Abstract
Riboswitches have gained attention as tools for synthetic biology, since they enable researchers to reprogram cells to sense and respond to exogenous molecules. In vitro evolutionary approaches produced numerous RNA aptamers that bind such small ligands, but their conversion into functional riboswitches remains difficult. We previously developed a computational approach for the design of synthetic theophylline riboswitches based on secondary structure prediction. These riboswitches have been constructed to regulate ligand-dependent transcription termination in Escherichia coli. Here, we test the usability of this design strategy by applying the approach to tetracycline and streptomycin aptamers. The resulting tetracycline riboswitches exhibit robust regulatory properties in vivo. Tandem fusions of these riboswitches with theophylline riboswitches represent logic gates responding to two different input signals. In contrast, the conversion of the streptomycin aptamer into functional riboswitches appears to be difficult. Investigations of the underlying aptamer secondary structure revealed differences between in silico prediction and structure probing. We conclude that only aptamers adopting the minimal free energy (MFE) structure are suitable targets for construction of synthetic riboswitches with design approaches based on equilibrium thermodynamics of RNA structures. Further improvements in the design strategy are required to implement aptamer structures not corresponding to the calculated MFE state.
INTRODUCTION
Riboswitches are non-coding RNA molecules regulating gene expression in response to binding of small ligand molecules. As they have no need for additional protein factors, riboswitches represent a valuable tool for gene regulation in the field of synthetic biology. Since their first discovery in 2002 (1–3), around 20 riboswitch classes have been described (4,5). Located mostly in the 5΄-UTR of prokaryotic mRNAs, riboswitches bind ligands with high affinity and specificity. Regulation of gene expression includes mechanisms such as transcription termination, translation initiation and control of mRNA self-cleavage (6,7). Despite these mechanistic differences, all riboswitches share a common layout consisting of two domains: the aptamer region binds the target molecule and acts as the sensory domain, while the regulatory or effector domain influences gene expression (7,8). This modular nature of riboswitches holds the potential for constructing synthetic riboswitches and reprogramming cells by replacing the aptamer domain with another aptamer–ligand-pair (9). In vitro selection procedures like SELEX enable researchers to isolate aptamer sequences that in theory can target any desired ligand molecule, rendering synthetic riboswitches an ideal tool to manipulate gene expression at the level of transcription or translation (10–12).
However, most of the selected aptamer-ligand pairs are not suitable for artificial riboswitches, as many ligands are not cell-permeable or toxic for the target organism, or their antibiotic character limits their use to eukaryotic organisms. As a consequence, only few in vitro selected aptamers have been incorporated in synthetic riboswitches, e.g. for neomycin (13–15), theophylline (16–20) and tetracycline (21–23). Other strategies use combinations of natural aptamer domains and expression platforms as chimeric riboswitches (24,9) or modified aptamers recognizing ligand analogues orthogonally to their natural counterparts (25,26). For the vast majority of in vitro selected aptamers, the conversion into new synthetic riboswitches remains difficult, since the fusion to the expression platforms resulting in functional regulatory devices is rarely straight-forward. Since most natural effector domains have specific sequence requirements, they do not function well in combination with aptamer domains other than their natural counterparts they co-evolved with. As a consequence, the creation of functional riboswitches often involves adaptation of the effector domain, e.g. by randomization of sequence parts and subsequent in vitro or in vivo selection and screening processes (27,28). Hence, several recent studies focus on RNA regulators that do not contain an aptamer domain and act in trans, like toehold switches (29) or small trans activating RNAs (30,31). Yet, synthetic riboswitches acting in cis have several important advantages over trans-acting RNA regulators. Trans-regulating RNAs clearly show a concentration-dependent efficiency and usually are required at high copy numbers in the cell. Further, they can have considerable side effects, when mRNAs other than the target transcript are bound as well. Riboswitches do not exhibit such off-target effects and show a high speed of response, as their regulation mode occurs exclusively in cis.
Recently, the implementation of aptamers into synthetic cis-acting riboswitches has been improved by several rational approaches. Computational methods based on secondary structure and folding prediction circumvent the need for large sequence libraries (19,24,32). In a de novo design strategy, a theophylline aptamer sequence was combined with a downstream located synthetic terminator sequence and fused to the 5΄-UTR of a reporter gene (19). This resulted in functional synthetic riboswitches in Escherichia coli, regulating transcription termination in a theophylline-dose-dependent way. The riboswitches have been employed for conditional gene expression of β-galactosidase and eGFP reporters and operate under the control of inducible (PBAD) as well as constitutive promoters (Prrn) (19,33). Here, we investigate the applicability of this design principle and replace the input (aptamer) domain of these synthetic riboswitches by in vitro selected aptamers for tetracycline and streptomycin (34,35). Furthermore, we designed a logic AND gate consisting of theophylline- and tetracycline-dependent riboswitches and identified important pitfalls that should be considered for successful computer-based riboswitch construction.
MATERIALS AND METHODS
Chemicals
Oligonucleotides were obtained from biomers.net, dNTPs from Jena Biosciences and [γ-32P]-ATP from Hartmann Analytic. LB medium was purchased from AppliChem. ONPG, streptomycin sulphate salt, doxycycline hydrochloride, tetracycline hydrochloride and theophylline were obtained from Sigma Aldrich. Ampicillin and arabinose were purchased from Carl Roth.
In silico design of synthetic riboswitches
A rational design algorithm was used to predict riboswitch sequences for the streptomycin (35) and tetracycline (34) aptamer as previously described for the design of synthetic theophylline riboswitches (19). Spacer sequences and lengths were randomized, and terminator 3΄-parts adapted accordingly. Calculations of free energy values and secondary structure predictions were performed with the RNAfold program of the Vienna RNA package (36).
Plasmid construction
Riboswitch DNA constructs were generated by overlap extension polymerase chain reaction (PCR) and fused into the 5΄-UTR of a bgaB reporter gene in pBAD2-bgaBshift (19) by site-directed mutagenesis PCR. Serial arrangements of Tet–RS constructs were generated by site-directed mutagenesis and fused to the bgaB reporter gene in pBAD2_bgaBshift, according to Wachsmuth et al. (33). Sequences of riboswitch constructs used in this study are shown in Supplementary Data.
Resistance of E. coli Top10 cells to tetracycline was achieved by co-expression of the ribosomal protection protein TetM together with tetracycline riboswitches. For this purpose, the on-site cat gene of pACYCDuet™-1 (Novagen/EMD Millipore) was replaced with a tetM gene from Enterococcus faecalis (37) by restriction cloning with SpeI and XmaI, resulting in a new pACYC-tetM vector (Supplementary Data). Primers used for this cloning strategy are shown in Supplementary Data.
Quantitative analysis of β-galactosidase activity
Quantitative analysis of β-galactosidase activity was performed in 2–3 independent experiments according to Wachsmuth et al. (19), using tetracycline- or streptomycin-resistant E. coli Top10 (F- mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu) 7697 galU galK rpsL (StrR) endA1 nupG λ-) with starter cultures grown between 16 and 24 h to obtain identical OD600. Activation rates were calculated separately for each construct as Miller units (MU) in the presence of ligand divided by MU in absence of ligand.
Structural probing of RNA
Streptomycin aptamer and Strep-RS7 RNAs were generated by in vitro transcription with T7 RNA polymerase and subsequent polyacrylamide (PAA) gel purification and ethanol precipitation. RNA 5΄-ends were dephosphorylated using antarctic phosphatase (NEB) and labeled with [γ-32P]-ATP (Hartmann Analytic) using T4 polynucleotide kinase (NEB) according to manufacturer's protocols. Structural analysis of 10 000 cpm 5΄-end labeled RNA per reaction was performed via in-line probing (38) in the presence of 50 mM Tris-HCl (pH 8.5), 12 mM MgCl2, 0.1 mg/ml tRNA from E. coli MRE 600 (Roche Diagnostics) and 0–10 mM streptomycin. All purification steps were carried out on 12.5% or 15% denaturing PAA gels. Imaging of 32P-labeled RNA was performed with a Typhoon 9410 Variable Imager (GE Healthcare).
RESULTS
In silico design of synthetic riboswitches
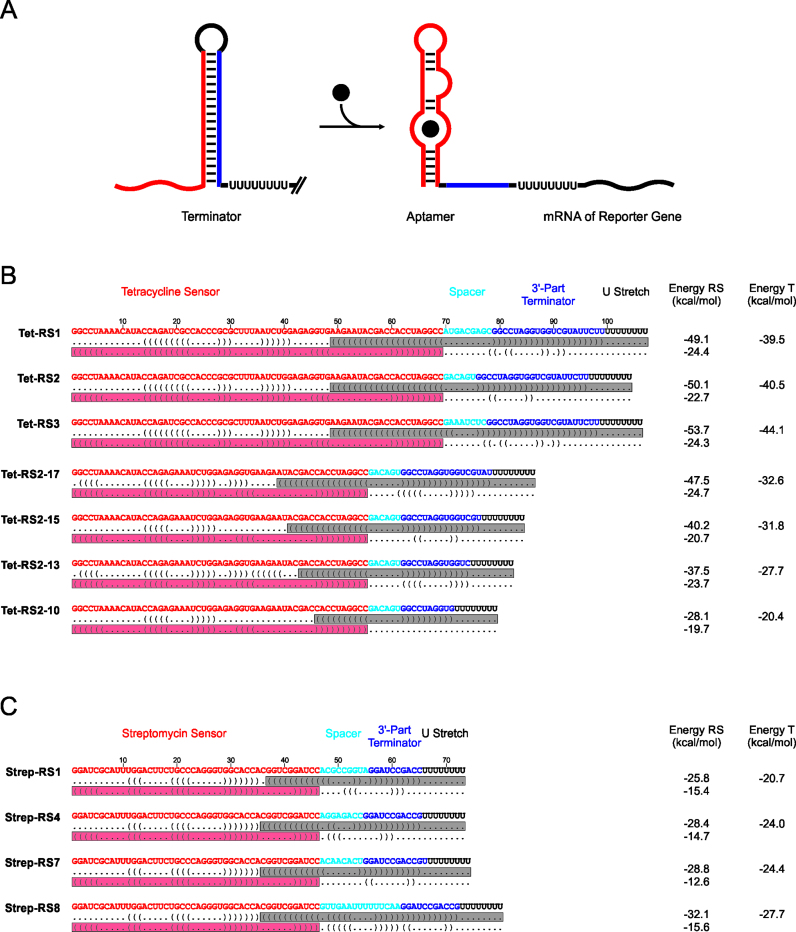
In a previous study, synthetic riboswitches regulating transcription termination in a theophylline-dependent way in E. coli were constructed with a computational approach based on secondary structure prediction (19). To investigate the usability of this design strategy with different SELEX-derived aptamer elements, we applied this approach for the generation of synthetic riboswitches based on tetracycline and streptomycin aptamers (34,35) (Figure 1).
Figure 1.
Synthetic riboswitches for regulation of transcription termination in Escherichia coli based on de novo design. (A) Design principle. In the absence of ligand, the 3΄-part of the aptamer domain (red) forms a terminator hairpin with the reverse complementary part (blue), leading to transcription termination. Binding of the corresponding ligand (full black circle) stabilizes the aptamer conformation and disrupts the terminator structure, enabling transcription of the reporter gene. (B) Tetracycline-dependent riboswitch (RS) candidates from secondary structure predictions. Sequences include the tetracycline aptamer ‘cb32sh’ in red (40), a connecting spacer region in cyan, the complementary part for the aptamer in blue and a black U-stretch. Given below each sequence are the terminator and aptamer conformations in dot-bracket annotation (grey and red bars, respectively). Calculated free energy values of the competing structures and the terminator elements are indicated. (C) Riboswitch candidates for the streptomycin aptamer ‘motif 1’ (35), including predicted free energy values. Color code is according to (B).
According to the recently described design parameters (19,33), riboswitch candidates were constructed based on randomly generated spacer sequences with lengths between 6 and 20 nucleotides (nts). Intrinsic terminator hairpins were formed by combining these spacers with sequences that exhibit perfect complementarity to the 3΄-part of the aptamer. The terminators were completed by an immediately downstream located stretch of eight U residues (U8). For the tetracycline aptamer, the start position of the complementary region was shifted from 34 to 59 and from 23 to 36 for the streptomycin aptamer. In brief, the design approach consists of a candidate generation and an in silico evaluation step. Initial prediction and filtering resulted in 25 tetracycline and 11 streptomycin riboswitch candidates, respectively. For these tetracycline candidates, spacer lengths varied from 6 to 18 and start positions of the complementary region ranged from position 43 to 49. The corresponding numbers for streptomycin are 6 to 15 and 33 to 36. The most promising candidates were cloned into a pBAD2-bgaBshift vector (19) and tested in E. coli Top10 cells for their ability to regulate gene expression of a downstream bgaB reporter.
Design and activity test of tetracycline riboswitch reporter constructs
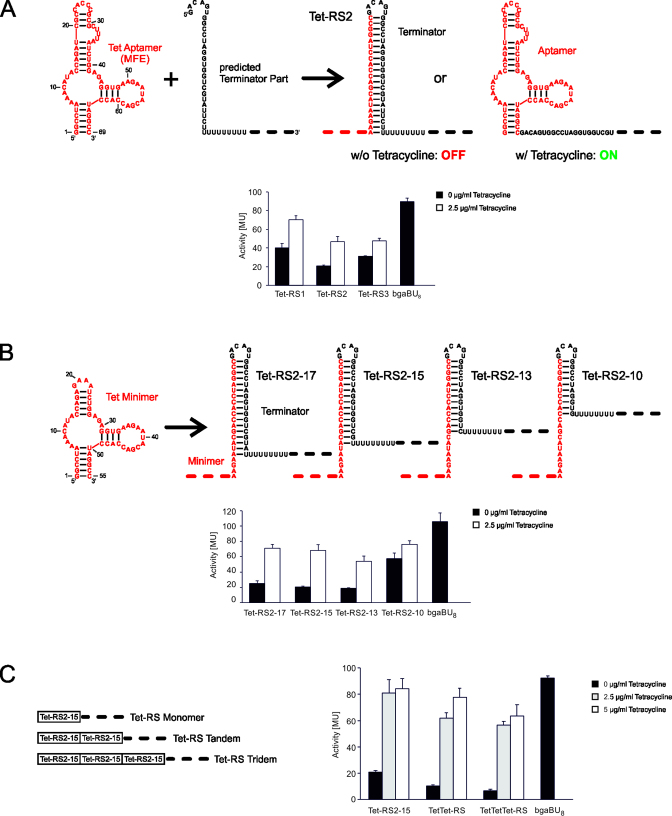
For the in silico design of synthetic riboswitches responding to tetracycline, the well-characterized aptamer cb32sh (34) was chosen as sensing platform. Conformation and ligand-binding of this aptamer have been studied extensively by structure probing (39), ITC (40), EPR (41) and fluorescence spectroscopy (42), showing a strong similarity to the minimal free energy (MFE) structure predicted by RNAfold (36,43) (Figure 2A).
Figure 2.
Activity tests of synthetic tetracycline riboswitches. (A) Design and activity of in silico predicted constructs. In the presence of 2.5 μg/ml tetracycline, expression of β-galactosidase was induced in E. coli, resulting in an increase of Miller units (MU). bgaBU8 represents the appropriate positive control (19,33), consisting of a plasmid expressing the reporter gene under the same promoter but lacking the aptamer and the terminator hairpin. Only the U stretch is present, as this can have an effect on the reporter gene expression. (B) Optimized Tet-RS2 constructs. The terminator hairpin stability of Tet-RS2 was modified by deleting residues from the 3΄-end of the hairpin. In the activity test, these modifications led to an increased response ratio for Tet-RS2-17, Tet-RS2-15 and Tet-RS2-13, while in Tet-RS2-10, the terminator hairpin is too unstable to compete with the aptamer fold. Accordingly, the construct is in a constitutive ON state, independent of tetracycline binding. Furthermore, this behavior is a direct indication that the riboswitch constructs regulate at the level of transcription (19). (C) Serial arrangements of two and three Tet-RS2-15 repeats increase the ligand-dependent responsiveness. The existence of two or three intrinsic terminator elements leads to a reduction of background gene expression in the absence of tetracycline. While the monomeric Tet-RS2-15 shows an absolute activation of 80 MU that is not further increased by a higher tetracycline concentration, tandem and tridem show a somewhat lower absolute response (60–80 MU), with some increase at the higher ligand concentration. As a result, the activation ratios for the tandem and tridem are considerably increased compared to the monomeric riboswitch form.
Based on this MFE structure, the prediction yielded a set of potential riboswitch candidates (Figure 1B). Tet-RS1, Tet-RS2 and Tet-RS3 were cloned into the 5΄-UTR of a bgaB reporter and tested for tetracycline-dependent gene regulation. In E. coli, all three candidates respond to tetracycline in the medium (2.5 μg/ml) and show a 1.8-, 2.2- and 1.5-fold increase in β-galactosidase activity, respectively (Figure 2A). Hence, these constructs represent functional riboswitches.
To achieve a higher ligand response ratio, the tetracycline riboswitches were further optimized, based on our experimental experience with the theophylline riboswitches (19,33). In these studies, we demonstrated that constructs can be improved by adjusting the stability of the terminator hairpin relative to the aptamer domain (19,33). Due to the size (69 nts) and stability (ΔG = −19.8 kcal/mol) of the tetracycline aptamer, terminator hairpins of the corresponding riboswitches have to be more stable than terminators in theophylline riboswitches (theophylline aptamer: 42 nts, ΔG = −12.2 kcal/mol) in order to compete with the aptamer structure. As Tet-RS2 showed the best response ratio in the initial β-galactosidase activity test, this construct was further optimized by destabilization of the corresponding terminator hairpin. A similar approach involving base pair deletions successfully converted a non-functional theophylline-specific riboswitch into a functional construct (33). However, in the case of Tet-RS2, it was not possible to delete complete base pairs at the top of the terminator stem, since these nucleotides are part of the tetracycline aptamer (Figure 2A). Instead, the hairpin was shortened by stepwise deletion of 3΄-terminal sequences of the aptamer-complementary region, resulting in Tet-RS2-17, Tet-RS2-15, Tet-RS2-13 and Tet-RS2-10 (numbers indicate remaining base pairs in the terminator hairpin; Figure 2B). For cloning reasons, a shorter version of the tetracycline aptamer was used, where nucleotides 19–36 have been replaced by a GAAA tetraloop, resulting in the cb32sh minimer (40). This tetracycline minimer displays the same folding and binding characteristics as the parental aptamer and does not affect riboswitch functionality, as a β-galactosidase activity test showed almost identical MU values and activation rates as in a comparable full-length construct (Supplementary Data).
Quantitative analysis of β-galactosidase activity of these terminator hairpin deletion variants revealed that Tet-RS2-17, Tet-RS2-15 and Tet-RS2-13 display similar background activities as Tet-RS2 in the absence of tetracycline, but show significantly higher MU values in the presence of the ligand, leading to increased response ratios of 2.5-, 3.4- and 3.0-fold, respectively (Figure 2B). Obviously, a medium terminator stem length of 15 bp leads to the most efficient tetracycline-dependent riboswitch regulation. A further shortening of the hairpin down to 10 bp abolishes terminator function and, consequently, riboswitch-mediated regulation of bgaB gene expression, resulting in constitutive gene expression, even in the absence of tetracycline (Tet-RS2-10, Figure 2B). These results are in agreement with those observed for synthetic theophylline riboswitches (19,33).
A further way to optimize the ligand response is to combine several repeats of the riboswitch construct 5΄ of the gene to be regulated. For tandem and tridem repeats of theophylline riboswitches, we could demonstrate that the serial arrangement of transcription terminators leads to a reduced background in gene expression, and, consequently, to an increase in the ligand-dependent response ratio (33). To investigate whether this is also true for tetracycline-dependent riboswitches, we generated serial repeats with two and three copies of Tet-RS2-15 upstream of the bgaB reporter gene (Figure 2C). Similar to the theophylline-riboswitch repeats, both constructs had a further decreased background activation in the absence of tetracycline, with values of 10 (TetTet-RS) and 7 MU (TetTetTet-RS), compared to 21 MU for the monomeric riboswitch (Figure 2C). Ligand-dependent gene expression was only slightly reduced in the riboswitch repeats, resulting in improved response ratios of 7.5x (TetTet-RS) and 9.5x (TetTetTet-RS).
To demonstrate the tetracycline specificity of the synthetic Tet-RS constructs, the structurally related drug doxycycline was used in a control experiment with Tet-RS2-15. Doxycycline has the same antibiotic properties as tetracycline, but is not bound by the aptamer and therefore represents an ideal negative control molecule (44,40,23). As expected, Tet-RS2-15 showed no doxycycline-dependent activation of bgaB (Supplementary Data).
These results demonstrate that the in silico design approach followed by structure-guided experimental optimization can be applied for non-theophylline aptamers as well, showing that functional transcriptional riboswitches can be rationally designed not only in the context of different promoters or reporter genes, but also for different aptamers.
Construction and testing of streptomycin riboswitches
In a second experimental setup, a streptomycin aptamer (35) was subjected to the riboswitch design approach (Figure 1C). From the resulting 11 candidates, four were cloned into the pBAD2_bgaBshift vector and tested in the presence of 1 mg/ml streptomycin in E. coli Top 10 cells carrying an intrinsic streptomycin resistance based on an altered ribosomal target site (S12 protein, rpsL StrR). Hence, as in the case of the above mentioned tetracycline resistance, the ligand is not destroyed or transported out of the cell.
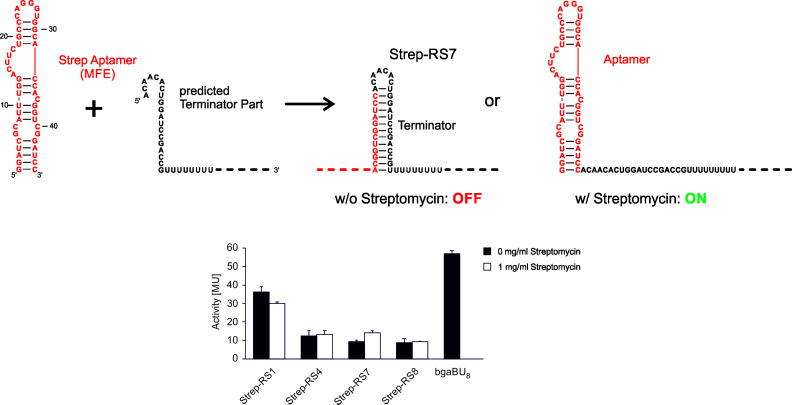
Riboswitch candidates Strep-RS1, Strep-RS4 and Strep-RS8 showed no response at all to streptomycin, and for Strep-RS7, only a very moderate 1.5-fold upregulation of the reporter gene expression was observed (Figure 3). While Strep-RS1 showed a weak constitutive reporter gene expression (30–40 MU), Strep-RS4 and Strep-RS8 are in a permanent OFF state (10–15 MU). These values correlate with the corresponding terminator stabilities (Figure 1C), where Strep-RS1 carries the least stable hairpin element (ΔG = −20.7 kcal/mol versus −24.0 kcal/mol and −27.7 kcal/mol for Strep-RS4 and Strep-RS8, respectively). The terminator of the slightly active Strep-RS7 has an intermediate stability (ΔG = −24.4 kcal/mol).
Figure 3.
Design and activity test of synthetic streptomycin riboswitches. Measurement of β-galactosidase activity in MU was performed in the presence (1 mg/ml) or absence of streptomycin. While constructs Strep-RS 1, 4 and 8 showed no ligand-dependent response, Strep-RS7 exhibited a ligand-dependent 1.5-fold increase in gene expression. bgaBU8: positive control without riboswitch (see legend Figure 2).
As variation of the streptomycin concentration in the medium from 0.05 mg/ml up to 5 mg/ml had no effect on the reporter gene activation (data not shown), the single weakly responding construct Strep-RS7 was selected for structure-guided optimization. Our investigations on theophylline- and tetracycline-dependent riboswitch constructs indicate that the terminator stability is one of the major factors affecting the regulatory potential of the constructs (33). Hence, similar to our strategy for Tet-RS2 constructs, the terminator stem of Strep-RS7 was destabilized by the introduction of mismatch positions and 3΄-terminal deletions (Supplementary Data). However, these modifications did not improve the functionality of Strep-RS7, but completely destroyed the modest response ratio observed for the original construct. It seems that in this case the riboswitch functionality is more dependent on the aptamer platform and its interaction with the ligand than on the terminator part of the riboswitch, as an impaired ligand interaction would result in non-functional riboswitches. To address this question, the structure and ligand-binding capability of the isolated streptomycin aptamer were analyzed by structure probing.
Secondary structure analysis of the streptomycin aptamer
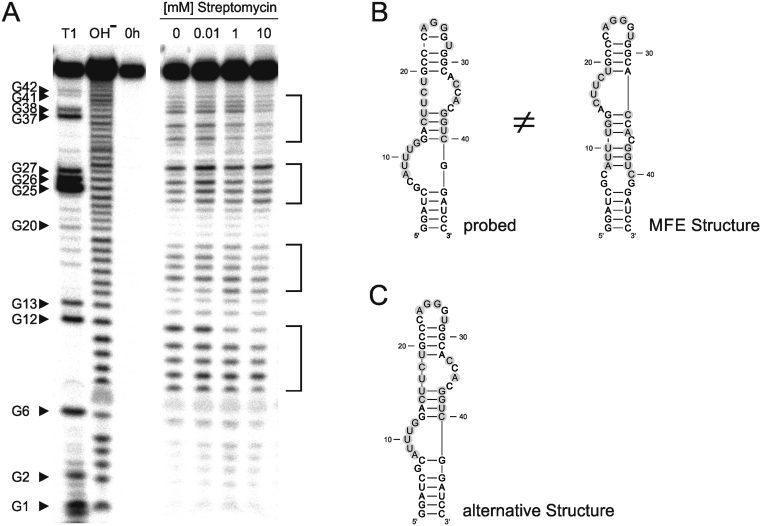
The secondary structure of the streptomycin aptamer, representing the sensory domain in our riboswitch constructs, was investigated by in-line probing (38). Starting from a PCR product with 5΄-located T7 promoter, the streptomycin aptamer RNA was transcribed in vitro, gel-purified and 5΄-end labeled with [γ-32P]-ATP. In-line probing was performed in the presence of 0–10 mM streptomycin and analyzed by PAA gel electrophoresis. Positions of G residues were assigned to the aptamer sequence using partial RNase T1 digestion and alkaline hydrolysis. Regions of high flexibility within the RNA structure are visible as prominent bands in the gel image and indicated by square brackets (Figure 4A).
Figure 4.
Secondary structure analysis of the 5΄-labeled streptomycin aptamer transcript. (A) In-line probing analysis. T1: Partial digest with RNase T1. OH−: Alkaline hydrolysis. 0 h: Transcript without incubation. Lanes 0–10: Transcript was probed in the presence of indicated streptomycin concentrations for 44 h. Regions of high cleavage activity are indicated by brackets. (B) The secondary structure according to probing results is highly similar to streptomycin aptamer ‘motif 1’ (35) and differs from the in silico predicted structure of minimal free energy (MFE). (C) From the folding predictions of RNAsubopt (36), a slightly less stable structure with a free energy of −11.0 kcal/mol is highly similar to the probed one, indicating that the approach used the wrong fold for riboswitch design. Regions of high cleavage activity from structure probing in (A) are depicted as grey circles.
The RNA cleavage pattern indicates a secondary structure that is in agreement with the previously published structure (Figure 4B) (35). However, the in silico design and secondary structure prediction of the synthetic riboswitches was based on a MFE structure (Figure 4B) which shows a strong discrepancy to the probed structure. Based on RNAfold, the underlying design approach includes the MFE structure in the riboswitch prediction, since it is the most stable from a set of all possible secondary structures and has the lowest ΔG value. In contrast, the actual probed structure is more closely related to a slightly less stable structure (Figure 4C) and was also obtained when the whole construct Strep-RS7 was probed (data not shown).
Hence, the aptamer as an isolated form as well as in the riboswitch context exhibits the same folding that is not compatible with the predicted MFE structure. For future designs, it will be important to implement naturally occurring non-MFE-like structures of aptamers as a folding constraint in order to generate functional riboswitches.
Construction of heterogeneous tandem riboswitches
Natural riboswitches do not invariably consist of one aptamer and one expression platform. Tandem riboswitches often contain two aptamer domains and show increased sensitivity and sometimes cooperative binding for their cognate ligand (45–49). Similar tandem combinations were also generated from synthetic riboswitches, incorporating signals from two different ligands (50–53). Furthermore, even tridem arrangements of synthetic transcriptional theophylline riboswitches were designed, showing a considerable improved response ratio and a dose-dependent linear activation rate (33).
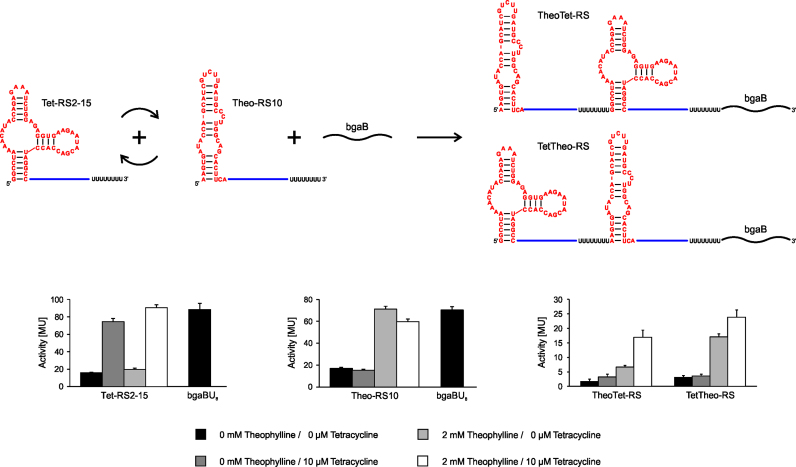
To investigate the regulatory potential of our designed riboswitches as a logic AND gate, heterogeneous tandem riboswitches were produced by combining a theophylline-sensing riboswitch Theo-RS10 (19; Supplementary Data) with tetracycline-binding Tet-RS2-15, both representing the best performing riboswitches of the two classes. Theo-RS10 as well as Tet-RS2-15 show both a highly specific response to their respective ligands, while they are not activated by the second target molecule (Figure 5). These ligand-specific riboswitch modules were arranged in both 5΄-3΄-orders, resulting in tandem constructs TheoTet-RS and TetTheo-RS (Figure 5). Both tandem arrangements should activate gene expression of the bgaB reporter only in the simultaneous presence of both theophylline and tetracycline.
Figure 5.
Design principle and activity test of tandem riboswitches. The individual components Tet-RS2-15 and Theo-RS10 show a highly specific activation only in the presence of their cognate ligand. This is not affected by the presence of the second ligand. In the absence of ligands, both tandem riboswitches TheoTet-RS and TetTheo-RS show a minimal activation of β-galactosidase expression. In the presence of theophylline, the order of the riboswitch modules has an impact on background activity, indicating that in TetTheo-RS, the Tet module is affected in its OFF state, leading to a rather high background activation.
In comparison with the individual riboswitches, the β-galactosidase background activity of both tandem constructs is considerably low and does not exceed 5 MU in the absence of both ligands. As a consequence of a background close to zero, activation rates are relatively high: in the presence of both ligands, the activation rates are 10.4-fold for TheoTet-RS and 7.7-fold for TetTheo-RS (Figure 5). TheoTet-RS indeed functions as a logic AND gate and shows low reporter gene activities in the presence of the individual ligands. Here, theophylline leads to an increase to 3 MU, while tetracycline induces the system to 6 MU compared to a zero-ligand background of 1–2 MU (Figure 5). The presence of both ligands activates the system, resulting in 17 MU. In the case of TetTheo-RS, the tandem is also fully activated in the presence of both ligands (24 MU; Figure 5). While tetracycline alone does not induce gene expression (3–4 MU, representing the background activity of the Theo-RS part), the addition of theophylline only leads already to partial activation (17 MU, representing the background activity of the Tet-RS part). Hence, the TetTheo-RS construct shows the same tendency as TheoTet-RS, however, the background activation of the Tet-RS2-15 part (in the presence of just theophylline) is significantly increased. Obviously, the Tet terminator seems to be too weak to efficiently shut off transcription before the downstream located Theo riboswitch is transcribed. This indicates a strong positioning effect in these constructs where the sequential order of the individual riboswitch elements has an impact on the ligand-dependent response.
As only the TheoTet construct exhibited a considerable functionality as logic ‘AND’ gate, a further structure-guided optimization of this tandem riboswitch was attempted. To reduce the Tet-RS2-dependent response background, we replaced the loop region in the corresponding terminator hairpin by a stabilizing GAAA tetraloop, resulting in TheoTet-RS2-15loop (Supplementary Data). This tetraloop sequence was previously used to increase the termination efficiency in a theophylline riboswitch (19). In a second construct, the 10 bp terminator hairpin (Figure 2B) stabilized by additional GC base pairs was introduced. However, both variants led to a complete repression of gene expression, independent of the ligands (Supplementary Data).
It is possible that the close proximity of both riboswitch modules interferes with correct folding of the individual units, so that binding of theophylline to its aptamer structure affects binding of tetracycline to the other corresponding aptamer. An increased distance between the two riboswitch elements should allow an independent folding of the individual regions and, consequently, abolish such interference. Accordingly, computer-generated sequences of 15, 20 and 30 nts length were designed that form an unstructured linker between the riboswitch modules and that do not interact with these elements. One of each linker sequence was inserted between the riboswitch modules in TheoTet-RS and tested for an improved response ratio (Supplementary Data). Yet, instead of improving riboswitch functionality by increasing β-galactosidase activity in the presence of both ligands, constructs TheoTet-RS-15 nt and TheoTet-RS-20 nt also responded to theophylline alone, whereas TheoTet-30 nt switched off gene expression altogether (Supplementary Data).
In conclusion, the tandem constructs clearly show that it is possible to generate Boolean logic gates from synthetic transcriptional riboswitch elements. As observed for tandem theophylline riboswitches, it seems that the downstream located module has the strongest impact on the response ratio (33).
DISCUSSION
Over the last years, riboswitches have increasingly gained attention in the field of synthetic biology, as they allow conditional control of gene expression by external ligands, representing a versatile tool for reprogramming of cells (54–56). The combination of in vitro generated aptamer domains specific for a whole plethora of target molecules with suited regulatory domains opens the principle possibility to generate custom-designed orthogonal devices regulating gene expression in pro- and eukaryotes (57,55,10,58,59). However, the application of such aptamers as synthetic riboswitches is a rather difficult task. First, suitable effector domains have to be generated or - in the case of natural compounds - identified. Batey et al. successfully constructed chimeric riboswitches by combining different aptamers with expression platforms of natural riboswitches (24,9). Natural aptamer domains can be used as well and developed into orthogonal riboswitches binding ligand analogues not present in the cell (25,26).
Our approach is based on a complete de novo design coupled with structure-guided experimental optimization, where artificial aptamers are fused to a designed transcription terminator element. This approach was very successful with a theophylline aptamer-based riboswitch in the context of different promoters and reporter genes (33,19). Here, we show that this approach can be applied to design tetracycline-dependent riboswitches acting on transcription. While individual tetracycline riboswitches are functional, they exhibit regulatory properties that are different from those of the in silico predicted theophylline riboswitches. For both Theo-RS and Tet-RS constructs, the reporter gene induction led to a comparable enzymatic activity of β-galactosidase of 60 (Theo-RS10shift) (19) to 70 MU (Tet-RS1; Figure 2A). The background activation in the absence of the ligands, however, is different. Without theophylline, the Theo-RS construct results in 10 MU of β-galactosidase activity, while the individual Tet-RS devices show a background enzyme activity of 20 to 40 MU (Figure 2A). Accordingly, the response ratios for tetracycline constructs are only in a range of 1.5- to 2.2-fold, whereas Theo-RS10shift is responding in a 6.5-fold ratio (19).
Due to the larger size of the tetracycline aptamer compared to the theophylline-binding structure, the Tet-RS constructs require a somewhat different composition. The free energy of the tetracycline aptamer is much lower than that for the theophylline aptamer (−19.8 versus −12.2 kcal/mol). Accordingly, the terminator hairpin has to exhibit an increased stability that can compete with the aptameric structure to allow a ligand-dependent re-folding of the RNA. As a result, the terminator hairpins are longer in sequence than those of theophylline riboswitches. In particular, they consist of 21 to 23 base pairs and cover a large region of the aptamer 3΄-part (Figures 1B and 2). In the theophylline constructs, the optimization of the response ratio correlates with ΔG values in an intermediate range, relative to the stability of the aptamer structure. Weak terminator hairpins are unable to disrupt aptamer formation, whereas too stable terminators inhibit a refolding of the riboswitch into the ON state (33,19). This basic principle is also true for tetracycline-dependent riboswitch constructs. A successive shortening of the aptamer reverse complementary sequence reduces the number of base pairs in the terminator structures. This experimental fine-tuning increases the response ratio up to 3.4-fold in Tet-RS2-15. A further truncation of the terminator in Tet-RS2-10, however, completely abolishes ligand-dependent gene activation. Here, the hairpin stability is too weak to compete with the aptamer fold. As a result, Tet-RS2-10 represents a constitutive ON state (Figure 2B). This behavior of Tet-RS2-10 indicates that these tetracycline riboswitches regulate gene expression at the level of transcription, as it was also shown for Theo-RS constructs (19,33). Furthermore, riboswitches carrying the miniaturized version of the tetracycline aptamer (Tet-RS1M, Supplementary Data) show a functionality identical to that of constructs with the original aptamer (Tet-RS1). This is due to the fact that the minimeric form of the tetracycline aptamer has a stability that is highly similar to that of the full-length aptamer (−17.6 kcal/mol versus −19.8 kcal/mol). In both constructs, the overlapping region is identical and spans 21 bases of the aptamers 3΄-ends (Figure 2). Hence, the terminator hairpin competes in both riboswitch forms with the same structure as well as sequence part.
For the theophylline-dependent riboswitches, a serial arrangement of two and three copies resulted in a greatly improved response ratio, as the increased number of transcriptional terminators dramatically reduced background activation in the absence of the ligand (33). The same principle holds true for the Tet-RS arrangements (Figure 2C), where we could observe a more than 3-fold increase in the responsiveness of the TetTetTet-RS construct compared to the monomer. The absolute gene expression levels (MU), however, are slightly reduced the more riboswitch repeats are introduced. As the functional principle of intrinsic terminators indicates that RNA polymerase is sensitive to stable secondary structures in the nascent transcript (60), it is possible that the now highly structured 5΄-UTR reduces the rate of transcription to some degree. Likewise, the riboswitch structures might diminish ribosome assembly at the neighboring ribosomal binding site. Yet, the serial arrangement of transcriptional riboswitches seems to be a general strategy to improve the ligand-dependent response, although these constructs need higher ligand concentrations for full activation, as all aptamer domains have to bind their ligand for disrupting their terminator structures.
However, not all design principles suited for theophylline riboswitches can be transferred to tetracycline riboswitches. For example, insertion of a GAAA tetraloop into the terminator hairpin of Theo-RS10 lead to a better ON-OFF ratio due to its helix-stabilizing effect (33). In a Tet-RS2-15, a corresponding loop completely inactivates the riboswitch (Supplementary Data). Obviously, the long terminator hairpin with a GAAA tetraloop is too stable to allow a refolding of the tetracycline aptamer, leading to a constitutive OFF state. Similarly, the introduction of additional GC base pairs in the terminator hairpin inhibits the ON state formation. These results demonstrate that an intrinsic terminator operates context-dependent and cannot be simply transferred into different sequence or structural environments (61,47,62). As a consequence, the applicability of computer-based predictions still has to be tested and optimized experimentally.
While the functionality of the synthetic riboswitches carrying a tetracycline aptamer show that our approach is not restricted to theophylline-sensing regulatory elements, expanding this design principle to riboswitches based on a streptomycin aptamer is difficult. Except Strep-RS7, the designed constructs did not respond to the presence of streptomycin, suggesting the predicted interaction between aptamer and terminator conformations is impaired in vivo (Figure 3). While the predicted most stable aptamer structure has a free energy of −12.5 kcal/mol, the stability of the selected terminator hairpins varies from −20.7 (Strep-RS1) to −27.7 kcal/mol (Strep-RS8; the contribution of the bound ligand was not taken into account). In Strep-RS1, carrying the weakest terminator element, the reporter gene was constitutively switched on, resulting in 30 to 40 MU (Figure 3). In contrast, all riboswitches with more stable terminators were in a constitutive OFF state, and only Strep-RS7 showed a slight regulatory potential, and a further destabilization of the terminator did not improve the response. Hence, although a considerable range of terminator stabilities was investigated, no substantial regulatory potential was observed. Even though this result seems to contradict the universality of our design pipeline, there are several possible scenarios explaining the observation. First, the structure probing data do not support the existence of the predicted MFE structure, and the candidates adopt an alternative—less stable—structure of the calculated folding tree (Figure 4). However, the terminator elements were designed to compete with the MFE structure and obviously do not allow a refolding of the aptamer into the ligand-binding competent state. Only the terminator of Strep-RS7 shows some compatibility. In Strep-RS1, on the other hand, the designed hairpin is either not stable enough to be formed in vivo or is not functional as a terminator, resulting both a constitutive ON state. Here, in addition to free energy considerations, incorporating the aptamer fold that has been reported on the basis of structure probing and/or crystallization should allow a further improvement in the design of synthetic riboswitches (35,63).
A second reason for the malfunction of the Strep-RS constructs might be the relatively low affinity of the streptomycin aptamer with a Kd of ∼1 μM (35). In contrast, the aptamers for theophylline and tetracycline bind their ligands with Kd values of 0.32 μM and 770 pM, respectively (64,40). Hence, these aptamers might be more suitable for an in vivo application in synthetic riboswitches than the streptomycin aptamer. A third possibility lies in the basic principle of aptamer generation that selects for a tight binding potential of RNA but not for its suitability for riboswitch regulation (58). The Süß lab introduced neomycin aptamers as regulatory translational roadblocks into the 5΄-UTR of a GFP reporter gene in yeast. The in vitro selected aptamer R23 showed no ligand-induced regulation, while an in vivo screened aptamer N1 allowed a neomycin-dependent translation (14,65). Aptamer R23 has a stable pre-formed binding pocket even in the absence of neomycin and shows no structural rearrangement upon ligand binding. Aptamer N1, however, exhibits a more dynamic behavior upon ligand interaction. The structure of the unbound aptamer seems to be less stable, allowing the scanning ribosomes to proceed and reach the translation start site, and translation is turned on. In contrast, the robust structure of the unbound R23 aptamer obviously is too stable for being disrupted by the ribosomes, and translation ceases (14,41,66). It is possible that the streptomycin aptamer also forms such a pre-folded stable structure in the Strep-RS constructs, inhibiting a refolding of the riboswitches upon ligand binding. Yet, the weakly ligand-responding Strep-RS7 indicates that the riboswitch-suitability of an aptamer seems to be context-dependent, and that a competing terminator hairpin of equal stability could destabilize the pre-formed aptamer structure in the absence of the ligand. In translational road block riboswitches, this is not possible, as there is no additional structural element competing with the aptamer fold. However, due to the observed low ligand response, the formation of this alternative folding seems to be quite inefficient in Strep-RS7 and aptamers with a pronounced refolding could represent better candidates also for riboswitches with a terminator-forming expression module. In summary, it is not clear yet whether the failure to design streptomycin-dependent riboswitches represents the limitations of the current computational design approach or the streptomycin aptamer with a stable pre-formed structure in the absence of the ligand is simply not suited for riboswitch construction.
Nevertheless, the functionality of the Tet-RS constructs demonstrates that our in silico design can be applied to different aptamers, even though only one of the Strep-RS constructs shows a moderate functionality. Hence, for aptamers that do not adopt the calculated MFE structure (like the streptomycin aptamer), the design strategy has to be modified so that it is not restricted to the MFE structure but uses a defined aptamer fold as a constraint. With such a modification, it should be possible to generate further riboswitch elements with sensor platforms forming less stable structures than predicted by RNAfold. Here, knowledge of the aptamer structure by crystallography or, even better, by structural data of the RNA in solution (NMR, SHAPE, in-line probing) is required. Yet, structure-guided experimental optimization as shown above is still necessary to obtain efficient activation ratios in these synthetic regulators.
The fact that the tetracycline aptamer could be combined with terminator constructs to form functional riboswitches offered the opportunity to generate heterogeneous riboswitch tandems acting on transcription. While the 2- and 3-fold arrangement of theophylline riboswitches was shown to dramatically increase the response ratio and leads to a linear dose-dependent gene activation (33), the combination of synthetic riboswitches sensing different ligands leads to a very different regulatory principle, acting as a logic ‘AND’ gate that is fully activated only in the presence of both ligands. Similar tandem riboswitches have been generated with different expression platforms regulating translation, like a cis-acting ribozyme that destroys the mRNA (50,51), a ribosomal binding site sequestor (52) or a sequence capable of ribosomal frameshift (53). Our TheoTet-RS combinations, however, act on transcription, allowing a faster response, as an early step of gene expression is regulated. While these tandems demonstrate that the individual riboswitch elements can be freely combined, the results indicate that additional parameters affecting the functionality have to be considered. First, the sequential order of the individual Theo- and Tet-RS lead to different response efficiencies. TheoTet-RS shows a ligand-dependent response similar to synthetic ‘AND’ gates acting on translation (50), with a low background activation in the presence of only one ligand (Figure 5). In contrast, TetTheo-RS, representing the reverse riboswitch order, shows an increase background activation in the presence of theophylline. Here, the Tet-RS element should be in the OFF state and terminate transcription. While the observed background activity corresponds to that of the individual Tet-RS2-15 (Figure 2), the same riboswitch seems to shut off transcription more efficiently when being located downstream of Theo-RS. Obviously, both riboswitch elements have an impact on the functionality of their neighboring construct, and an increase of the distance by introducing linker regions between both riboswitches could not reduce this effect (Supplementary Data). Hence, the individual components of these tandem arrangements cannot be regarded as independent building blocks, and it will be important to expand the design algorithm to include possible interactions between the modules that disturb correct folding into individual aptamer and terminator conformations.
Taken together, our results show that rational design of synthetic riboswitches can be expanded to different aptamers, although further improvements of the approach are required to allow the use of non-MFE-like structures and different tandem arrangements. While there are alternative strategies available, like adaptation of natural platforms and screening of libraries of randomized RNA sequences, the de novo construction is essential to identify important design parameters in order to understand the functionality of riboswitches and their required ligand-dependent refolding in detail. Design strategies regulating gene expression via trans-acting RNAs like toehold switches (29) and small trans activating RNAs (30) are also very promising approaches. However, OFF target effects of the trans-acting RNAs can impair the usability of these systems. In addition, stoichiometric effects have to be considered, as a certain concentration of the trans-acting RNA is required for finding and binding its target. Finally, features identified in a rational design strategy not only facilitate the in silico construction of functional riboswitches, but will also be transferable into the design of these alternative regulatory RNA constructs, deepening our understanding of RNA-based gene regulation in nature as well as in synthetic biology.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Daniel N. Wilson and Agata L. Starosta for kindly providing plasmid pET46-tetM, Frauke Olthoff for cloning of pACYC-tetM and tetracycline riboswitch RS1, and Stefanie Heidenreich for cloning of tetracycline riboswitches RS2-10 and RS2-15. Furthermore, we thank Julia Weigand and Beatrix Süß for helpful discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft [MO 634/9-1 to M.M., STA 850/15-1 to P.F.S.]; European Commission under the Environment Theme of the 7th Framework Program for Research and Technological Development [323987 to S.F.]. Funding for open access charge: DFG, Leipzig University [MO 634/9-1 to M.M., STA 850/15-1 to P.F.S.]; We acknowledge support from the German Research Foundation (DFG) and Universität Leipzig within the program of Open Access Publishing.
Conflict of interest statement. None declared.
REFERENCES
- 1. Winkler W.C., Nahvi A., Breaker R.R.. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002; 419:952–956. [DOI] [PubMed] [Google Scholar]
- 2. Nahvi A., Sudarsan N., Ebert M.S., Zou X., Brown K.L., Breaker R.R.. Genetic control by a metabolite binding mRNA. Chem. Biol. 2002; 9:1043. [DOI] [PubMed] [Google Scholar]
- 3. Winkler W.C., Cohen-Chalamish S., Breaker R.R.. An mRNA structure that controls gene expression by binding FMN. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:15908–15913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Breaker R.R. Riboswitches and the RNA world. Cold Spring Harb. Perspect. Biol. 2012; 4:a003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Serganov A., Nudler E.. A decade of riboswitches. Cell. 2013; 152:17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nudler E., Mironov A.S.. The riboswitch control of bacterial metabolism. Trends Biochem. Sci. 2004; 29:11–17. [DOI] [PubMed] [Google Scholar]
- 7. Barrick J.E., Breaker R.R.. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol. 2007; 8:R239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Breaker R.R. Prospects for riboswitch discovery and analysis. Mol. Cell. 2011; 43:867–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ceres P., Trausch J.J., Batey R.T.. Engineering modular ‘ON’ RNA switches using biological components. Nucleic Acids Res. 2013; 41:10449–10461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Groher F., Suess B.. Synthetic riboswitches - A tool comes of age. Biochim. Biophys. Acta. 2014; 1839:964–973. [DOI] [PubMed] [Google Scholar]
- 11. Rode A.B., Endoh T., Sugimoto N.. Tuning riboswitch-mediated gene regulation by rational control of aptamer ligand binding properties. Angew. Chem. Int. Ed. Engl. 2015; 54:905–909. [DOI] [PubMed] [Google Scholar]
- 12. Topp S., Gallivan J.P.. Emerging applications of riboswitches in chemical biology. ACS Chem. Biol. 2010; 5:139–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Werstuck G., Green M.R.. Controlling gene expression in living cells through small molecule-RNA interactions. Science. 1998; 282:296–298. [DOI] [PubMed] [Google Scholar]
- 14. Weigand J.E., Sanchez M., Gunnesch E.-B., Zeiher S., Schroeder R., Suess B.. Screening for engineered neomycin riboswitches that control translation initiation. RNA. 2008; 14:89–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Klauser B., Atanasov J., Siewert L.K., Hartig J.S.. Ribozyme-based aminoglycoside switches of gene expression engineered by genetic selection in S. cerevisiae. ACS Synth. Biol. 2015; 4:516–525. [DOI] [PubMed] [Google Scholar]
- 16. Desai S.K., Gallivan J.P.. Genetic screens and selections for small molecules based on a synthetic riboswitch that activates protein translation. J. Am. Chem. Soc. 2004; 126:13247–13254. [DOI] [PubMed] [Google Scholar]
- 17. Suess B., Fink B., Berens C., Stentz R., Hillen W.. A theophylline responsive riboswitch based on helix slipping controls gene expression in vivo. Nucleic Acids Res. 2004; 32:1610–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Topp S., Gallivan J.P.. Guiding bacteria with small molecules and RNA. J. Am. Chem. Soc. 2007; 129:6807–6811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wachsmuth M., Findeiß S., Weissheimer N., Stadler P.F., Mörl M.. De novo design of a synthetic riboswitch that regulates transcription termination. Nucleic Acids Res. 2013; 41:2541–2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hsu H.-T., Lin Y.-H., Chang K.-Y.. Synergetic regulation of translational reading-frame switch by ligand-responsive RNAs in mammalian cells. Nucleic Acids Res. 2014; 42:14070–14082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Suess B., Hanson S., Berens C., Fink B., Schroeder R., Hillen W.. Conditional gene expression by controlling translation with tetracycline-binding aptamers. Nucleic Acids Res. 2003; 31:1853–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Weigand J.E., Suess B.. Tetracycline aptamer-controlled regulation of pre-mRNA splicing in yeast. Nucleic Acids Res. 2007; 35:4179–4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Beilstein K., Wittmann A., Grez M., Suess B.. Conditional control of mammalian gene expression by tetracycline-dependent hammerhead ribozymes. ACS Synth. Biol. 2015; 4:526–534. [DOI] [PubMed] [Google Scholar]
- 24. Ceres P., Garst A.D., Marcano-Velazquez J.G., Batey R.T.. Modularity of select riboswitch expression platforms enables facile engineering of novel genetic regulatory devices. ACS Synth. Biol. 2013; 2:463–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dixon N., Duncan J.N., Geerlings T., Dunstan M.S., McCarthy J.E.G., Leys D., Micklefield J.. Reengineering orthogonally selective riboswitches. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:2830–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Robinson C.J., Vincent H.A., Wu M.-C., Lowe P.T., Dunstan M.S., Leys D., Micklefield J.. Modular riboswitch toolsets for synthetic genetic control in diverse bacterial species. J. Am. Chem. Soc. 2014; 136:10615–10624. [DOI] [PubMed] [Google Scholar]
- 27. Lynch S.A., Desai S.K., Sajja H.K., Gallivan J.P.. A high-throughput screen for synthetic riboswitches reveals mechanistic insights into their function. Chem. Biol. 2007; 14:173–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fowler C.C., Brown E.D., Li Y.. A FACS-based approach to engineering artificial riboswitches. Chembiochem. 2008; 9:1906–1911. [DOI] [PubMed] [Google Scholar]
- 29. Green A.A., Silver P.A., Collins J.J., Yin P.. Toehold switches: de-novo-designed regulators of gene expression. Cell. 2014; 159:925–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chappell J., Takahashi M.K., Lucks J.B.. Creating small transcription activating RNAs. Nat. Chem. Biol. 2015; 11:214–220. [DOI] [PubMed] [Google Scholar]
- 31. Meyer S., Chappell J., Sankar S., Chew R., Lucks J.B.. Improving fold activation of small transcription activating RNAs (STARs) with rational RNA engineering strategies. Biotechnol. Bioeng. 2016; 113:216–225. [DOI] [PubMed] [Google Scholar]
- 32. Espah Borujeni A., Mishler D.M., Wang J., Huso W., Salis H.M.. Automated physics-based design of synthetic riboswitches from diverse RNA aptamers. Nucleic Acids Res. 2016; 44:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wachsmuth M., Domin G., Lorenz R., Serfling R., Findeiß S., Stadler P.F., Mörl M.. Design criteria for synthetic riboswitches acting on transcription. RNA Biol. 2015; 12:221–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hanson S., Berthelot K., Fink B., McCarthy J.E.G., Suess B.. Tetracycline-aptamer-mediated translational regulation in yeast. Mol. Microbiol. 2003; 49:1627–1637. [DOI] [PubMed] [Google Scholar]
- 35. Wallace S.T., Schroeder R.. In vitro selection and characterization of streptomycin-binding RNAs: recognition discrimination between antibiotics. RNA. 1998; 4:112–123. [PMC free article] [PubMed] [Google Scholar]
- 36. Lorenz R., Bernhart S.H., Honer Zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L.. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011; 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dönhöfer A., Franckenberg S., Wickles S., Berninghausen O., Beckmann R., Wilson D.N.. Structural basis for TetM-mediated tetracycline resistance. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:16900–16905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Regulski E.E., Breaker R.R.. In-line probing analysis of riboswitches. Methods Mol. Biol. 2008; 419:53–67. [DOI] [PubMed] [Google Scholar]
- 39. Hanson S., Bauer G., Fink B., Suess B.. Molecular analysis of a synthetic tetracycline-binding riboswitch. RNA. 2005; 11:503–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Müller M., Weigand J.E., Weichenrieder O., Suess B.. Thermodynamic characterization of an engineered tetracycline-binding riboswitch. Nucleic Acids Res. 2006; 34:2607–2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wunnicke D., Strohbach D., Weigand J.E., Appel B., Feresin E., Suess B., Müller S., Steinhoff H.-J.. Ligand-induced conformational capture of a synthetic tetracycline riboswitch revealed by pulse EPR. RNA. 2011; 17:182–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Förster U., Weigand J.E., Trojanowski P., Suess B., Wachtveitl J.. Conformational dynamics of the tetracycline-binding aptamer. Nucleic Acids Res. 2012; 40:1807–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gruber A.R., Lorenz R., Bernhart S.H., Neubock R., Hofacker I.L.. The Vienna RNA websuite. Nucleic Acids Res. 2008; 36:W70–W74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Berens C., Thain A., Schroeder R.. A tetracycline-binding RNA aptamer. Bioorg. Med. Chem. 2001; 9:2549–2556. [DOI] [PubMed] [Google Scholar]
- 45. Mandal M., Lee M., Barrick J.E., Weinberg Z., Emilsson G.M., Ruzzo W.L., Breaker R.R.. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science. 2004; 306:275–279. [DOI] [PubMed] [Google Scholar]
- 46. Sudarsan N., Hammond M.C., Block K.F., Welz R., Barrick J.E., Roth A., Breaker R.R.. Tandem riboswitch architectures exhibit complex gene control functions. Science. 2006; 314:300–304. [DOI] [PubMed] [Google Scholar]
- 47. Welz R., Breaker R.R.. Ligand binding and gene control characteristics of tandem riboswitches in Bacillus anthracis. RNA. 2007; 13:573–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Poiata E., Meyer M.M., Ames T.D., Breaker R.R.. A variant riboswitch aptamer class for S-adenosylmethionine common in marine bacteria. RNA. 2009; 15:2046–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhou H., Zheng C., Su J., Chen B., Fu Y., Xie Y., Tang Q., Chou S.-H., He J.. Characterization of a natural triple-tandem c-di-GMP riboswitch and application of the riboswitch-based dual-fluorescence reporter. Sci. Rep. 2016; 6:20871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Win M.N., Smolke C.D.. Higher-order cellular information processing with synthetic RNA devices. Science. 2008; 322:456–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nomura Y., Zhou L., Miu A., Yokobayashi Y.. Controlling mammalian gene expression by allosteric hepatitis delta virus ribozymes. ACS Synth. Biol. 2013; 2:684–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Goodson M.S., Harbaugh S.V., Chushak Y.G., Kelley-Loughnane N.. Integrating and amplifying signal from riboswitch biosensors. Meth. Enzymol. 2015; 550:73–91. [DOI] [PubMed] [Google Scholar]
- 53. Anzalone A.V., Lin A.J., Zairis S., Rabadan R., Cornish V.W.. Reprogramming eukaryotic translation with ligand-responsive synthetic RNA switches. Nat. Methods. 2016; 13:453–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wittmann A., Suess B.. Engineered riboswitches: Expanding researchers' toolbox with synthetic RNA regulators. FEBS Lett. 2012; 586:2076–2083. [DOI] [PubMed] [Google Scholar]
- 55. Chang A.L., Wolf J.J., Smolke C.D.. Synthetic RNA switches as a tool for temporal and spatial control over gene expression. Curr. Opin. Biotechnol. 2012; 23:679–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kushwaha M., Rostain W., Prakash S., Duncan J.N., Jaramillo A.. Using RNA as molecular code for programming cellular function. ACS Synth. Biol. 2016; 5:795–809. [DOI] [PubMed] [Google Scholar]
- 57. Berens C., Suess B.. Riboswitch engineering - making the all-important second and third steps. Curr. Opin. Biotechnol. 2015; 31C:10–15. [DOI] [PubMed] [Google Scholar]
- 58. Schneider C., Suess B.. Identification of RNA aptamers with riboswitching properties. Methods. 2016; 97:44–50. [DOI] [PubMed] [Google Scholar]
- 59. Peters G., Coussement P., Maertens J., Lammertyn J., Mey M.de. Putting RNA to work: translating RNA fundamentals into biotechnological engineering practice. Biotechnol. Adv. 2015; 33:1829–1844. [DOI] [PubMed] [Google Scholar]
- 60. Zhang J., Landick R.. A Two-Way Street: Regulatory Interplay between RNA Polymerase and Nascent RNA Structure. Trends Biochem. Sci. 2016; 41:293–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lesnik E.A., Sampath R., Levene H.B., Henderson T.J., McNeil J.A., Ecker D.J.. Prediction of rho-independent transcriptional terminators in Escherichia coli. Nucleic Acids Res. 2001; 29:3583–3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Blouin S., Chinnappan R., Lafontaine D.A.. Folding of the lysine riboswitch: importance of peripheral elements for transcriptional regulation. Nucleic Acids Res. 2011; 39:3373–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tereshko V., Skripkin E., Patel D.J.. Encapsulating streptomycin within a small 40-mer RNA. Chem. Biol. 2003; 10:175–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Jenison R.D., Gill S.C., Pardi A., Polisky B.. High-resolution molecular discrimination by RNA. Science. 1994; 263:1425–1429. [DOI] [PubMed] [Google Scholar]
- 65. Weigand J.E., Schmidtke S.R., Will T.J., Duchardt-Ferner E., Hammann C., Wohnert J., Suess B.. Mechanistic insights into an engineered riboswitch: a switching element which confers riboswitch activity. Nucleic Acids Res. 2011; 39:3363–3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Reuss A.J., Vogel M., Weigand J.E., Suess B., Wachtveitl J.. Tetracycline determines the conformation of its aptamer at physiological magnesium concentrations. Biophys. J. 2014; 107:2962–2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.