Abstract
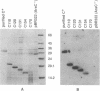
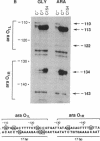
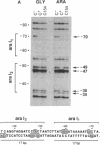
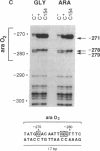
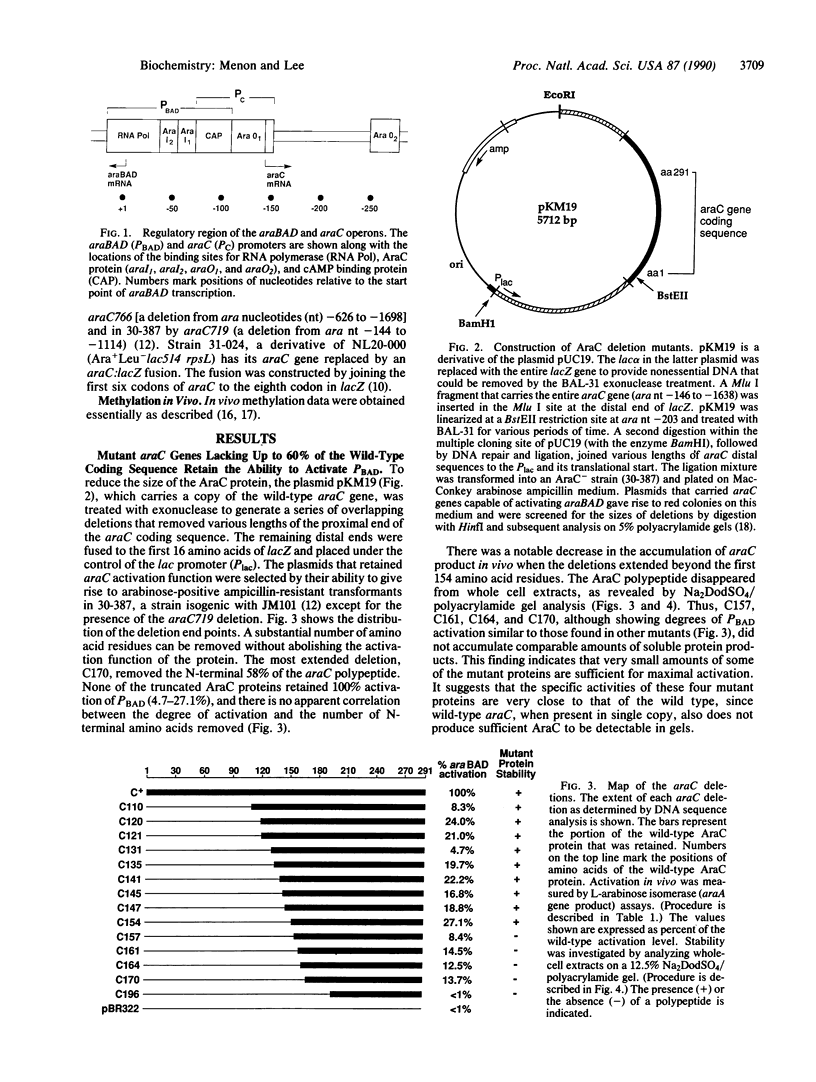
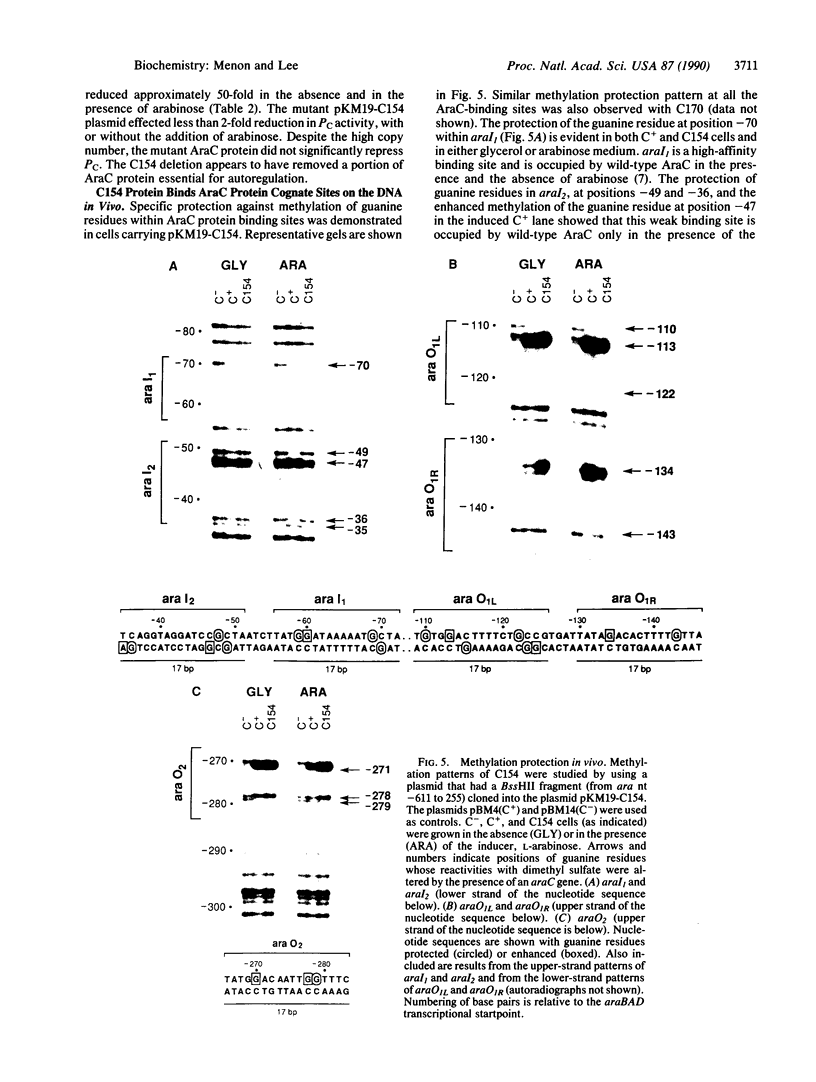
The araC gene of Escherichia coli encodes a protein that binds the inducer L-arabinose to activate the transcription of three ara operons. In a study to determine the functional domains within the AraC protein, we have generated a set of overlapping deletions from the proximal end of the araC gene. We found that the removal of up to nearly 60% of the coding sequence of this protein still allows transcriptional activation of the ara operons in vivo, up to 27% that of the wild type. These truncated proteins, however, no longer require arabinose for induction. The ligand-induced conformational change apparently either releases or unmasks an existing functional domain within AraC, rather than generating a new conformation that is required for activation of the promoter of araBAD. Since the truncated protein of the mutant C154 (which lacks 153 amino acid residues from the N terminus) retains DNA binding specificity, the DNA-recognition domain is localized in the C-terminal half of the AraC protein. Truncated proteins were unable to repress araBAD or araC in vivo, even though they were able to bind all ara operators. We propose that the N-terminal half of AraC is essential for the formation of the DNA loops that are responsible for repression of araBAD and for autoregulation of araC.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aiba H., Krakow J. S. Isolation and characterization of the amino and carboxyl proximal fragments of the adenosine cyclic 3' ,5'-phosphate receptor protein of Escherichia coli. Biochemistry. 1981 Aug 4;20(16):4774–4780. doi: 10.1021/bi00519a038. [DOI] [PubMed] [Google Scholar]
- Borowiec J. A., Gralla J. D. High-resolution analysis of lac transcription complexes inside cells. Biochemistry. 1986 Sep 9;25(18):5051–5057. doi: 10.1021/bi00366a012. [DOI] [PubMed] [Google Scholar]
- Borowiec J. A., Zhang L., Sasse-Dwight S., Gralla J. D. DNA supercoiling promotes formation of a bent repression loop in lac DNA. J Mol Biol. 1987 Jul 5;196(1):101–111. doi: 10.1016/0022-2836(87)90513-4. [DOI] [PubMed] [Google Scholar]
- Brown C. E., Hogg R. W. A second transport system for L-arabinose in Escherichia coli B-r controlled by the araC gene. J Bacteriol. 1972 Aug;111(2):606–613. doi: 10.1128/jb.111.2.606-613.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J. Regulation of the regulatory gene for the arabinose pathway, araC. J Mol Biol. 1976 Jul 5;104(3):557–566. doi: 10.1016/0022-2836(76)90120-0. [DOI] [PubMed] [Google Scholar]
- Cass L. G., Wilcox G. Mutations in the araC regulatory gene of Escherichia coli B/r that affect repressor and activator functions of AraC protein. J Bacteriol. 1986 Jun;166(3):892–900. doi: 10.1128/jb.166.3.892-900.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn T. M., Hahn S., Ogden S., Schleif R. F. An operator at -280 base pairs that is required for repression of araBAD operon promoter: addition of DNA helical turns between the operator and promoter cyclically hinders repression. Proc Natl Acad Sci U S A. 1984 Aug;81(16):5017–5020. doi: 10.1073/pnas.81.16.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynan W. S., Tjian R. The promoter-specific transcription factor Sp1 binds to upstream sequences in the SV40 early promoter. Cell. 1983 Nov;35(1):79–87. doi: 10.1016/0092-8674(83)90210-6. [DOI] [PubMed] [Google Scholar]
- Englesberg E., Irr J., Power J., Lee N. Positive control of enzyme synthesis by gene C in the L-arabinose system. J Bacteriol. 1965 Oct;90(4):946–957. doi: 10.1128/jb.90.4.946-957.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francklyn C. S., Lee N. AraC proteins with altered DNA sequence specificity which activate a mutant promoter in Escherichia coli. J Biol Chem. 1988 Mar 25;263(9):4400–4407. [PubMed] [Google Scholar]
- Geisler N., Weber K. Isolation of amino-terminal fragment of lactose repressor necessary for DNA binding. Biochemistry. 1977 Mar 8;16(5):938–943. doi: 10.1021/bi00624a020. [DOI] [PubMed] [Google Scholar]
- Godowski P. J., Rusconi S., Miesfeld R., Yamamoto K. R. Glucocorticoid receptor mutants that are constitutive activators of transcriptional enhancement. Nature. 1987 Jan 22;325(6102):365–368. doi: 10.1038/325365a0. [DOI] [PubMed] [Google Scholar]
- Guarente L. Regulatory proteins in yeast. Annu Rev Genet. 1987;21:425–452. doi: 10.1146/annurev.ge.21.120187.002233. [DOI] [PubMed] [Google Scholar]
- Hahn S., Schleif R. In vivo regulation of the Escherichia coli araC promoter. J Bacteriol. 1983 Aug;155(2):593–600. doi: 10.1128/jb.155.2.593-600.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton E. P., Lee N. Three binding sites for AraC protein are required for autoregulation of araC in Escherichia coli. Proc Natl Acad Sci U S A. 1988 Mar;85(6):1749–1753. doi: 10.1073/pnas.85.6.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenberg S. M., Giguere V., Segui P., Evans R. M. Colocalization of DNA-binding and transcriptional activation functions in the human glucocorticoid receptor. Cell. 1987 Apr 10;49(1):39–46. doi: 10.1016/0092-8674(87)90753-7. [DOI] [PubMed] [Google Scholar]
- Huo L., Martin K. J., Schleif R. Alternative DNA loops regulate the arabinose operon in Escherichia coli. Proc Natl Acad Sci U S A. 1988 Aug;85(15):5444–5448. doi: 10.1073/pnas.85.15.5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lichenstein H. S., Hamilton E. P., Lee N. Repression and catabolite gene activation in the araBAD operon. J Bacteriol. 1987 Feb;169(2):811–822. doi: 10.1128/jb.169.2.811-822.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin K., Huo L., Schleif R. F. The DNA loop model for ara repression: AraC protein occupies the proposed loop sites in vivo and repression-negative mutations lie in these same sites. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3654–3658. doi: 10.1073/pnas.83.11.3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathanson N. M., Schleif R. Paucity of sites mutable to constitutivity in the araC activator gene of the L-arabinose operon of Escherichia coli. J Mol Biol. 1975 Jul 25;96(1):185–199. doi: 10.1016/0022-2836(75)90190-4. [DOI] [PubMed] [Google Scholar]
- Novotny C. P., Englesberg E. The L-arabinose permease system in Escherichia coli B/r. Biochim Biophys Acta. 1966 Mar 28;117(1):217–230. doi: 10.1016/0304-4165(66)90169-3. [DOI] [PubMed] [Google Scholar]
- Ohlendorf D. H., Anderson W. F., Matthews B. W. Many gene-regulatory proteins appear to have a similar alpha-helical fold that binds DNA and evolved from a common precursor. J Mol Evol. 1983;19(2):109–114. doi: 10.1007/BF02300748. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T., Sturtevant J. M., Ptashne M. The lambda repressor contains two domains. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1608–1612. doi: 10.1073/pnas.76.4.1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raibaud O., Schwartz M. Positive control of transcription initiation in bacteria. Annu Rev Genet. 1984;18:173–206. doi: 10.1146/annurev.ge.18.120184.001133. [DOI] [PubMed] [Google Scholar]