Abstract
Photodetachment photoelectron spectroscopy is used to probe the electronic structure of mono-, di-, and trinucleotide anions in the gas phase. A weak and well defined threshold band was observed in the photoelectron spectrum of 2′-deoxyguanosine 5′-monophosphate at a much lower ionization energy than the other three mononucleotides. Density function theory calculations revealed that this unique spectral feature is caused by electron-detachment from a π orbital of the guanine base on 2′-deoxyguanosine 5′-monophosphate, whereas the lowest ionization channel for the other three mononucleotides takes place from the phosphate group. This low-energy feature was shown to be a “fingerprint” in all the spectra of dinucleotides and trinucleotides that contain the guanine base. The current experiment provides direct spectroscopic evidence that the guanine base is the site with the lowest ionization potential in oligonucleotides and DNA and is consistent with the fact that guanine is most susceptible to oxidation to give the guanine cation in DNA damage.
Keywords: DNA oxidation, nucleotide
The ionization of nucleotide plays very important roles in the chemistry and biology of DNA. Induced by electrophiles or ionizing radiation, the electron-deficient site (hole) on the nucleotide and its migration may lead directly to DNA damage (1–6). In most cases, guanine is the initial oxidation site, or the electron-loss center ultimately moves through the DNA π stack to end up at a guanine base (7). This observation is attributed to the low ionization potential (IP) of guanine relative to the other DNA bases (8–10). Thus, the electronic structure of nucleotides and their ionization properties are essential for understanding the mechanism of DNA damage. Extensive experimental and theoretical studies have focused on these topics (11–19). In particular, LeBreton and coworkers (9, 20–24) have investigated the ionization of nucleic acid bases, sugar residue, and the phosphate in the gas phase by using UV photoelectron spectroscopy (PES) and then combining these studies and molecular orbital calculations to address the electronic structure of the nucleotides. Gas-phase studies probe the intrinsic electronic properties of the nucleotides and provide important experimental data to compare with and verify theoretical results and methods.
Matrix-assisted laser desorption ionization (25) and electrospray ionization (26) techniques provide powerful tools for characterizing biological molecules in the gas phase. The gas-phase conformations of a series of free mononucleotide, dinucleotide, and trinucleotide ions were investigated by using matrix-assisted laser desorption ionization and ion-mobility experiment by Bowers and coworkers (27–30). Our group has developed an experimental technique that couples an electrospray ionization source with a magnetic-bottle photoelectron spectrometer (31) to investigate multiply charged anions and solution species in the gas phase (32, 33). Here, we report a PES study on a series of singly charged nucleotide anions in the gas phase, including all 4 2′-deoxy-5′-mononucleotide anions, all 16 dinucleotide anions, and 8 selected trinucleotide anions. PES spectra were taken at 157 nm, and the electron-detachment energies for each species were measured. It is interesting that a weak and well defined threshold band was observed in the spectra of all the species containing one or more guanine base, yielding a much lower electron-detachment energy than the other nucleotides. Density function theory calculations revealed that this unique spectral feature is caused by the electron detachment from a π orbital localized on the guanine base. Our observation provides direct spectroscopic evidence that the guanine site indeed has the lowest IP in the oligonucleotides and thus should serve as the reducing agent or the ultimate oxidation site in DNA damage.
Materials and Methods
PES. Details of the experimental method have been given elsewhere (31). Briefly, 1 mM sample solutions were prepared by dissolving the nucleotides or their sodium salts in a water/methanol mixed solvent (1:4 volume ratio) and were introduced into the gas phase by electrospray. Deprotonated nucleotide anions generated from the electrospray source were guided into a quadruple ion trap, in which ions were accumulated for 0.1 sec before being pulsed into the extraction zone of a time-of-flight mass spectrometer. During the PES experiment, the anions of interest were mass-selected and decelerated before being intercepted by a probe laser beam in the photodetachment zone of a magnetic-bottle photoelectron analyzer. In the current study, 157-nm (7.866-eV) photons from an F2 excimer laser were used because of the relatively high electron-binding energies of the singly charged nucleotide anions. Photoelectrons were collected at nearly 100% efficiency by the magnetic bottle and analyzed in a 4-m-long electron-flight tube. Photoelectron time-of-flight spectra were collected and then converted to kinetic energy spectra, calibrated by the known spectra of I– and O–. The electron-binding energy spectra (EB) presented here were obtained by subtracting the kinetic energy (KE) spectra from the detachment photon energies (EB = hν – KE). The electron kinetic energy resolution (ΔKE/KE) was ≈2% (i.e., ≈10 meV for 0.5 eV electrons), as measured from the spectrum of I– at 355 nm.
Theoretical Methods. The nwchem computational chemistry program (34) was used to determine the geometrical and electronic structures. All species were optimized by using density functional theory (35, 36) and the hybrid B3LYP (Becke three-parameter hybrid functional combined with Lee–Yang–Parr correlation functional) exchange-correlation functional (37, 38). We used the triple ζ valence basis set with long-range polarization functions optimized for density functional theory (39) by adding diffuse functions derived from an even-tempered expansion of the outermost functions in the original basis set. Thus, an additional diffuse s function was added to each hydrogen atom (exponent = 0.04573), and diffuse s and p functions were added to carbon, oxygen, nitrogen, and phosphorus atoms (exponents 0.04441 and 0.02922 on carbon, 0.08142 and 0.04812 on oxygen, 0.06035 and 0.04073 on nitrogen, and 0.04026 and 0.02813 on phosphorous). Frequencies were calculated for all species to confirm that the geometries were minima on the potential energy surface. The Extensible Computational Chemistry Environment (ecce) (40) was used to set up, launch, and evaluate the results from nwchem. We computed the theoretical vertical electron-detachment energy (VDE) of an anion as the difference in total energy between the optimized anion and the neutral molecule calculated at the anion geometry.
Results and Discussion
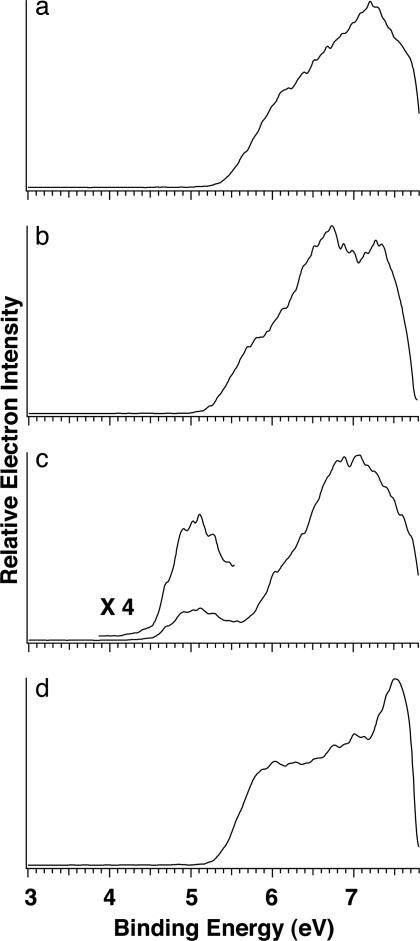
Photoelectron Spectra and Electron-Binding Energies of Mononucleotides. The 157-nm spectra of the four mononucleotide anions, 2′-deoxyadenosine 5′-monophosphate (dAMP–), 2′-deoxycytidine 5′-monophosphate (dCMP–), 2′-deoxyguanosine 5′-monophosphate (dGMP–), and 2′-deoxythymidine 5′-monophosphate (dTMP–), are shown in Fig. 1. The spectral patterns for dAMP–, dCMP–, and dTMP– are similar: each has a spectral onset at ≈5.4 eV with broad and continuous spectral features, suggesting high density of electronic states for the nucleotides. However, the spectrum of dGMP– is distinctly different: it displays a weak and well separated band at much lower binding energies, whereas the higher binding energy part of this spectrum is similar to those of dAMP– or dCMP–. The adiabatic electron-detachment energy (ADE) and VDE for each mononucleotide were determined and are given in Table 1. Note that the ADE of dGMP– is lower than that of dAMP– by almost 1 eV. Because of the lack of a well resolved threshold band for dAMP–, dCMP–, and dTMP–, their VDEs only could be estimated.
Fig. 1.
Photoelectron spectra of deprotonated 2′-deoxy-5′-deoxymononucleotide anions dAMP– (a), dCMP– (b), dGMP– (c), and dTMP– (d) at 157 nm (7.866 eV).
Table 1. The experimental ADEs and VDEs and the calculated VDEs for the four mononucleotides.
| Mononucleotide | ADE* | VDE (experimental) | VDE (calculated) |
|---|---|---|---|
| dAMP- | 5.55 ± 0.10 | 6.05 ± 0.50† | 5.23 |
| dCMP- | 5.30 ± 0.10 | 5.80 ± 0.50† | 5.24 |
| dGMP- | 4.61 ± 0.10 | 5.05 ± 0.10 | 4.90 |
| dTMP- | 5.40 ± 0.10 | 5.85 ± 0.50† | 5.36 |
All energies are in eV.
The ADEs were measured by drawing a straight line along the leading edge of the threshold band and then adding a constant to the intersection with the binding energy axis to take into account the instrumental resolution at the given energy range.
Because of the lack of separated threshold band, the VDEs should be considered as rough estimates.
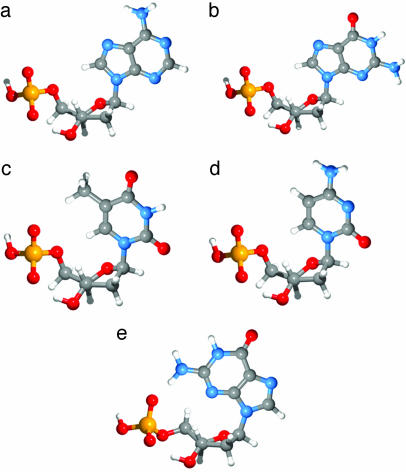
Structures and Energetics of the Mononucleotides. Initial conformational analysis was done on a set of 1′-pyrrolo-2′-deoxyribose-5′-monophosphate anions to determine the low-energy conformations for the deoxyribose monophosphate moiety. The lowest-energy conformation was the C3′-endo, corresponding closely to the conformation in the B-DNA double helix. Fig. 2 shows the low-energy conformations for the corresponding mononucleotide anions; those for the dAMP–, dCMP–, and dTMP– anions are the lowest energy conformation in the gas phase corresponding to the “anti” conformation in DNA. Two dGMP– structures are shown in Fig. 2: the anti conformation (Fig. 2b) is higher in energy (by 11.2 kcal/mol) than the “syn” conformation (Fig. 2e), in which the guanine base is rotated to allow the formation of an internal H bond between the amide group of the base and the deprotonated phosphate. Fig. 3 shows the calculated lowest-energy conformations for the four mononucleotides with the highest occupied molecular orbital (HOMO) isosurface of the Kohn–Sham molecular orbital (41) for each anion. The current results are consistent with the recent ion-mobility experiment and amber calculations (27). The calculated VDEs are also listed in Table 1, but they all seem to be smaller than the experimental values.
Fig. 2.
The lowest-energy conformations for anti-dAMP– (a), anti-dGMP– (b), anti-dTMP– (c), anti-dCMP– (d), and syn-dGMP– (e).
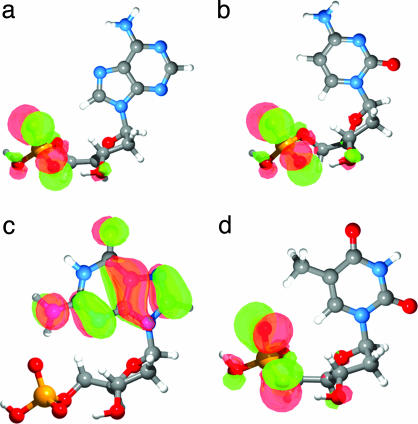
Fig. 3.
The lowest-energy conformation and HOMO isosurface for anti-dAMP– (a), anti-dCMP– (b), syn-dGMP– (c), and anti-dTMP– (d).
Kohn–Sham molecular orbital energies (42) for the global minima of the 2′-deoxyribonucleotides are shown in Table 2. The anti conformation of dGMP– is included for comparison. All anions have occupied orbitals with negative energies (i.e., these are all stable species in the gas phase). The closely lying molecular orbitals for all of the four species are qualitatively in agreement with the congested PES spectra (Fig. 1). Note that their relative energies are also consistent with the experimental ionization energies. In particular, the low binding energies of dGMP– are reproduced. It is most important that the calculations show that the HOMO of dGMP– is on the base, whereas the HOMOs for the other three nucleotides are localized on the phosphate, as shown in Fig. 3. Thus, the distinct low-energy band in the spectrum of dGMP– is caused by ionization of the guanine base, whereas the onset of the spectra for dAMP–, dCMP–, and dTMP– is caused by electron detachment from the phosphate. Among the four free nucleic acid bases, guanine is known to possess the lowest IP (9, 10). The current results show that the low IP of guanine is fully displayed in the intact mononucleotides even when dGMP– is in the anti conformation (as seen in Table 2).
Table 2.
Molecular orbital energies (eV) for the 10 highest-energy occupied Kohn–Sham orbitals (B3LYP/TZVP+) and their location for the four anti-mononucleotides and syn-dGMP–
| Molecular orbital | Anti-dAMP- | Anti-dTMP- | Anti-dCMP- | Anti-dGMP- | Syn-dGMP- |
|---|---|---|---|---|---|
| HOMO | -3.58 (PO4) | -3.82 (PO4) | -3.67 (PO4) | -3.52 (PO4) | -3.29 (Base) |
| HOMO-1 | -3.88 (PO4) | -3.97 (Base) | -3.86 (Base-PO4) | -3.81 (PO4) | -3.82 (PO4) |
| HOMO-2 | -4.04 (Base-PO4) | -4.11 (PO4) | -3.98 (PO4) | -3.89 (Base) | -4.08 (Sugar) |
| HOMO-3 | -4.06 (Base-PO4) | -4.26 (PO4) | -4.12 (PO4) | -3.99 (Sugar-PO4) | -4.25 (PO4) |
| HOMO-4 | -4.30 (Sugar) | -4.50 (Sugar) | -4.30 (Sugar) | -4.26 (Sugar-PO4) | -4.57 (PO4) |
| HOMO-5 | -4.76 (PO4) | -4.86 (All) | -4.54 (Base-Sugar) | -4.71 (PO4) | -4.69 (All) |
| HOMO-6 | -4.83 (All) | -4.99 (PO4) | -4.77 (Base) | -4.87 (All) | -4.89 (Base) |
| HOMO-7 | -4.93 (All) | -5.17 (All) | -4.84 (PO4) | -5.17 (All) | -4.99 (All) |
| HOMO-8 | -5.07 (All) | -5.43 (All) | -5.00 (All) | -5.18 (All) | -5.15 (All) |
| HOMO-9 | -5.38 (Sugar-PO4) | -5.56 (Base) | -5.11 (Base) | -5.34 (All) | -5.22 (All) |
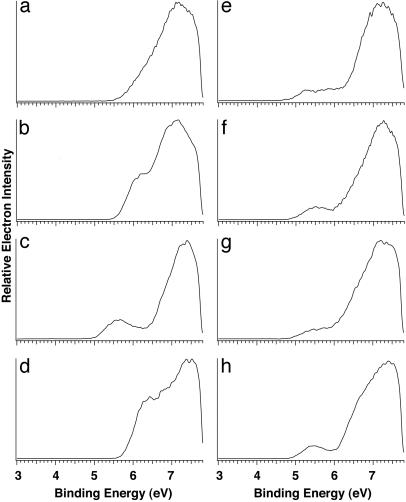
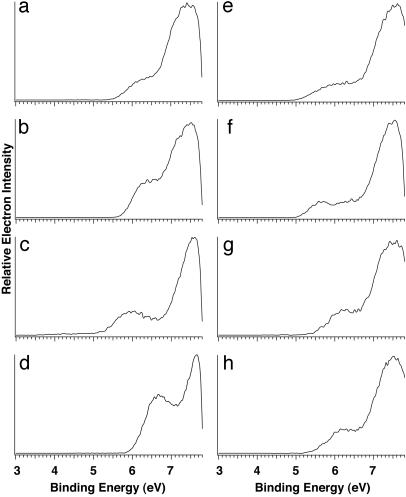
The Lowest IP Site in Oligonucleotides. Photoelectron spectra for all 16 dinucleotide anions were taken at 157 nm (8 of which are shown in Fig. 4; the rest can be found in Fig. 6, which is published as supporting information on the PNAS web site). Fig. 4 Left shows the spectra of the four dinucleotides with two identical bases: dAA– (5′-ApA-3′), dCC–, dGG–, and dTT–. The appearances for these spectra are similar to those of the corresponding mononucleotides except that the dinucleotides have higher electron-binding energies. A distinct and low-binding-energy feature was observed again for dGG–. In fact, a similar low-binding-energy band appears in the spectra of all those dinucleotides that contain a G base. Fig. 4 Right presents four of them (dAG–, dCG–, dGA–, and dTG–), and the spectra of dGC– and dGT– are shown in Fig. 6. All these G-containing dinucleotides have similar ADEs around 5 eV, lower than the ADEs of the non-guanine-containing species by between 0.6 and 0.8 eV (Table 3 and Table 4, which is published as supporting information on the PNAS web site).
Fig. 4.
Photoelectron spectra of deprotonated dinucleotide anions dAA– (a), dCC– (b), dGG– (c), dTT– (d), dAG– (e), dCG– (f), dGA– (g), and dTG– (h) at 157 nm (7.866 eV).
Table 3. The experimental ADEs for di- and trinucleotides.
| Species | ADE |
|---|---|
| Dinucleotide | |
| dAA- | 5.72 ± 0.10 |
| dCC- | 5.65 ± 0.10 |
| dGG- | 5.08 ± 0.10 |
| dTT- | 5.80 ± 0.10 |
| dAG- | 4.87 ± 0.10 |
| dCG- | 4.98 ± 0.10 |
| dGA- | 4.92 ± 0.10 |
| dTG- | 4.96 ± 0.10 |
| Trinucleotide | |
| dAAA- | 5.62 ± 0.10 |
| dCCC- | 5.78 ± 0.10 |
| dGGG- | 5.27 ± 0.10 |
| DTTT- | 6.04 ± 0.10 |
| dGGC- | 5.11 ± 0.10 |
| dGCG- | 5.13 ± 0.10 |
| dGCC- | 5.48 ± 0.10 |
| dCGC- | 5.44 ± 0.10 |
All energies are given in eV.
The similar PES spectral features for the di- and mononucleotides suggest that they have similar HOMOs (i.e., the low-binding-energy features in the G-containing dinucleotides are all caused by ionization from the guanine base, at which the HOMO is localized). Note that the intensity of the low-energy band in dGG– is stronger than those dinucleotides that contain only one G base, suggesting that both G bases in dGG– contribute to the first PES band. Because of their increased complexity and sizes, we could not afford to carry out high-level density function theory calculations on the dinucleotides. Fortunately, the gas-phase conformations for all of the 16 dinucleotides have been examined by ion-mobility experiments and amber calculations (28, 29). Three families of conformations with similar energies were identified: (i) “stacked” (in which the two bases are stacked, producing the most compact structures); (ii) “H bond” (this is also a relatively compact family of structures in which H bonds are formed between the two bases); and (iii) “open” (in which the two bases are far apart and are not interacting directly with each other, producing the most open structures). At room temperature, the ion-mobility experiment showed that these structures can freely convert to each other. Because our experiment was carried out at room temperature, the PES spectra of the dinucleotides should be considered to be the average of all these different structures. It is interesting to note that, regardless of the structures, the G base remains the lowest IP site in all the dinucleotides.
Fig. 5 shows the PES spectra of eight singly charged trinucleotide anions. All the trinucleotides have two phosphate groups, one of which is the negative charge carrier. The characteristic low-binding-energy band, the “fingerprint” of guanine, was observed again in all trinucleotides that contain the G bases. The spectrum of dGGG– is similar to that of dGG–, with a well defined and distinct low-energy band. The current data indicate that the HOMO of the G-containing trinucleotides should again be located on the G bases, giving rise to their low IPs. We note that the spectrum of dAAA– also displayed a weak low-binding-energy feature, with an ADE higher than that of dGGG– but lower than that of dCCC– and dTTT–, suggesting that the HOMO for dAAA– may also be localized on the A bases. The lower-binding-energy part of the four spectra in Fig. 5 Right was very broad, which was most likely caused by the coexistence of multiple isomers. Ionmobility experiments for three trinucleotides (dGTT–, dTGT–, and dTTG–) suggested that they each have two isomers, an “open” one and a “folded” one, which convert to each other at room temperature (30). In addition to these two conformers, the negative charge carrier can be on different phosphate groups, which can give conformers with different ADEs.
Fig. 5.
Photoelectron spectra of deprotonated singly charged trinucleotide anions dAAA– (a), dCCC– (b), dGGG– (c), dTTT– (d), dGGC– (e), dGCG– (f), dGCC– (g), and dCGC– (h) at 157 nm (7.866 eV).
In a real biological environment, the bases form Watson–Crick pairs and stack up in the double helix of DNA, whereas the phosphate groups are stabilized by counter ions and solvent molecules. Electron-transfer processes occur along the base stack. The current PES experimental results reveal that the guanine bases are the lowest IP sites in a variety of environments in the free oligonucleotide anions, providing direct spectroscopic support that they should be the lowest IP sites in DNA.
Conclusions
We report a photoelectron spectroscopic study on mononucleotide, dinucleotide, and trinucleotide singly charged anions in the gas phase to probe their intrinsic electronic structure and ionization properties. We observed a distinct low electron-binding energy band for all the nucleotides that contain one or more guanine bases. Density function theory calculations revealed that this unique spectral feature is caused by the ionization of a π orbital from the guanine base. This low ionization feature becomes a fingerprint for guanine-containing oligonucleotides and confirms that the guanine base is the lowest IP site among all the DNA bases. The current results provide direct spectroscopic evidence that guanine serves as the reducing agent in DNA and is the favorite site for oxidation and DNA damage.
Supplementary Material
Acknowledgments
We thank Prof. M. T. Bowers for valuable discussions about the structures of the nucleotides. This work was supported by the U.S. Department of Energy, Office of Basic Energy Sciences, Chemical Science Division. The experiment and calculations were performed at the W. R. Wiley Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the Department of Energy Office of Biological and Environmental Research and located at Pacific Northwest National Laboratory, which is operated for the Department of Energy by Battelle.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: IP, ionization potential; PES, photoelectron spectroscopy; VDE, vertical electron-detachment energy; ADE, adiabatic electron-detachment energy; dAMP–, 2′-deoxyadenosine 5′-monophosphate; dCMP–, 2′-deoxycytidine 5′-monophosphate; dGMP–, deoxyguanosine 5′-monophosphate; dTMP–, deoxythymidine 5′-monophosphate; HOMO, highest occupied molecular orbital.
References
- 1.Singer, B. & Grunberger, D. (1983) Molecular Biology of Mutagens and Carcinogens (Plenum, New York).
- 2.Melvin, T., Botchway, S. W., Parker, A. W. & O'Neill, P. (1996) J. Am. Chem. Soc. 118, 10031–10036. [Google Scholar]
- 3.Gorner, H. & Gurzadyan, G. G. (1993) Photochem. Photobiol. A Chem. 71, 155–160. [DOI] [PubMed] [Google Scholar]
- 4.Becker, D. & Sevilla, M. D. (1993) in Advances in Radiation Biology, eds. Lett, J. T. & Adler, H. (Academic: New York), Vol. 17, pp. 121–180. [Google Scholar]
- 5.Burrows, C. J. & Muller J. G. (1998) Chem. Rev. (Washington, D.C.) 98, 1109–1151. [DOI] [PubMed] [Google Scholar]
- 6.Boon, E. M. & Barton, J. K. (2002) Curr. Opin. Struct. Biol. 12, 320–329. [DOI] [PubMed] [Google Scholar]
- 7.Melvin, T., Plumb, M. A., Botchway, S. W., O'Neill, P. & Parker A. W. (1995) Photochem. Photobiol. 61, 584–591. [DOI] [PubMed] [Google Scholar]
- 8.Pullman, B. & Pullman, B. (1958) Proc. Natl. Acad. Sci. USA 44, 1197–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin, J., Yu, C., Peng, S., Akiyami, I., Li, K., Lee, L. K. & LeBreton, P. R. (1980) J. Phys. Chem. 84, 1006–1012. [Google Scholar]
- 10.Steenken, S. & Jovanovic, S. V. (1997) J. Am. Chem. Soc. 119, 617–618. [Google Scholar]
- 11.Jovanovic, S. V. & Simic, M. G. (1986) J. Phys. Chem. 90, 974–978. [Google Scholar]
- 12.Sugiyama, H. & Saito, I. (1996) J. Am. Chem. Soc. 118, 7063–7068. [Google Scholar]
- 13.Yoshioka, Y., Kitagawa, Y., Takano, Y., Yamaguchi, K., Nakamura, T. & Saito, I. (1999) J. Am. Chem. Soc. 121, 8712–8719. [Google Scholar]
- 14.Hendricks, J. H., Lyapustina, S. A., de Clercq, H. L., Snodgrass, J. T. & Bowen, K. H. (1996) J. Chem. Phys. 104, 7788–7791. [Google Scholar]
- 15.Desfrancois, C., Abdoul-Carime, H. & Schermann, J. P. (1996) J. Chem. Phys. 104, 7792–7794. [Google Scholar]
- 16.Schiedt, J., Weinkauf, R., Neumark, D. M. & Schlag, E. W. (1998) Chem. Phys. 239, 511–524. [Google Scholar]
- 17.Li, X., Cai, Z. & Sevilla, M. (2002) J. Phys. Chem. A 106, 1596–1603. [Google Scholar]
- 18.Dolgounitcheva, O., Zakrzewski, V. G. & Ortiz, J. V. (2002) Int. J. Quantum Chem. 90, 1547–1554. [Google Scholar]
- 19.Richardson, N. A., Gu, J., Wang, S., Xie, Y. & Schaefer, H. F. (2004) J. Am. Chem. Soc. 126, 4404–4411. [DOI] [PubMed] [Google Scholar]
- 20.Tasaki, K., Yang, X., Urano, S., Fetzer, S. & LeBreton, P. R. (1990) J. Am. Chem. Soc. 112, 538–548. [Google Scholar]
- 21.Kim, H. S., Yu, M., Jiang, Q. & LeBreton, P. R. (1993) J. Am. Chem. Soc. 115, 6169–6183. [Google Scholar]
- 22.Kim, H. S. & LeBreton, P. R. (1994) Proc. Natl. Acad. Sci. USA 91, 3725–3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fernando, H., Papadantonakis, G. A., Kim, N. S. & LeBreton, P. R. (1998) Proc. Natl. Acad. Sci. USA 95, 5550–5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu, Q. & LeBreton, P. R. (2000) J. Am. Chem. Soc. 122, 12824–12834. [Google Scholar]
- 25.Hillenkamp, F., Karas, M., Beavis, R. C. & Chait, B. T. (1991) Anal. Chem. 63, A1193–A1202. [DOI] [PubMed] [Google Scholar]
- 26.Fenn, J. B., Mann, N., Weng, C. K. & Wong S. F. (1990) Mass Spectrom. Rev. 9, 37–70. [Google Scholar]
- 27.Gidden, J. & Bowers, M. T. (2003) J. Phys. Chem. B 107, 12829–12837. [Google Scholar]
- 28.Gidden, J., Bushnell, J. E. & Bowers, M. T. (2001) J. Am. Chem. Soc. 123, 5610–5611. [DOI] [PubMed] [Google Scholar]
- 29.Gidden, J. & Bowers, M. T. (2002) Eur. Phys. J. D 20, 409–419. [Google Scholar]
- 30.Gidden, J. & Bowers, M. T. (2003) J. Am. Soc. Mass. Spectrom. 14, 161–170. [DOI] [PubMed] [Google Scholar]
- 31.Wang, L. S., Ding, C. F., Wang, X. B. & Barlow, S. E. (1999) Rev. Sci. Instrum. 70, 1957–1966. [Google Scholar]
- 32.Wang, L. S. & Wang, X. B. (2000) J. Phys. Chem. A 104, 1978–1990. [Google Scholar]
- 33.Wang, X. B., Yang, X. & Wang, L. S. (2002) Int. Rev. Phys. Chem. 21, 473–498. [Google Scholar]
- 34.High Performance Computational Chemistry Group (2002) nwchem, A Computational Chemistry Package for Parallel Computers (Pacific Northwest National Laboratory, Richland, WA), Version 4.5.
- 35.Andzelm, J. (1991) in Density Functional Methods in Chemistry, eds. Labanowski, J. & Andzelm, J. (Springer, New York), p. 155.
- 36.Ziegler, T. (1991) Chem. Rev. (Washington, D.C.) 91, 651–667. [Google Scholar]
- 37.Becke, A. D. (1993) J. Chem. Phys. 98, 5648–5652. [Google Scholar]
- 38.Lee, C., Yang, W. & Parr, R. G. (1988) Phys. Rev. B Condens. Matter 37, 785–789. [DOI] [PubMed] [Google Scholar]
- 39.Godbout, N., Salahub, D. R., Andzelm, J. & Wimmer, E. (1992) Can. J. Chem. 70, 560–571. [Google Scholar]
- 40.Black, G., Chase, J., Didier, B., Feller, D., Gracio, D., Jones, D., Keller, T., Matsumoto, S., Mendoza, E., Olander, M., et al. (1999) ecce, A Problem Solving Environment for Computational Software (Pacific Northwest National Laboratory, Richland, WA), Version 1.5.
- 41.Baerends, E. J. & Gritsenko, O. V. (1997) J. Phys. Chem. A 101, 5383–5403. [Google Scholar]
- 42.Stowasser, R. & Hoffmann, R. (1999) J. Am. Chem. Soc. 121, 3414–3420. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.