Abstract
Extracellular vesicles (EVs) have been shown to play an important role in intercellular communication as carriers of DNA, RNA and proteins. While the intercellular transfer of miRNA through EVs has been extensively studied, the stability of extracellular miRNA (ex-miRNA) once engulfed by a recipient cell remains to be determined. Here, we identify the ex-miRNA-directed phenotype to be transient due to the rapid decay of ex-miRNA. We demonstrate that the ex-miR-223-3p transferred from polymorphonuclear leukocytes to cancer cells were functional, as demonstrated by the decreased expression of its target FOXO1 and the occurrence of epithelial-mesenchymal transition reprogramming. We showed that the engulfed ex-miRNA, unlike endogenous miRNA, was unstable, enabling dynamic regulation and a return to a non-invasive phenotype within 8 h. This transient phenotype could be modulated by targeting XRN1/PACMAN exonuclease. Indeed, its silencing was associated with slower decay of ex-miR-223-3p and subsequently prolonged the invasive properties. In conclusion, we showed that the ‘steady step’ level of engulfed miRNA and its subsequent activity was dependent on the presence of a donor cell in the surroundings to constantly fuel the recipient cell with ex-miRNAs and of XRN1 exonuclease, which is involved in the decay of these imported miRNA.
INTRODUCTION
Extracellular vesicles (EVs), including exosomes, microvesicles, ectosomes, microparticles, are small, lipid bilayer membrane vesicles that are secreted by all cell types and are found in all biological fluids (1). Initially discovered as garbage bags for removal of unneeded material from cells, EVs are now recognized as major players in intercellular communication. EVs contain and shuttle bioactive molecules including DNA, mRNA as well as non-coding microRNAs (miRNAs) from one cell to another, leading to the reprogramming of the recipient cells (2,3). This genetic transfer is particularly important in cancer where the level of circulating miRNAs is dramatically increased in the blood of patients and correlated with tumor progression (4). It is now well appreciated that tumor-derived exosomes can hijack surrounding endothelial cells to support tumor angiogenesis, create an immunosuppressive micro-environment and mobilize polymorphonuclear leukocyte neutrophils (PMN) and stromal cells to promote a tumor niche. Although critical for therapeutic intervention, the exact mechanisms mediating the roles of EVs in cancer have not yet been fully elucidated. Because of their involvement in promoting disease progression, it is currently proposed that the uptake of EVs may induce persistent modulation of recipient cells (5).
Among the EV bioactive cargoes, miRNAs are an important component of gene regulation, eliciting either decay or translational repression of target mRNAs (6). With the potential to regulate more that 30% of human genes, miRNAs are critical in biology and particularly in every step of tumor development from initiation, progression, to metastasis, behaving either as tumor suppressors or oncogenic miRNAs (7). Control of these master regulators thus represents a fundamental aspect of gene regulation. While mechanisms of miRNA upregulation and processing have been well documented, little is known about how miRNAs are downregulated, a necessary facet of dynamic expression. Compelling studies have shown that miRNAs are highly stable in most cell types, with a half-life ranging from 28 to 220 h (8,9). Similarly to endogenous miRNA, ex-miRNA trapped within circulating EVs show remarkable stability (10). Based on these features, it has been postulated but not yet proven that ex-miRNA once engulfed into surrounding cells, are able to promote persistent pro-tumoral activities.
Nevertheless, this concept, i.e. stability of ex-miRNA, does not explain the dynamic cell plasticity observed during development and in human malignancies. To better define the function of ex-miRNA we decided to focus our attention on metastasis, a spatio-temporal process in which cancer cells undergo epithelial-mesenchymal transition (EMT) and its reversal, mesenchymal-epithelial transition (MET), as these cells escape from the primary tumor and take seed in a secondary organ (11,12). Recapitulating the driving role of inflammation in metastasis (13), we provide here the first evidence that the invasive properties of tumor cells are dependent on the secretion of EVs by PMN. MiRNA-223-3p enriched in PMN-derived exosomes activated in tumor cancer cells an EMT program that was reversed upon removal of inflammatory cells. One of molecular mechanisms underlying this transient tumor cell re-programming stems from the previously unrecognized instability of EV-derived miRNA mediated by the exonuclease XRN1. These results clearly provide new insight into the intricate cell communication occurring in the tumor micro-environment and into the emerging role of ex-miRNA in disease progression.
MATERIALS AND METHODS
Cell culture
The A549, NCI-H1975, NCI-H1299, SK-LU-1 and HeLa cells were cultured according to the recommendations of the ATCC. A549 (human lung adenocarcinoma epithelial cell line, ATCC, number CCL-185), NCI-H441 (human lung adenocarcinoma epithelial cell line, ATCC, HTB-174) and HeLa (human cervical adenocarcinoma epithelial cell line, ATCC, number CCL-2) cells were grown in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 50 U/ml penicillin and 50 μg/ml streptomycin (Life Technologies). PLB-985 (human myeloid cell line HL-60 subclone prone to PMN differentiation, gift from Pr Sylvie Chollet-Martin, Paris, France), NCI-H1975 (human lung adenocarcinoma epithelial cell line, ATCC, number CRL-5908) and NCI-H1299 (human lung carcinoma epithelial cell line, ATCC, number CRL-5803) cells were grown in RPMI-1640 supplemented with 10% FBS, 1 mM sodium pyruvate, 50 U/ml penicillin and 50 μg/ml streptomycin (Thermo Fisher Scientific). SK-LU-1 (human lung adenocarcinoma epithelial cell line, ATCC, number HTB-57) cells were grown in Minimum Essential Media supplemented with 10% FBS, 1 mM sodium pyruvate, 100 μM MEM Non-Essential Amino Acids, 50 U/ml penicillin and 50 μg/ml streptomycin (Life Technologies). All the cells were maintained for <25 passages at 37°C in a 5% CO2 and humidified atmosphere. All cell lines were authenticated after the first amplification or subcloning (here, stable miR-223-3p clone selection) by determining the genetic characteristics using polymerase chain reaction (PCR)-single-locus-technology (Promega, PowerPlex 21 PCR Kit) and certified (Eurofins, Eurofins Genomics, Ebersberg, Germany). Testing for mycoplasma contamination was performed monthly using PlasmoTest™-Mycoplasma Detection Kit (Invivogen).
Patient recruitment and PMN isolation
After approval by the local Ethics Committees (Nice University Hospital Center), written informed consent was obtained from all participants (healthy volunteers), after explaining the nature of the study. The study was performed according to the guidelines of the Declaration of Helsinki.
Human PMN were extracted from whole blood of healthy donors by Ficoll gradient centrifugation to remove peripheral blood mononuclear cells followed by gelatin sedimentation to remove red blood cells. Residual red blood cells were then lysed with isotonic ammonium chloride solution. After washing in Hanks' balanced saline solution without Ca2+ or Mg2+, the cells were counted and resuspended at 5.107 PMN/ml. PMN (95% pure) with 98% viability (assessed by trypan blue exclusion) were used within 1 h after isolation.
Reagents actinomycin D treatment
Cells were treated with actinomycin D (Act. D), to block RNA transcription, at 10 μg/ml. At indicated times, cells were lysed for miRNA analysis as described below. The activity of actinomycin D was confirmed by a decrease in MYC and the stability of GADPH mRNA as previously described (10). For miRNA analysis, we added spike-in cel-miR-39, -54 and -238 (miScript miRNA mimic, MSY0000010, MSY0000054, MSY0000293) before RNA extraction for unbiased normalization.
Supernatant production (SPN) and extracellular vesicle (EVs) purification
PMN and PLB-985 cells were incubated either in DMEM or RPMI-1640, both supplemented with 10% EV-free FBS for supernatant production (SPN). SPN was isolated from PMN conditioned media after 300 g centrifugation to remove cells and 2000 g to remove cell debris. SPN were then ultra-centrifugated at 100 000 g for 70 min at 4°C. After centrifugation, EV-free SPN was collected and stored for cell culture experiments. The pellet containing EVs was either resuspended in phosphate-buffered saline (PBS) for culture cell experiments or in lysis buffer for protein analysis (1).
Immunoblotting
Immunoblotting was performed as described previously (14). Whole cell lysates were extracted from cells using Laemmli lysis buffer (12.5 mM Na2HPO4, 15% glycerol, 3% sodium dodecyl sulphate). The protein concentration was measured with the DC Protein Assay (BIO-RAD) and 30 μg of total protein was loaded onto 7.5 or 11% sodium dodecyl sulphate-polyacrylamide gel electrophoresis and transferred onto polyvinylidene difluoride membranes (Millipore). After 1 h of blocking with 5% bovine serum albumin or non-fat milk prepared in TBS-Tween buffer, the blots were incubated overnight at 4°C with antibodies against FOXO1 (clone C29H4, 1:1000, Cell Signaling Technologies, ref #2880), XRN1 (1:1000, Bethyl Laboratories, ref A-300-443A), E-CADHERIN (clone 4A2C7, 1:500, Invitrogen, ref 33–4000), SNAIL (clone C15D3, 1:1000, Cell Signaling Technologies, ref #3879), SLUG (clone A-7, 1:400, Santa Cruz Biotechnology, ref sc-166476), VIMENTIN (clone SP20, prediluted, 1:100, Abcam, ab27608) and b-ACTIN (clone AC-40, 1:10 000, Sidma-Aldrich, ref A3853) used as loading control. After 1 h of incubation with a horseradish peroxidase-conjugated secondary antibody (1:6000, Santa Cruz Biotechnologies), protein bands were visualized using an enhanced chemiluminescence detection kit (Millipore) with the imaging system, Syngene Pxi4 (Syngene).
RNA isolation and Reverse Transcription qPCR (RT-qPCR) analysis
Total RNA extraction was performed using TRI-Reagent (Sigma-Aldrich) and QIAGEN RNAeasy kit. Isolation of RNA from cultured cells or human samples was performed as described previously (15,16). The RNA concentration was measured by NanoDrop 2000 (Thermo Fisher Scientific), the purity (A260/A280) of RNA was around 2.0 and the yield was in a range of 200–500 ng/μl. Then, mRNA were analyzed on a Bioanalyzer (Agilent) and only samples with RIN > 9 were used for further experiments.
For miRNA
Taqman miRNA assays (Applied Biosystems) were used to quantify the expression levels of mature hsa-miR-223-3p, hsa-miR-143-3p, U6 snRNA, cel-miR-39, cel-miR-54 and cel-miR-238. Specific miRNA-primed reverse transcription was carried out on 10 ng of total RNA using MultiScribe™ Reverse Transcriptase at 50 U/μl (Applied Biosystems) in a final volume of 15 μl. No DNase treatment was performed for miRNA analysis. RNA were treated with Rnase Inhibitor using a TaqMan® MicroRNA Reverse Transcription Kit (ref 4366597, Applied Biosystems): 30 min at 16°C, 30 min at 42°C, 5 min at 85°C and a pause at 4°C. The cDNA strand was synthesized using specific reverse transcription primers for U6 snRNA, miR-223-3p, miR-143-3p U6 snRNA, cel-miR-39, cel-miR-54 or cel-miR-238 (Thermo Fisher Scientific). The expression levels were analysed by RT-qPCR. RT-qPCR was performed in triplicate 10 μl per reaction on diluted cDNA (1 μl) with TaqMan® Fast Advanced Master Mix (2×), that contain AmpliTaq® Fast DNA Polymerase, using a StepOne™ Plus Real-Time PCR system (Applied Biosystems): 20 s at 95°C, [1 s at 95°C, 20 s at 60°C] for 40 cycles. The relative miR-223-3p and miR-143-3p expression levels were calculated for each sample after normalization against the endogenous expression level of the reference miRNA, U6 snRNA, using the ΔΔCt method for comparison of relative fold-expression differences with Applied Biosystems StepOne™ Software v2.3. For actinomycin D treatment, the cel-miRNA were used as normers instead of U6 snRNA.
For mRNA
multiplex oligo(dT)-primed reverse transcription was carried out on 500 ng DNase-treated total RNA using MultiScribe™ Reverse Transcriptase at 50 U/μl (Applied Biosystems) in a final volume of 20 μl. RNA were treated with DNase I (1 U) 15 min at room temperature, then DNase I was inactivated with ethylenediaminetetraacetic acid (2.5 mM) and 10 min at 65°C. RNA were also treated with RnaseOUT—Ribonuclease Inhibitor (ref 10777-019, Applied Biosystems) in high capacity cDNA Reverse Transcription Kit (ref 4368813, Applied Biosystems): 10 min at 25°C, 120 min at 37°C, 5 min at 85°C and a pause at 4°C. RT-qPCR was performed in triplicate with 10 μl per reaction on diluted cDNA (1 μl, 1/5) with Fast SYBR® Green Master Mix (2×), that contained AmpliTaq® Fast DNA Polymerase, using a StepOne™ Plus Real-Time PCR system (Applied Biosystems): 20 s at 95°C, [3 s at 95°C, 30 s at 60°C] for 40 cycles, 15 s at 95°C, 1 s at 60°C, 15 s at 95°C with temperature increment +0.3°C (for Melt Curve). PCR Primers (Sigma-Aldrich) used for quantification of FOXO1, MYC, GAPDH and RPLP0 genes were: FOXO1 forward (ACGAGTGGATGGTCAAGAGC) and reverse (CTGCACACGAATGAACTTGC); MYC forward (ATGAAAAGGCCCCCAAGGTA) and reverse (CGTTTCCGCAACAAGTCCTCT); GAPDH forward (ACCATGGAGAAGGCTGGGGC) and reverse (TGGACTGTGGTCATGAGTCC); RPLP0 forward (GCATCAGTACCCCATTCTATCAT) and reverse (AGGTGTAATCCGTCTCCACAGA), at a concentration of 250 nM. All targets were analyzed for efficiency and used in the range of linearity. The relative FOXO1, MYC, GAPDH expression levels were calculated for each sample after normalization against the endogenous expression level of the reference gene, RPLP0, using the ΔΔCt method for comparison of relative fold-expression differences with Applied Biosystems StepOne™ Software v2.3. cDNA were stored in DNase/RNase-free water at −20°C.
miRNA and siRNA transient transfection
Cells were plated at 200 000 cells/well in 6-well plates. After 24 h, cells were transfected with the mirVana miRNA mimic Control or miR-223-3p at 17 nM or with the mirVana miRNA antagomiR Control or miR-223-3p at 68 nM or with siRNA negative Control or Silencer Pre-designed siRNA XRN1 at 50 nM using Lipofectamine RNAiMAX (Life Technologies) according to the manufacturer's instructions. Forty-eight hours after transfection, cells were lysed for RNA or protein analysis as described below.
miRNA stable transfection
Lentiviral particles for hsa-miR-223 (Cat. #: PMIRH223PA-1) were purchased from System Biosciences. Lentivirus transduction was performed according to the manufacturer's instructions. Briefly, A549 cells were seeded onto a 24-well plate and cultured for 24 h to achieve 50% confluence. A549 cells were then transfected with lentivirus and washed the next day. The efficiency of infection was measured under a fluorescent microscope one week after the transfection and the cells were sorted using a flow cytometry. The sorted cells were used in the following experiments.
Dual luciferase reporter assay
HeLa cells were seeded at 10 000 cells/well in 96-well plates. After 24 h incubation, cells were transfected with the mirVana miRNA mimic Control or miR-223-3p. Twenty-four hours after transfection, cells were transfected with different constructs of psiCHECK™-2-FOXO1 3΄UTR (1.1; 1.2; 1.3 and 1.4). Forty-eight hours after transfection, cells were assayed by both firefly and renilla luciferase using the dual luciferase assay system (Promega) according to manufacturer's instructions. All transfection experiments were conducted in triplicate and repeated three times independently.
Cell invasion assays
In vitro cell invasion assays were performed using transwell chambers coated with matrigel (pore size of 8 μm; Corning). Cells were resuspended in serum-free medium, and 500 μl of the cell suspension (100 000 cells/transwell) was added to the upper chamber. A total of 750 μl of 5% FBS medium was added to the bottom wells of the chambers. After 16 h, cells were removed from the upper face of the filters using cotton swabs. After fixation in 4% paraformaldehyde and staining in a dye solution containing 0.4% crystal violet and 20% ethanol, cells that adhered to the lower membrane of the inserts were counted. Images of five different fields (200× magnification) were taken for each membrane, and the number of invaded cells was counted. All counts of the same experiments were divided by the mean of the control condition in order to calculate the migration index and the relative increase/decrease induced by treatments.
Statistical analysis
Quantitative data were described and presented graphically as medians and interquartiles or means and standard deviations. The distribution normality was tested with the Shapiro's test and homoscedasticity with a Bartlett's test. For two categories, statistical comparisons were performed using the Student's t-test or the Mann–Whitney's test. For three and more categories, analysis of variance (ANOVA) or non-parametric data with Kruskal–Wallis (KW) was performed to test variables expressed as categories versus continuous variables. If this test was significant, we used the Tukey's test or the Nemenyi's test to compare these categories.
Disease-free-survival was defined as the interval between the date of diagnosis and the date of discovery of the first evidence after treatment of any tumor (local, regional, metastatic or second primary) or death from any cause. Patients were recorded at the time of death or at the last follow-up. These data were estimated and presented graphically using the Kaplan–Meier method. The survival curves were compared using the log-rank test.
All statistical analyses were performed by the biostatistician using R.3.2.2 software and Prism6 program from GraphPad software. Tests of significance was two-tailed and considered significant with an alpha level of P < 0.05. (graphically: * for P < 0.05, ** for P < 0.01, *** for P < 0.001).
RESULTS
Ex-miR-223-3p contained in EVs and engulfed into tumor cells is active in recipient cancer cells
Here, we developed a model that mimics tumor–stroma interactions in which miRNA, specifically expressed in PMN (17,18), but not in epithelial cancer cells (Figure 1A), were transferred into lung cancer cells via either cell–cell contact or EV transfer.
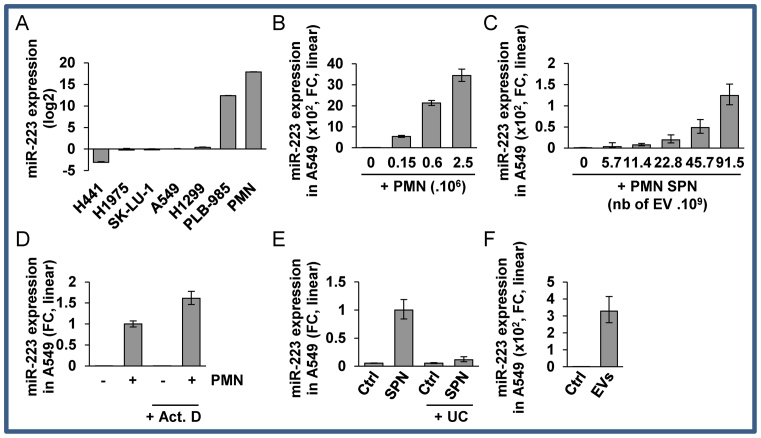
Figure 1.
Ex-miR-223-3p is engulfed into recipient cells (A) Relative quantification analysis of ex-miRNA-223-3p expression in H441, H1975, SK-LU-1, A549, H1299, PLB-985 and PMN cells. Cell lines were harvested at 70% confluency. PMN were isolated from blood as described in the ‘Materials and Methods’ section. (B) Relative quantification of the expression of ex-miR-223-3p in A549 cells after overnight co-culture with increasing numbers of PMN and extensive washes. (C) Relative quantification of the expression of ex-miR-223-3p in A549 cells following incubation with increasing amounts of SPN. (D) Relative quantification of the expression of ex-miR-223-3p in A549 cells following incubation with SPN or SPN depleted of EVs by ultra-centrifugation (UC). (E) Relative quantification of the expression of ex-miR-223-3p in A549 cells incubated overnight with EVs isolated from SPN of PLB-985 cells. (F) Relative quantification of the expression of ex-miR-223-3p in A549 cells co-cultured overnight with PMN and treated with actinomycin D (Act. D) at 10 μg/ml. Cells were extensively washed and harvested. (A–E) For all experiments, the levels of miR-223-3p were normalized using U6 snRNA. (F) Spike-in were used for normalization (see the ‘Materials and Methods’ section). (A–F) Results are representative of three biological replicates, ‘centre values’ as mean and error bars as s.d.
First, in order to show that PMN could transfer material to lung cancer cells, we stained the PMN with DiIC16, a lipid dye probe. After removal of excess dye, either stained PMN or their conditioned SPN were co-cultured with A549 cells (Supplementary Figure S1A and B). We observed lipid transfer from PMN to cancer cells that was cell-cell contact independent since the transfer could be reproduced with SPN only (Supplementary Figure S1B). Second, together with this lipid transfer, we then confirmed that PMN-specific miRNA miR-223-3p (17,18) were absent in cancer cells (Figure 1A) and transferred following incubation with PMN into lung cancer cells in a dose dependent manner (Figure 1B; Supplementary Figure S2A and B). This transfer was recapitulated in different cancer or immortalized cell types (Supplementary Figure S2D–F). Furthermore, we also observed the transfer of miR-143-3p, a PMN-specific miRNA expressed at lower levels than miR-223-3p (Supplementary Figure S2G), from PMN to cancer cells (Supplementary Figure S2H), underlying that transfer is not restricted to miR-223-3p.
At this stage, it was of interest to show that transfer could also occur in a dose-dependent manner with SPN only (Figure 1C). To determine the component of SPN that was associated with transfer, we tested if it was specifically dependent on EVs. First, we detected little, if any, miRNA transfer when EV-depleted SPN was added to lung cancer cells (Figure 1D). Thus, we isolated EVs from granulocytic PLB-985 cell lines and we showed that their size profile was remarkably similar to the PMN SPN profile (Supplementary Figure S1C and D) and that they displayed classical enrichment for CD63, CD82 and leukocyte-specific CD53 (Supplementary Figure S1E). Then, we showed that the EVs were indeed responsible for lipid (Supplementary Figure S1F) and miRNA (Figure 1E) transfer between PMN to cancer cells.
Moreover, to ascertain that PMN or their supernatant did not transfer a factor responsible for miR-223-3p expression in recipient cells, we analyzed miR-223-3p accumulation during actinomycin D-mediated transcriptional inhibition in cancer cells (Figure 1F). The efficiency of transcription inhibition was confirmed by RT-qPCR of the unstable c-MYC proto-oncogene (Supplementary Figure S2I). As expected, a significant reduction in the c-MYC mRNA levels was detected following 8 h of transcriptional shutoff, while the stable GAPDH mRNA was still present at a high level (Supplementary Figure S2I). Of note, we noticed that U6, which is frequently used as a reference miRNA, was sensitive to actinomycin D in A549 cells (Supplementary Figure S2J), so to analyse miRNA expression, we added spike-in cel-miR-39, -54 and -238 before RNA extraction for unbiased normalization. We showed that miR-223-3p accumulation was not impaired by actinomycin D confirming that miR-223-3p was indeed transferred from PMN (Figure 1F).
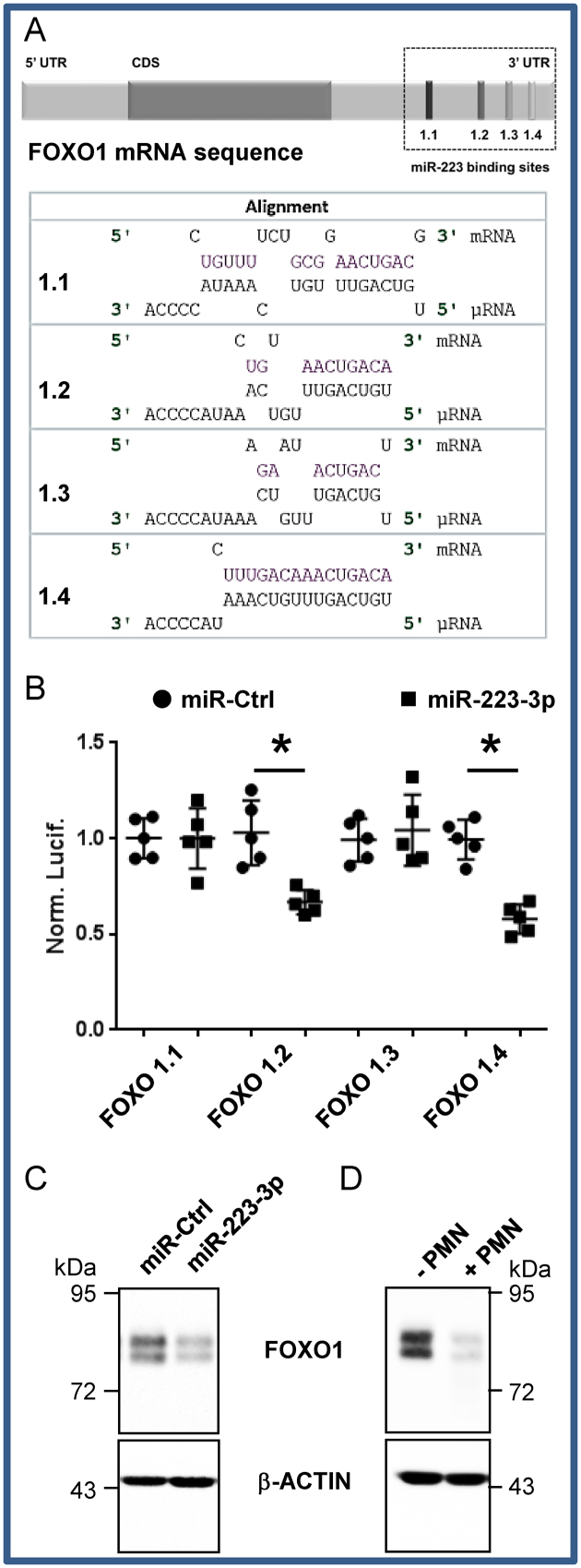
We then checked whether the ex-miR-223-3p transferred into recipient cells were functional by analyzing the expression of FOXO1, a bona fide miR-223-3p target (19) that contains four potential miR-223-3p binding sites in its 3΄UTR (Figure 2A). We cloned the 450 bp region surrounding each potential binding sites and found that two of them were functionally targeted, as assessed by post-transcriptional repression by miR-223-3p in a site-specific luciferase reporter assay (Figure 2B). Finally, the FOXO1 protein was silenced in A549 cells either treated with miR-223-3p (Figure 2C) or co-cultured with PMN (Figure 2D) indicating that miR-223-3p molecules transferred from PMN to lung cancer cells were indeed functional in the recipient cells.
Figure 2.
FOXO1 is a bona fide target of miR-223-3p (A) In silico prediction of miR-223-3p and FOXO1 3΄UTR mRNA interaction. (B) Each potential binding site was evaluated independently using the dual luciferase assay as presented in the ‘Materials and Methods’ section. Each dot corresponds to a biological replicate. The Renilla/Firefly ratios were normalized on paired to miR-Ctrl conditions (Norm. Lucif.). Data represent the quantification of three biological replicates, ‘centre values’ as mean and error bars as s.d. * for P < 0.05. (C) FOXO1 expression in A549 cells after 48 h transfection with miR-223-3p. (D) FOXO1 expression in A549 cells co-cultured overnight with PMN. (C and D) FOXO1 expression was analysed by immunoblotting. β-ACTIN served as reference for loading.
Engulfed ex-miR-223-3p is active in tumor cells and promotes EMT and invasion
Because miRNA target numerous mRNA in cells, following only one target may not resume all the observed phenotypic changes, so we thus focused on a global ‘gain of function phenotype’ that was miR-223-3p dependent. We show here that transfection of miR-223-3p into A549 cells induced impaired cell adhesion and an EMT phenotype, as suggested by transcriptome analysis with GO_Cellular_Component via numerous potential targets of miR-223-3p (Supplementary Table S1 and Supplementary Figure S3A and B), and as verified by the accumulation of mesenchymal markers like SNAIL, SLUG and VIMENTIN proteins (Supplementary Figure S3C), and E-cadherin relocation and actin remodeling (Supplementary Figure S3D).
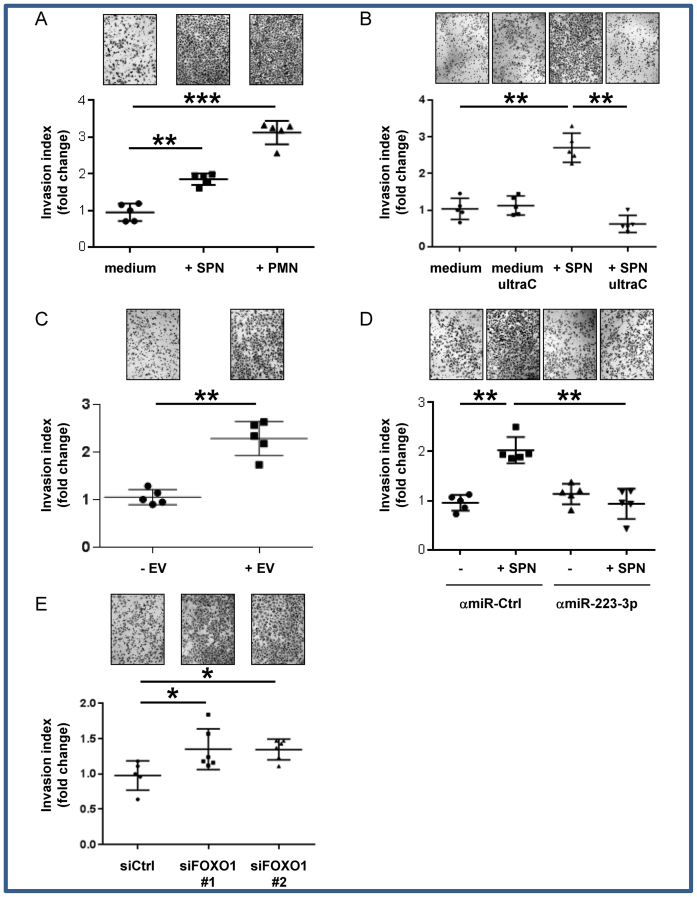
Accordingly, PMN or SPN of PMN both enhanced the invasion of A549 or H1975 cells through matrigel chambers (Figure 3A and Supplementary Figure S4A respectively). Of note, this PMN-driven EMT of A549 cells was independent of reactive oxygen species (Supplementary Figure S4B) but dramatically rescued by EV removal from the SPN (Figure 3B), suggesting the relevant paracrine contribution of EVs. Consistently, we confirmed that A549 cells incubated with EVs isolated from granulocytic PLB-985 cells had an increased invasive behavior (Figure 3C). The effect of EVs on A549 cell invasion was recapitulated by the transfection of miR-223-3p (Supplementary Figure S4C) and completely lost after depletion of miR-223-3p by transfection with specific miRNA-antagomir-223-3p in PMN-treated cells (Figure 3D). Furthermore, we tested the effect of FOXO1 silencing on migration and showed that siFOXO1 modestly induced invasion (Figure 3E). This result showed that miR-223-3p-induced invasion is not entirely explained by FOXO1 silencing suggesting a multi-target additive effect that promotes EMT and invasion.
Figure 3.
Engulfed miR-223-3p transfer induces enhanced invasive properties (A) In vitro invasion assay of A549 cells co-cultured either with PMN or their conditioned supernatant produced in serum-free medium. (B) In vitro invasion assay of A549 cells co-cultured with conditioned supernatant depleted of EVs by ultra-centrifugation (UltraC). (C) In vitro invasion assay of A549 cells co-cultured with EVs isolated from PLB-985 cells. (D) In vitro invasion assay of antagomiR-223-3p (amiR-223-3p) transfected A549 cells co-cultured with SPN of PMN (E) In vitro invasion assay of A549 cells transiently transfected either with Ctrl siRNA (siCtrl) or FOXO1 siRNA (siFOXO). (A–E) Each dot corresponds to a biological replicate. * for P < 0.05, ** for P < 0.01, *** for P < 0.001.
Engulfed ex-miR-223-3p rapid decay after donor cell removal is associated with transient EMT
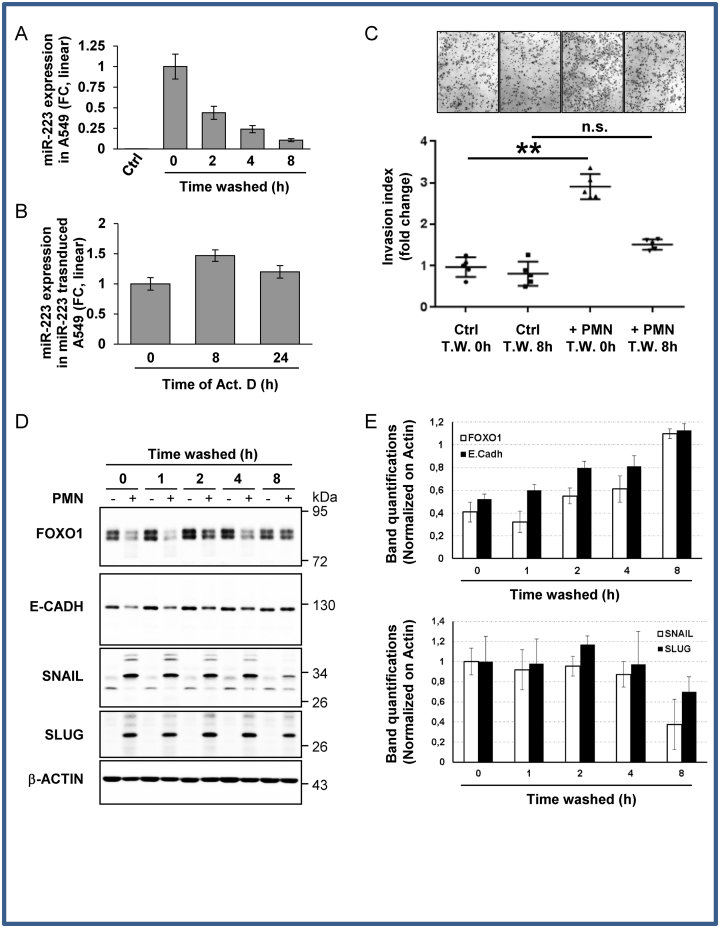
It has been demonstrated that tumor epithelial cells could be reprogrammed to adopt a transient mesenchymal phenotype by miRNA transfer (20). However, the molecular mechanisms that govern this epithelial plasticity remain poorly understood. Since heterotypic cell interactions are highly dynamic within the tumor micro-environment, we questioned the stability of the ex-miRNA once the donor cell had been removed. Ex-miRNAs are present in blood plasma and serum in a remarkably stable form. However, this resistance, especially for miR-223-3p, was mainly due to membrane protection rather than RISC-associated protection by nucleases (21). Here, we discovered, in the different tested tumor recipient cell lines, that the level of exosomal delivered miR-223-3p rapidly decreased with a half-life of 1.2 h (+/− 0.4 h) (Figure 4A), in contrast to the 24 h half-life of stably transfected miR-223-3p in actinomycin D-treated A549 cells (Figure 4B). These data reveal different fates for ex-miRNA and cellular miRNA. Along with the decline in ex-miRNA found in different cell lines (Supplementary Figure S5A–C), the miR-223-3p driven responses, i.e. FOXO1 silencing, decreased epithelial markers (E-cadherin) and enhanced mesenchymal markers (SNAIL, SLUG), as well as the invasive behavior, could be gradually rescued overtime up to complete recovery within 8 h after PMN removal (Figure 4C–E). FOXO1 recovery was not related to an increase in transcription suggesting that FOXO1 rescue was dependent on miRNA decay (Supplementary Figure S5D). In addition, we showed that the decrease was not restricted to miR-223-3p since miR-143-3p also decreased in A549 cells after donor cell removal (Supplementary Figure S5E).
Figure 4.
Engulfed miR-223-3p is quickly decayed in recipient cells after donor cell removal. (A) Time-dependent decay of ex-miRNA-223-3p expression in A549 cells. Following overnight co-culture with PMN, A549 cells were harvested at the indicated time periods after PMN removal. The level of miR-223-3p was measured by RT-qPCR and normalized using U6 snRNA. Results are representative of three biological replicates, ‘centre values’ as mean and error bars as s.d. (B) RT-qPCR analysis of miRNA-223-3p expression in stably miR-223 transduced A549 cells treated with actinomycin D (Act. D) at 10 μg/ml for the indicated period of time. Spikes-in were used for normalization (see the ‘Materials and Methods’ section). (C) In vitro invasion assay of A549 cells post washes. A549 cells co-cultured with (+PMN) or without PMN (Ctrl) overnight were collected at the indicated time post initial washes (Time Washed, T.W.) and seeded, in the upper part of transwells. The number of cells attached to the bottom of a Matrigel-coated membrane after 16 h was quantified after crystal violet staining. Data represent the quantification of five biological replicates, ‘centre values’ as mean and error bars as s.d. (D) Immunoblot analysis of FOXO1 and EMT marker expression levels upon PMN removal from A459 cells co-cultured with (+) or without (−) PMN. β-ACTIN served as reference for loading. (E) Quantifications of gels from three independents experiments are presented. * for P < 0.05, ** for P < 0.01.
Engulfed ex-miR-223-3p stability and EMT longevity depend on XRN1 levels
To uncover the origin of this unexpected instability of ex-miRNA, we focused on the mechanism of decay of miRNA, which remains unclear in mammals. Some studies have highlighted the instability of some specific animal miRNAs (10,22). A potential decay mechanism described in Arabidopsis implicated the 3΄-to-5΄ exonucleases SDN (23) and more recently Dis3 in Drosophila (24) and the 5΄-to-3΄ exoribonucleases XRN-1 and XRN-2 in Caenorhabditis elegans (25–27).
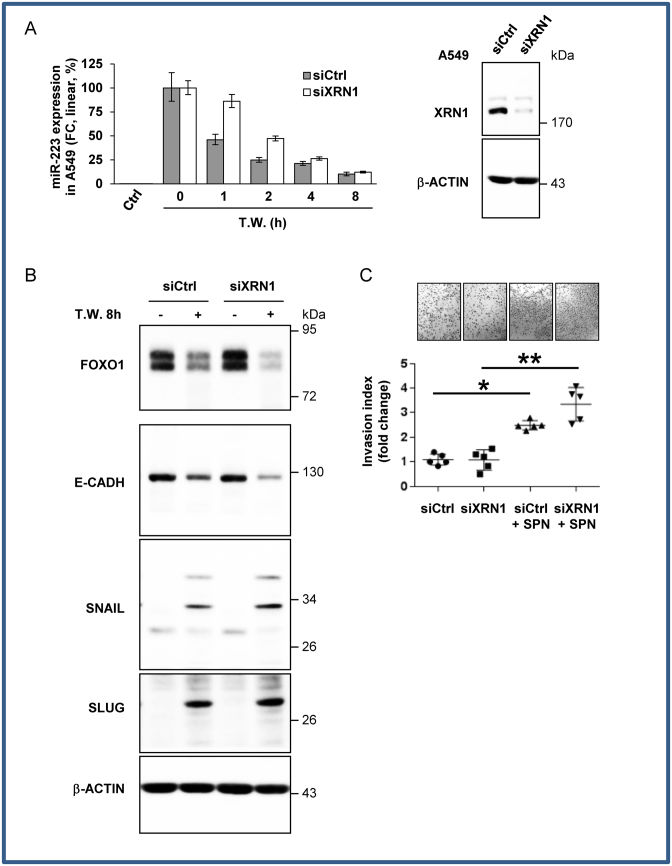
After siRNA screening (Supplementary Figure S6) for these exonuclease isoforms in humans, XRN1 appeared to have a major impact on ex-miR-223-3p decay in our model, increasing its half-life to 2.45 h +/− 0.3 (Figure 5A). Consequently, XRN1 knock-out also affected fluctuations in the miR-223-3p targets up to 4 h upon PMN removal. FOXO1 expression levels were lower in siXRN1-transfected cells than in siCtrl-transfected cells after 4 h (Figure 5B). This finding was also corroborated by increased expression of the mesenchymal SNAIL and SLUG markers and decreased expression of E-cadherin in siXRN1-treated cells incubated with SPN (Figure 5B). Along with these observations, we showed that the siXRN1-transfected cells had a higher invasion capacity than siCtrl-treated cells in the presence of supernatant of PMN (Figure 5C). Furthermore, we checked if the observed effects were not associated with phenotypic changes due to siXNR1 depletion in cancer cells. We showed that siXRN1 neither perturbed proliferation of A549 cells (Supplementary Figure S7A) nor modified the invasive properties (Supplementary Figure S7B). Moreover, we showed that XRN1 was not sensitive to actinomycin D treatment within 24 h, suggesting that the ‘endogenous’ miR-223-3p stability in stable-miR-223-3p cell lines could not be explained by depletion of XRN1 during actinomycin D treatment (Supplementary Figure S7C).
Figure 5.
XRN1 regulates ex-miRNA decay in recipient cells (A) Relative quantification analysis of ex-miRNA-223-3p in A549 cells. siXRN1-transfected A549 cells co-cultured with PMN overnight were harvested at the indicated periods of time post PMN removal (Time Washed, T.W.). Results are representative of three biological replicates. In the right panel, immunoblot analysis of XRN1 expression. b-ACTIN served as an equal loading control. (B) Immunoblot analysis of FOXO1 and EMT marker expression levels. β-ACTIN served as an equal loading control. (C) In vitro invasion assay of siXRN1-transfected A549 cells. A549 cells co-cultured with SPN of PMN, produced in serum-free medium, were seeded in the upper part of transwells. The number of cells attached to the bottom of a Matrigel-coated membrane after 16 h was quantified after crystal violet staining. Data represent the quantification of five biological replicates, ‘centre values’ as mean and error bars as s.d. * for P < 0.05, ** for P < 0.01.
DISCUSSION
Regulation of miRNA levels in normal physiology as in cancer is often exclusively studied at the transcription and processing steps. However, we show here that an additional and essential level of control occurs via ex-miRNA decay. Indeed, we present evidence that PMN release EVs that contain functional miRNAs, which are transferred into cancer cells. Specifically, ex-miR-223-3p induces lung cancer cell reprogramming as demonstrated by induction of the EMT signature and the acquisition of a more invasive phenotype. While the transfer of EV-derived miRNAs from tumor cells to the inflammatory micro-environment has been well-documented (28), our study provides the first evidence of ex-miRNA trafficking from PMN to tumor cells. The role of miRNA in cellular EMT and tumor metastases has been well-established, irrespective of the miRNA nature, i.e. endogenous or derived from EVs (29,30). In agreement with this, we show that the presence of miR-223-3p, following both stable transfection or EVs transfer, activates EMT. Importantly, our results reveal that ex-miR-223-3p triggered an unexpected sequential EMT–MET mechanism. Mechanistically, we demonstrated that the ex-miR-223-3p are extremely labile miRNA, rapidly degraded by XRN1 following removal of EVs producing PMN.
There are several implications in our understanding of the unique proprieties of ex-miRNA. First, the remarkable instability of ex-miRNAs suggests the ex-miRNAs are the specific target of miRNA-binding proteins or nucleases that control their stability, thus fine-tuning their effectiveness. So far, very little information is currently available regarding the decay of miRNAs and even less for the ex-miRNA once engulfed into recipient cells. By silencing all known exonucleases, we identified XRN1 as the 5΄-3΄exoribonuclease 1 that specifically targets ex-miR-223-3p in human tumor cells. Knockdown of XRN1 counteracted EV-induced MET, consistent with ex-miR-223-3p stabilization. In mammals, Bail et al. showed that endogenous miRNA presented a discrepancy in terms of stability after actinomycin D treatment. Moreover, they showed the involvement of XRN1, and not XRN2, in the decay of endogenous miR-382 (10). Since miR-223-3p has been shown to be protected within the EV and quickly degraded by RNAse A after EV permeabilization in a cell-free system (21), our findings together with these results suggest that XRN1 is predominantly targeting unprotected miRNA in cells. This effect may arise from at least two different possibilities: either a given miRNA is lacking some post-transcriptional modification like 5΄-phosphate (31) allowing different pools of miRNA within the cell with one more susceptible for degradation or a miRNA is differentially ‘protected’ depending on its different degree of affinity for miRNA-associated proteins like AGO2. Unfortunately, our miRNA sequencing results did not clearly identified the ‘code signal’ for decay at the origin of the differential miRNA susceptibility (data not shown) and further studies that clarify this ‘code signal’ are of great interest.
Second, our study points to the observation that the ex-miRNA and endogenous miRNA do not function in a similar way, as originally expected. In the literature, miRNAs function as key oncogenic and tumor suppressor miRNA. It has been inferred that the stable expression of microRNA mimics the responses triggered by the engulfed ex-miRNA. However, we found that the uptake of ex-miR-223-3p activated rapid EMT followed by a delayed MET in tumor cells, in sharp contrast to the maintenance of EMT of miR-223-transfected cells. We reconciled these apparently contradictory results by showing that ex-miR-223-3p was highly sensitive to degradation whereas the ectopically expressed miR-223-3p were resistant. These findings point to the important notion that the efficacy of ex-miRNA in recipient cells is the net result of a dynamic balance between uptake and degradation.
Third, the sequential EMT–MET of lung cancer cells shown herein following ex-miRNA decay recapitulates the metastasis process. While there is no doubt that EMT is associated with worse prognosis, the cancer cells concurrently express multiple EMT markers, which render them difficult to transfer for diagnostic or prognostic purposes. A previous study suggests that miR-223-3p promotes invasion of breast cancer cells via regulation of MEF2C. In our experiments, we confirmed that both extracellular transferred- and ectopically expressed-miR-223-3p promote cell invasion, however these effects were independent of MEF2C, which was not expressed in our cellular models (data not shown). Our microarray analysis showed rather that miR-223-3p targets a large panel of genes, including FOXO1, which are involved in cell migration. In fact, the worst clinical outcomes of metastatic cancers were recently associated with a dynamic ‘variance gradient’ ranging from EMT phenotypes. In consequence, we propose a model in which the regulation of ex-miRNA decay is particularly important during tumor metastasis, where the plasticity of tumor cells between the epithelial and mesenchymal states requires selective and programmed decay of miRNAs far from their inflammatory site of production. Along this line, the observation that the chromosomal loss of XRN1 was associated with poor disease free survival (logrank, P-value = 0.0219, Hazard Ratio: 1.453, 95% CI: 1.065–2.168) in lung adenocarcinoma (The Cancer Genome Atlas TCGA-LUAD data collection) is of great interest (Supplementary Figure S8).
In conclusion, our results suggest that EVs released into the micro-environment contain miRNA that could help cancer cell invade the surrounding tissue. We showed that the ‘steady step’ level of engulfed miRNA and its subsequent activity is dependent of the presence of donor cells in the surrounding to constantly fuel the recipient cell with extracellular miRNAs and that the level of XRN1 exonuclease is involved in the decay of these imported miRNA.
AVAILABILITY/ACCESSION NUMBER
The raw sequence and processed data have been submitted to the NCBI Gene Expression Omnibus (GEO) under accession number GSE81370.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Katia Havet-Zahaf, Salomé Lalvée, Eric Selva, Julien Fayada, Pascal Grier, Ludovic Cervera for technical help. The authors acknowledge the technological expertise of the CytoMed, the IRCAN's Flow Cytometry Facility, the Nice Sophia-Antipolis Functional Genomics Platform and the Nice Hospital Related Biobank (BB 0033-00025).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
French Government (Agence Nationale de Recherche, ANR) through the ‘Investments for the Future’ LABEX SIGNALIFE [ANR-11-LABX-0028-01]; ANR Jeunes Chercheurs Jeunes Chercheuses [ANR-12- JSV1-0009- 01]; Infrastructure France-Génomique [ANR-10-INBS-09-03]; Fondation ARC pour la recherche sur le cancer ARC SL220110603478; CANC’AIR Genexposomic project, Canceropole PACA; Region PACA (France); Conseil Général 06; FEDER; Ministère de l’Enseignement Supérieur; Région Provence Alpes-Côte d’Azur; INSERM.
Conflict of interest statement. None declared.
REFERENCES
- 1. Thery C., Amigorena S., Raposo G., Clayton A.. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006; doi:10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
- 2. Rahman M.A., Barger J.F., Lovat F., Gao M., Otterson G.A., Nana-Sinkam P.. Lung cancer exosomes as drivers of epithelial mesenchymal transition. Oncotarget. 2016; 7:54852–54866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vlassov A.V., Magdaleno S., Setterquist R., Conrad R.. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim. Biophys. Acta. 2012; 1820:940–948. [DOI] [PubMed] [Google Scholar]
- 4. Sanfiorenzo C., Ilie M.I., Belaid A., Barlesi F., Mouroux J., Marquette C.H., Brest P., Hofman P.. Two panels of plasma microRNAs as non-invasive biomarkers for prediction of recurrence in resectable NSCLC. PLoS One. 2013; 8:e54596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang X., Yuan X., Shi H., Wu L., Qian H., Xu W.. Exosomes in cancer: small particle, big player. J. Hematol. Oncol. 2015; 8:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zamore P.D., Tuschl T., Sharp P.A., Bartel D.P.. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000; 101:25–33. [DOI] [PubMed] [Google Scholar]
- 7. Croce C.M., Calin G.A.. miRNAs, cancer, and stem cell division. Cell. 2005; 122:6–7. [DOI] [PubMed] [Google Scholar]
- 8. Gantier M.P., McCoy C.E., Rusinova I., Saulep D., Wang D., Xu D., Irving A.T., Behlke M.A., Hertzog P.J., Mackay F. et al. Analysis of microRNA turnover in mammalian cells following Dicer1 ablation. Nucleic Acids Res. 2011; 39:5692–5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang Z., Qin Y.W., Brewer G., Jing Q.. MicroRNA degradation and turnover: regulating the regulators. Wiley Interdiscip. Rev. RNA. 2012; 3:593–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bail S., Swerdel M., Liu H., Jiao X., Goff L.A., Hart R.P., Kiledjian M.. Differential regulation of microRNA stability. RNA. 2010; 16:1032–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Spaderna S., Schmalhofer O., Hlubek F., Berx G., Eger A., Merkel S., Jung A., Kirchner T., Brabletz T.. A transient, EMT-linked loss of basement membranes indicates metastasis and poor survival in colorectal cancer. Gastroenterology. 2006; 131:830–840. [DOI] [PubMed] [Google Scholar]
- 12. Thiery J.P., Acloque H., Huang R.Y., Nieto M.A.. Epithelial-mesenchymal transitions in development and disease. Cell. 2009; 139:871–890. [DOI] [PubMed] [Google Scholar]
- 13. Liu Y., Gu Y., Han Y., Zhang Q., Jiang Z., Zhang X., Huang B., Xu X., Zheng J., Cao X.. Tumor exosomal RNAs promote lung pre-metastatic niche formation by activating alveolar epithelial TLR3 to recruit neutrophils. Cancer Cell. 2016; 30:243–256. [DOI] [PubMed] [Google Scholar]
- 14. Brest P., Lapaquette P., Souidi M., Lebrigand K., Cesaro A., Vouret-Craviari V., Mari B., Barbry P., Mosnier J.F., Hébuterne X. et al. A synonymous variant in IRGM alters a bindingsite for miR-196 and causes deregulationof IRGM-dependent xenophagy in Crohn'sdisease. Nat Genet. 2011; 43:242–245. [DOI] [PubMed] [Google Scholar]
- 15. Pottier N., Maurin T., Chevalier B., Puissegur M.P., Lebrigand K., Robbe-Sermesant K., Bertero T., Lino Cardenas C.L., Courcot E., Rios G. et al. Identification of keratinocyte growth factor as a target of microRNA-155 in lung fibroblasts: implication in epithelial-mesenchymal interactions. PLoS One. 2009; 4:e6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Triboulet R., Mari B., Lin Y.L., Chable-Bessia C., Bennasser Y., Lebrigand K., Cardinaud B., Maurin T., Barbry P., Baillat V. et al. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science. 2007; 315:1579–1582. [DOI] [PubMed] [Google Scholar]
- 17. Allantaz F., Cheng D.T., Bergauer T., Ravindran P., Rossier M.F., Ebeling M., Badi L., Reis B., Bitter H., D’Asaro M. et al. Expression profiling of human immune cell subsets identifies miRNA-mRNA regulatory relationships correlated with cell type specific expression. PLoS One. 2012; 7:e29979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Haneklaus M., Gerlic M., O’Neill L.A., Masters S.L.. miR-223: infection, inflammation and cancer. J. Intern. Med. 2013; 274:215–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wu L., Li H., Jia C.Y., Cheng W., Yu M., Peng M., Zhu Y., Zhao Q., Dong Y.W., Shao K. et al. MicroRNA-223 regulates FOXO1 expression and cell proliferation. FEBS Lett. 2012; 586:1038–1043. [DOI] [PubMed] [Google Scholar]
- 20. Srivastava S.P., Koya D., Kanasaki K.. MicroRNAs in kidney fibrosis and diabetic nephropathy: roles on EMT and EndMT. Biomed. Res. Int. 2013; 2013:125469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li L., Zhu D., Huang L., Zhang J., Bian Z., Chen X., Liu Y., Zhang C.Y., Zen K.. Argonaute 2 complexes selectively protect the circulating microRNAs in cell-secreted microvesicles. PLoS One. 2012; 7:e46957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rissland O.S., Hong S.J., Bartel D.P.. MicroRNA destabilization enables dynamic regulation of the miR-16 family in response to cell-cycle changes. Mol. Cell. 2011; 43:993–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ramachandran V., Chen X.. Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science. 2008; 321:1490–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Towler B.P., Jones C.I., Viegas S.C., Apura P., Waldron J.A., Smalley S.K., Arraiano C.M., Newbury S.F.. The 3΄-5΄ exoribonuclease Dis3 regulates the expression of specific microRNAs in Drosophila wing imaginal discs. RNA Biol. 2015; 12:728–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bosse G.D., Ruegger S., Ow M.C., Vasquez-Rifo A., Rondeau E.L., Ambros V.R., Grosshans H., Simard M.J.. The decapping scavenger enzyme DCS-1 controls microRNA levels in Caenorhabditis elegans. Mol. Cell. 2013; 50:281–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chatterjee S., Fasler M., Bussing I., Grosshans H.. Target-mediated protection of endogenous microRNAs in C. elegans. Dev. Cell. 2011; 20:388–396. [DOI] [PubMed] [Google Scholar]
- 27. Chatterjee S., Grosshans H.. Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature. 2009; 461:546–549. [DOI] [PubMed] [Google Scholar]
- 28. Thery C., Ostrowski M., Segura E.. Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 2009; 9:581–593. [DOI] [PubMed] [Google Scholar]
- 29. Tang J., Li Y., Wang J., Wen Z., Lai M., Zhang H.. Molecular mechanisms of microRNAs in regulating epithelial-mesenchymal transitions in human cancers. Cancer Lett. 2016; 371:301–313. [DOI] [PubMed] [Google Scholar]
- 30. Kosaka N. Decoding the Secret of Cancer by Means of Extracellular Vesicles. J. Clin. Med. 2016; 5:e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Salzman D.W., Nakamura K., Nallur S., Dookwah M.T., Metheetrairut C., Slack F.J., Weidhaas J.B.. miR-34 activity is modulated through 5΄-end phosphorylation in response to DNA damage. Nat. Commun. 2016; 7:10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.