcription and translation in rapidly growing Escherichia coli. Front. Microbiol. 2015; 6:636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Zhang J., Fei J., Leslie B.J., Han K.Y., Kuhlman T.E., Ha T.. Tandem spinach array for mRNA imaging in living bacterial cells. Sci. Rep. 2015; 5:17295. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.