Abstract
The DEAH-box NTPase Prp43 disassembles spliceosomes in co-operation with the cofactors Ntr1/Spp382 and Ntr2, forming the NTR complex. How Prp43 is regulated by its cofactors to discard selectively only intron-lariat spliceosomes (ILS) and defective spliceosomes and to prevent disassembly of earlier and properly assembled/wild-type spliceosomes remains unclear. First, we show that Ntr1΄s G-patch motif (Ntr1GP) can be replaced by the GP motif of Pfa1/Sqs1, a Prp43΄s cofactor in ribosome biogenesis, demonstrating that the specific function of Ntr1GP is to activate Prp43 for spliceosome disassembly and not to guide Prp43 to its binding site in the spliceosome. Furthermore, we show that Ntr1΄s C-terminal domain (CTD) plays a safeguarding role by preventing Prp43 from disrupting wild-type spliceosomes other than the ILS. Ntr1 and Ntr2 can also discriminate between wild-type and defective spliceosomes. In both type of spliceosomes, Ntr1-CTD impedes Prp43-mediated disassembly while the Ntr1GP promotes disassembly. Intriguingly, Ntr2 plays a specific role in defective spliceosomes, likely by stabilizing Ntr1 and allowing Prp43 to enter a productive interaction with the GP motif of Ntr1. Our data indicate that Ntr1 and Ntr2 act as ‘doorkeepers’ and suggest that both cofactors inspect the RNP structure of spliceosomal complexes thereby targeting suboptimal spliceosomes for Prp43-mediated disassembly.
INTRODUCTION
The spliceosome assembles de novo for each new round of pre-mRNA splicing by the sequential recruitment of the U1, U2 and U4/U6.U5 small nuclear ribonucleoproteins (snRNPs) and numerous non-snRNP proteins to the intron (1). It undergoes a series of remodeling steps that are driven by eight conserved DExD/H-box ATPases or RNA helicases (2,3). Initially, U1 snRNP interacts with the 5΄ splice site (SS) of the intron. Stable recruitment of the U2 snRNP to the branch-site (BS) requires the action of the RNA helicase Prp5 (4) and yields the spliceosomal A complex. U2 snRNA forms a base-pairing interaction with the BS nucleotides, while several U2 SF3a and SF3b proteins stabilize U2 snRNP binding to the intron by interacting with intron nucleotides upstream and downstream of the BS (5–7). Subsequently, the U4/U6.U5 tri-snRNP associates with the A complex, resulting in the formation of the pre-catalytic B complex. Activation of the spliceosome, yielding the Bact complex, involves displacement of U4 snRNA, as well as all U4/U6-associated proteins, from the spliceosome (1). This allows U6 snRNA to form an intramolecular stem–loop and to interact with U2 snRNA. The resulting U2/U6/U5 pre-mRNA interaction network forms the heart of the spliceosome's catalytic center (8–12). Concomitant with these RNA rearrangements, more than twenty new proteins, including the Prp19 complex (NTC), are stably integrated into the Bact complex (13).
The pre-catalytic Bact complex is then transformed into the catalytically active B* complex by the Prp2 RNA helicase and its cofactor Spp2 (see Figure 1A), which also involves RNP remodeling, such as destabilization of the U2 snRNP SF3a and SF3b proteins (14–17). Upon recruitment of the step 1 factor Cwc25 to the B* complex, step 1 catalysis occurs, whereby the 2΄ hydroxyl group of the adenosine at the BS attacks the 5΄SS, generating the cleaved-off 5΄ exon and the intron lariat (IL)-3΄exon intermediates (18,19). Following a further remodeling step, which is mediated by the RNA helicase Prp16 (20), the C complex catalyzes step 2 of splicing, leading to exon ligation (21). The mature mRNA is then dissociated from the intron lariat spliceosome (ILS) by the RNA helicase Prp22 (22).
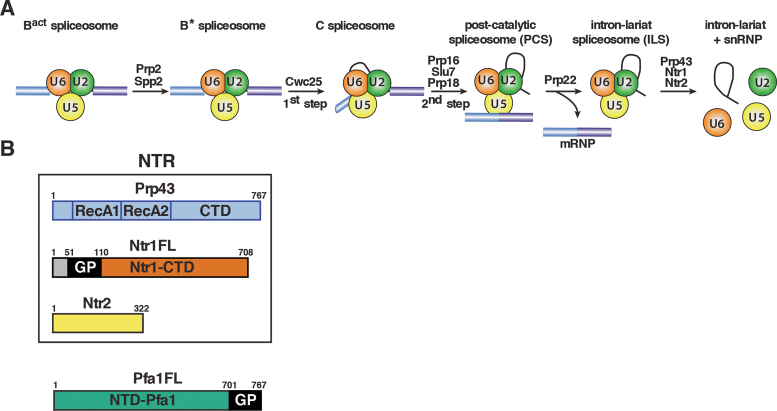
Figure 1.
Isolation of intron-lariat spliceosomes (ILSs) and schematic representation of the NTR complex and the G-patch cofactor Pfa1. (A) Activated spliceosomes (BactΔPrp2) assembled on Actin7 wild-type pre-mRNA in heat-inactivated splicing extracts from a prp2-1 yeast strain expressing a temperature-sensitive Prp2 mutant, were first purified. Purified BactΔPrp2 complexes were then incubated with recombinant Prp2 and Spp2, generating the B* spliceosome, and then Cwc25 was added to promote catalysis of step 1 of splicing and the formation of complex C. For catalysis of step 2, which generates post-catalytic spliceosomes (PCS), recombinant Prp16, Slu7 and Prp18 were added. Finally, for the purification of the ILS, the spliced mRNA was dissociated from the ILS by incubation of the PCS with Prp22 and ATP. Addition of ATP and recombinant Prp43, Ntr1 and Ntr2, leads to disassembly of the ILS into the intron-lariat, 20S U2 snRNP, 18S U5 snRNP and free U6 snRNA (26). (B) The NTR complex is composed of Prp43, Ntr1 and Ntr2. Full-length Ntr1 (Ntr1FL), includes the G-patch motif (GP) at its N-terminal region (residues 51–110), and the C-terminal domain (Ntr1-CTD), Pfa1 full-length (Pfa1FL) includes the G-patch motif (GP) at its C-terminal region (residues 701 to 767), and a N-terminal domain (NTD-Pfa1).
In the last phase of the spliceosome cycle, the IL spliceosome (ILS) is dismantled by the RNA helicase Prp43, which is necessary for recycling the snRNPs and splicing factors, and thereby to promote efficient splicing in the cell (23–26). Prp43 interacts with two cofactors, Ntr1 (also termed Spp382) and Ntr2, to form the NTR complex (nineteen-related complex) (see Figure 1B) (25,27,28). Ntr1 contains a glycine-rich G-patch motif (GP motif) in its N-terminal region (29). The GP motif comprises the major Prp43 interaction site, while Ntr1΄s C-terminal region binds Ntr2 (25). The GP motif is necessary and sufficient to stimulate Prp43΄s ATPase and RNA helicase activities in vitro (30–32). Prp43 is also essential for the processing of pre-ribosomal RNA (33–35), where it interacts with the GP motif protein, Pfa1 (also termed Sqs1) (Figure 1B) (28,36,37). Like for Ntr1, truncated Pfa1 variants including the GP motif are also sufficient to stimulate Prp43΄s ATPase and helicase activities in vitro (36).
Prp43 is not only required for the disassembly of ILSs, but it is also implicated in the discard pathway of spliceosomes associated with suboptimal/mutated pre-mRNA substrates, which become stalled at earlier stages of the spliceosomal cycle (38) and Prp43 promotes dissociation of these stalled spliceosomes (39). As several spliceosomal RNA helicases/ATPases such as Prp5, Prp2, Prp16 and Prp22 reject spliceosomes associated with suboptimal substrates (38,40), it is likely that Prp43 is also involved in the discard of early spliceosome assembly intermediates that differ from the ILS both biochemically and structurally. Currently, it is not well understood how Prp43 is regulated such that in a productive splicing cycle it only dismantles ILSs, while it dissociates also earlier spliceosomal complexes provided they exhibit assembly defects. There is evidence that both Ntr1 and Ntr2 cofactors are involved in Prp43-mediated discard of spliceosome intermediates stalled at earlier stages of the spliceosomal cycle (27,41). While it is generally believed that Ntr1 and Ntr2 together function as coordinators to target Prp43 to the ILS or defective spliceosomes, their respective role is only poorly understood in molecular terms.
In a purified yeast splicing system the NTR is necessary and sufficient to dissociate not only the IL RNA from the ILS, but also the snRNPs from each other, yielding free U6 snRNA, a 20S U2 and an 18S U5 snRNP (26). We have recently shown that the spliceosomal target of Prp43 is the pre-mRNA branchsite/U2 snRNP interaction (42). Our data revealed that Prp43 fused to the GP motif of Ntr1 is a minimal discard enzyme leading to spliceosome disassembly, while Ntr2 is not strictly required for disassembly (42). As the GP motif on its own is sufficient to stimulate Prp43΄s ATPase and helicase activities (32,43) and, when fused to Prp43, to promote Prp43-mediated disassembly of spliceosomes, this led to the conclusion that Ntr2 and the C-terminal domain (CTD) of Ntr1 play a role in controlling directly or indirectly the productive interaction of Ntr1΄s GP motif with Prp43 in the spliceosome thus, acting as doorkeepers that regulate the access of Prp43 to its target site in the spliceosome (42).
Here, we have dissected the roles of Ntr1 and Ntr2 in Prp43-mediated disassembly of spliceosomal complexes using purified components (42). Our studies demonstrate that the primary function of Ntr1GP is to activate Prp43 for spliceosome disassembly and not to aid Prp43 in the recognition of its binding site in the spliceosome. Moreover, they reveal a safeguarding role for Ntr1΄s CTD, which prevents Prp43-mediated disassembly of wild-type spliceosomes other than the ILS. In a similar manner Prp43 and Ntr1 can discriminate between wild-type and defective Bact spliceosomes. In both Bact spliceosomes, Ntr1-CTD impedes Prp43-mediated disassembly while the Ntr1GP, by activating Prp43, guarantees it. In defective spliceosomes Ntr2 plays a specific role, likely by stabilizing the Prp43/Ntr1 interaction and allowing Prp43 to enter a productive mode of action. Our data suggest that the NTR may proofread or monitor the RNP structure of spliceosomal complexes, thus leading to discard of suboptimal spliceosomes.
MATERIALS AND METHODS
Spliceosome purification and reconstitution
Yeast Bact ΔPrp2 or Act-brC Bact ΔPrp2 complexes were assembled in heat-inactivated extracts from the yeast strain prp2-1 (44) by incubating with the indicated pre-mRNA containing MS2 aptamers at 23°C for 45 min. Samples were centrifuged for 10 min at 9,000 rpm and loaded onto columns containing 200–450 μl of amylose matrix equilibrated with GK75 buffer (20 mM HEPES–KOH pH 7.9, 1.5 mM MgCl2, 75 mM KCl, 5% glycerol, 0.01% NP40). The matrix was washed twice with 10 ml GK75 buffer. Spliceosomes were eluted with 12 mM maltose in GK75 buffer and 400 μl was loaded onto linear 10–30% (v/v) glycerol gradients containing GK75 buffer. Samples were centrifuged for 16 h at 21 500 rpm in a TH660 rotor (Thermo Scientific) and harvested manually from the top in 23 fractions of 175 μl. Fractions were analyzed by Cherenkov counting in a scintillation counter. Peak fractions containing complexes were pooled and the glycerol concentration was adjusted to 5–10% with GK75 buffer without glycerol. To obtain B* spliceosomes, Bact ΔPrp2 complexes were supplemented with a 10-fold molar excess of recombinant proteins (Prp2, Spp2) and the reaction volume was adjusted to 400 μl with GK75 buffer; then 40 μl of 10× ‘rescue’ solution [200 mM KPO4 (pH 7.4), 10 mM MgCl2, 20 mM ATP, 10% PEG 8000] were added to the reaction that was incubated at 23°C for 45 min. To obtain ILSs (26), Bact ΔPrp2 complexes bound to the amylose matrix were supplemented with a 10-fold molar excess of recombinant proteins (Prp2, Spp2, Cwc25, Prp16, Slu7 and Prp18) and the reaction volume was adjusted to 400 μl with GK75 buffer; then 40 μl of 10× ‘rescue’ solution were added to the reaction. After thorough mixing, the reaction was incubated at 23°C for 45 min. Matrixes were subsequently washed three times with 10 column volumes of GK75 buffer. Then a 10-fold molar excess of recombinant Prp22 was added and the volume was adjusted to 400 μl with 1× ‘rescue’ solution prepared in GK75 buffer. After thorough mixing, the reaction was incubated at 23°C for 10 min. The supernatant (containing the released ILS) was collected, and GK75 buffer was added to the matrix to a final volume of 400 μl. After gentle mixing and repeated centrifugation for 1 min at 2000 rpm the supernatant was collected.
Spliceosome disassembly assays
To dismantle spliceosomes (Bact ΔPrp2, Act-brC Bact ΔPrp2, B*, Act-brC B*, or ILS) (42), samples were incubated with distinct combinations of a 10-fold molar excess over the spliceosome of recombinant Prp43, Ntr1FL, Ntr2, Pfa1FL or a 15-fold molar excess of the G-patch motifs of Ntr1 or Pfa1. The volume was adjusted to 400 μl with 1× ‘rescue’ solution prepared in GK75 buffer containing ATP. After thorough mixing, the mixture was incubated at 23°C for 10 min and then subjected to glycerol gradient centrifugation for 2 h at 60 000 rpm in a TH660 rotor (Thermo Scientific) and harvested manually from the top in 23 fractions of 175 μl. Each fraction was digested with Proteinase K followed by phenol–chloroform–isoamyl alcohol (PCI) extraction. RNA was precipitated with ethanol, and then analyzed by PAGE on 8% polyacrylamide, 8M urea gels (PAGE) and visualized by autoradiography or Northern blot analysis.
ATPase hydrolysis assays
The ATPase activity of Prp43 was monitored via a coupled enzymatic assay by detecting the decrease of NADH absorption at 340 nm at 25°C using an Ultrospec 2100 pro UV/Vis spectrophotometer (GE Healthcare) (45). In this method ATP hydrolysis rate is measured by monitoring ADP production via two enzymatically catalyzed reactions (45). Prp43 was used at concentrations between 0.25 and 2.5 μM. Pfa1GP was added in a 4-fold excess over Prp43 and Pfa1FL was added in a 1:1 ratio. Measurements with the RNA cofactor were performed in the presence of a five-fold excess of poly-A20 RNA (Sigma-Aldrich) over Prp43. Reactions were supplemented with 250 nM NADH, 500 nM phosphoenolpyruvate, 6–8.3 U/ml pyruvate kinase (Sigma-Aldrich) and 9–14 U/ml lactic dehydrogenase (Sigma-Aldrich) and performed in 25 mM Tris–HCl pH 7.5, 150 mM KCl and 3 mM MgCl2. ATP was added at a concentration between 0 and 2 mM. Experimental data were fitted according to the Michaelis-Menten equation using Sigma Plot (Systat Software, Inc.) to obtain kcat and KM values. Error bars are obtained from three distinct experiments and indicate the standard deviation.
Cloning and expression strategy
Recombinant Prp2, Spp2, Cwc25, Prp16, Slu7, Prp18, Prp22 and Ntr2 were purified as described in (26). Prp43 and the G-patch motif of Ntr1 (residues 51–110), Ntr1GP, was prepared as described in (32). Full-length PFA1 and NTR1 were amplified from Saccharomyces cerevisiae (ATCC 204508/S288c) genomic DNA. PFA1FL was cloned into pASG-IBA33 using the IBA StarGate cloning system. The G-patch motif of Pfa1 (residues 701–767), PFA1GP, was sub-cloned into pGEX-6P-1 using BamHI and SalI restriction sites. NTR1FL was cloned into pETM11 (Novagen). Pfa1FL was expressed in Escherichia coli BL21 (DE3) pLysS at 16°C for 18 h and induced with 200 μg/l anhydrotetracycline at an OD600 of 0.8. Purification of this C-terminally tagged His6-protein was carried out as described previously (36). The expression of N-terminally GST-tagged Pfa1GP was performed in Escherichia coli BL21 (DE3) for 16 h at 16°C and induction was initiated at an OD600 of 0.8 by addition of 0.5 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG). The cell lysis was performed with a Microfluidizer (Microfluidics, USA), in buffer containing 50 mM Tris–HCl pH 7.7, 500 mM NaCl and 2 mM MgCl2, in the presence of a protease inhibitor cocktail containing aprotinin, leupeptin, and pepstatin. After ultracentrifugation at 35 000 x g for 30 min, GST-Pfa1GP was purified by affinity-chromatography on glutathione Sepharose (Protino® GST/4B, Macherey & Nagel). GST-Pfa1GP was eluted with buffer containing additionally 30 mM reduced glutathione. GST-tag cleavage was performed with PreScission protease (GE Healthcare) added 1:100 (w/w). Pfa1GP was purified to homogeneity by gel filtration (Superdex 75, GE Healthcare) in 20 mM Tris/HCl pH 7.7, 400 mM NaCl, and 2 mM MgCl2. E. coli Rosetta II (DE3) cells, harboring the pETM11-His6-Ntr1 plasmid, were grown at 37°C to an OD600 of 0.8. Subsequently, the expression of His6-Ntr1 was induced by addition of 0.1 mM IPTG and the cells were then incubated at 17°C for 20 h. Cells were harvested by centrifugation and resuspended in buffer A [50 mM Tris (pH 7.5), 400 mM NaCl, 2 mM DTT, 2 mM MgCl2, 15% glycerol, 10 mM imidazole (pH 7.5), 0.05% NP-40] and EDTA-free protease inhibitor (Roche). Cells expressing His6-Ntr1 were then lysed with a Microfluidizer as above. The lysate was ultracentrifuged for 30 min at 15 500 × g. His6-Ntr1 was isolated from the soluble fraction by using nickel beads (Roche) with incubation for 3 h at 4°C with head-over-tail rotation. Beads were washed with buffer A and then with a combination of 20% buffer B / 80% buffer A, and eluted with buffer B [50 mM Tris (pH 7.5), 400 mM NaCl, 2 mM DTT, 2 mM MgCl2, 5% glycerol, 300 mM imidazole (pH 7.5)].
RESULTS
Prp43 dissociates the ILS in co-operation with full-length Ntr1
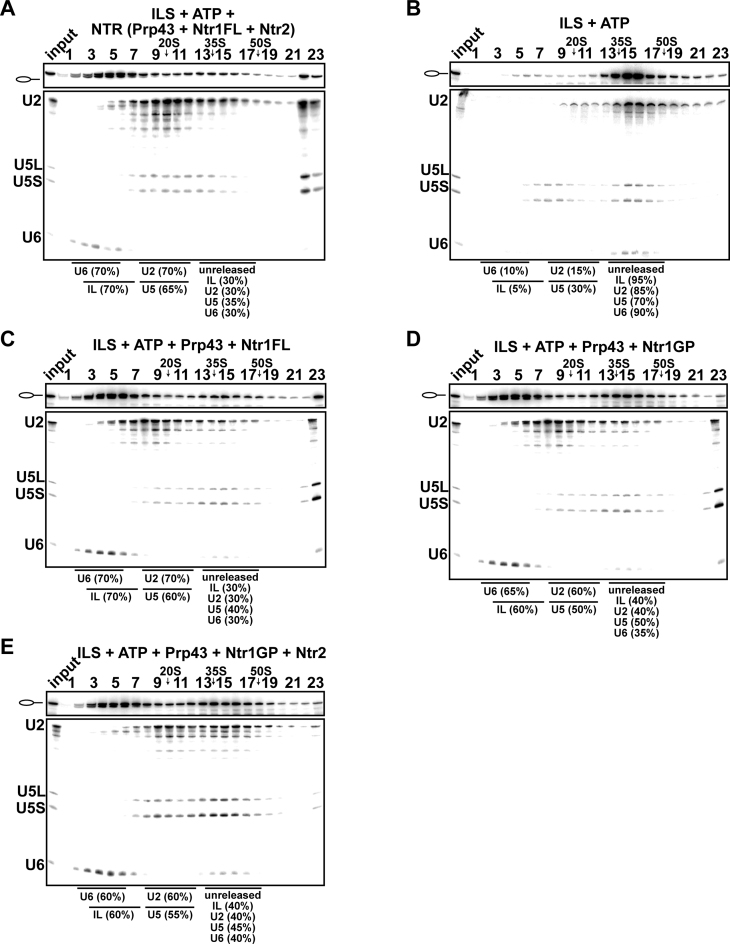
We previously showed that a fusion protein of Prp43 and the G-patch motif of Ntr1 (Prp43_Ntr1GP), efficiently dismantles purified ILS, yielding the same dissociation products as the NTR (42). This indicated that Prp43_Ntr1GP recognizes its natural target structure(s) in the ILS and disassembles spliceosomes even in the absence of Ntr2 and Ntr1΄s C-terminal domain (Ntr1-CTD) (42). Thus, the contribution played by the Ntr1-CTD and Ntr2 on ILS disassembly is unknown, raising questions about their roles in regulating spliceosome disassembly. To investigate this we purified the ILS as described previously (Figure 1A) (26,42) and examined first whether full-length Ntr1 (Ntr1FL) (Figure 1B) containing the G-patch motif (Ntr1GP) and the C-terminal domain (Ntr1-CTD) could promote dissociation of the ILS by Prp43. As previously shown, incubation of the ILS with ATP and recombinant Prp43 plus Ntr1FL and Ntr2 (forming the NTR complex, Figure 1B) (30,42) resulted in the release of ∼70% of the IL RNA and dissociation of the remaining spliceosomal RNP core into free U6 snRNA (70%), 18S U5 (65%) and 20S U2 snRNPs (70%), while in the absence of the NTR, only a very low level (∼5%) of the IL RNA was dissociated from the ILS (Figure 2A and B) (26,42). Likewise, when we incubated the ILS with Prp43 and Ntr1FL without Ntr2, ∼70% of the IL RNA was dissociated from the ILS (Figure 2C). This is consistent with previous results indicating that Ntr2 is not strictly needed for ILS disassembly (42). It is possible, however, that Ntr2 and Ntr1-CTD play a role in regulating the productive interaction of Ntr1GP with Prp43 in the ILS. Thus, to determine whether Ntr1GP can activate Prp43 in ILS disassembly also in isolation (i.e. when not fused to Prp43 and without the Ntr1-CTD), the ILSs were incubated with Prp43, ATP and the recombinant Ntr1GP, comprising amino acids 51 to 110 (Figure 1B). Figure 2D shows that ∼60% of the IL RNA was released from the ILS, indicating that the GP motif of Ntr1 alone is sufficient to activate Prp43 nearly as efficiently as Ntr1FL (compare Figure 2C and D). Disassembly of the ILS either by Ntr1FL or Ntr1GP yielded the same snRNP and RNA dissociation products (i.e., IL RNA, free U6 snRNA, 18S U5 and 20S U2 snRNPs) (Figure 2C and D) as those obtained with the NTR (Figure 2A), and the efficiency of disassembly was nearly identical. Ntr2, which was shown to bind Ntr1΄s C-terminal region (25), did not increase the efficiency of ILS disassembly when incubated with Prp43, ATP and Ntr1GP (Figure 2E). In summary, these results demonstrate that Ntr1GP alone (i.e. not fused to Prp43) is sufficient for Prp43-mediated ILS disassembly, while the Ntr1-CTD and Ntr2 do not play a major role in regulating the productive interaction of Ntr1GP with Prp43 in ILS disassembly.
Figure 2.
Full-length Ntr1 is sufficient to promote dissociation of the ILS by Prp43. 10–30% glycerol gradient sedimentation of purified ILS incubated in solution with (A) ATP plus NTR, (B) no recombinant protein, (C) Prp43 and full-length Ntr1 (Ntr1FL), (D) Prp43 and Ntr1 G-patch motif (Ntr1GP), (E) Prp43, Ntr1GP and Ntr2. U2, U5 and U6 snRNAs were visualized by northern blotting followed by autoradiography. RNA identities are indicated on the left. Quantifications were performed with ImageQuant software (Molecular Dynamics). ILS: numbers represent the percentage of intron-lariat RNA released in the peak fractions (sum of fractions 3–7) or associated with the ILS (unreleased, sum of fractions 13–17) relative to the intron-lariat RNA distributed in fractions 3–7 and 13–17, the sum of which was set to 100%. U2 and U5: numbers represent the percentage of U2 or U5 snRNAs released in the peak fractions (sum of fractions 8–12) or associated with the ILS (unreleased, sum of fractions 13–17) relative to the U2 or U5 snRNA distributed in the fractions 8–12 and 13–17, the sum of which was set to 100%. U6: numbers represent the percentage of U6 snRNA released in the peak fractions (sum of fractions 2–6) or associated with the ILS (unreleased, sum of fractions 13–17) relative to the U6 snRNA distributed in the fractions 2–6 and 13–17, the sum of which was set to 100%.
Prp43 dissociates the ILS in co-operation with the GP motif of Pfa1 but not with full-length Pfa1
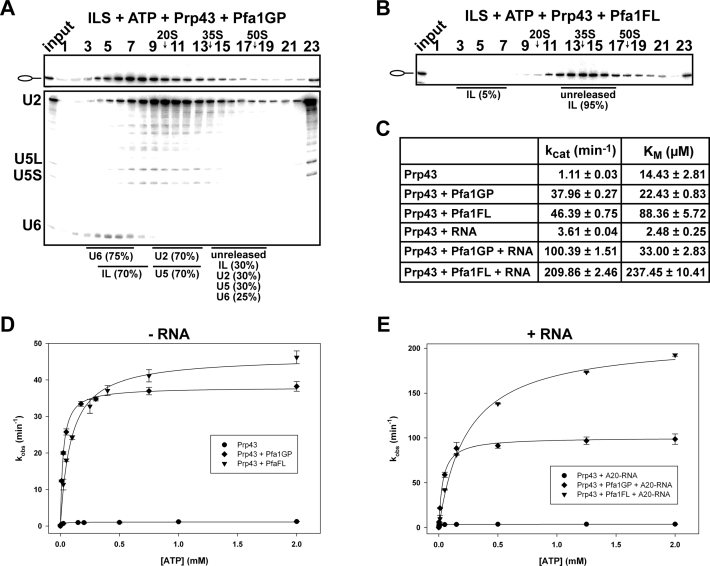
An intriguing question, relevant to the mechanism of helicase activation, is the extent to which the activation of Prp43 in spliceosome disassembly is due specifically to the GP motif of Ntr1, or whether the GP motif of another Prp43΄s cofactor would also be sufficient. Therefore, we incubated purified ILS with Prp43, ATP and the GP motif of Pfa1 comprising amino acids 701 to 767 (Pfa1GP) (Figure 1B). Surprisingly, Pfa1΄s GP motif stimulates the release of the IL RNA nearly as efficiently as Ntr1΄s GP (70% versus 60%) (Figures 3A and 2D). These results indicate that the primary function of a GP domain is to induce the ATPase/helicase activity of Prp43, as a prerequisite for the dissociation of the ILS, and not to regulate the access of Prp43 to its target site in the ILS or the pre-ribosome. This is consistent with earlier evidences suggesting that Prp43 recognizes its target site in the spliceosome alone and the GP motif of Ntr1 is required only to activate the ATPase/helicase activity of Prp43 and not for proper recognition of its binding site in the spliceosome (42).
Figure 3.
Prp43 dissociates the ILS in co-operation with the GP motif of Pfa1 but not with full-length Pfa1. 10–30% glycerol gradient sedimentation of purified ILS incubated in solution with ATP, Prp43 and (A) Pfa1 G-patch motif (Pfa1GP), (B) Pfa1 full length (Pfa1FL). Samples were analyzed and quantified as described in Figure 2. (C) Kinetic parameters (kcat and KM) of Prp43, Prp43 + Pfa1GP and Prp43 + Pfa1FL in absence or presence of poly-A20 RNA cofactor. (D) ATPase activity of Prp43 in the absence and presence of Pfa1GP and Pfa1FL, respectively, and without poly-A20 RNA. (E) ATPase activity of Prp43 as in (D) but with poly-A20 RNA. Experimental points were fitted with the Michaelis-Menten equation and error bars (S.D.) were obtained from three independent experiments. For detailed information see Methods.
Next we asked whether the full-length Pfa1 (Pfa1FL) (Figure 1B), which in vitro stimulates the ATPase/helicase activity of Prp43 (36), might also lead to the Prp43-ATP-dependent dissociation of ILS. However, this was not the case, as only ∼5% of the IL RNA was dissociated from the ILS (Figure 3B). To rule out the possibility that our Pfa1FL was nonfunctional, we tested its ability to stimulate Prp43΄s ATPase activity in the absence or presence of an RNA cofactor (Figure 3C–E). As shown in Figure 3C and E in the presence of RNA, a higher turnover number (2-fold increase in kcat) was observed for Pfa1FL versus Pfa1GP. On the other hand, a higher molar concentration of ATP was needed by Prp43 to hydrolyze the same amount of ATP in the presence of Pfa1FL than Pfa1GP, suggesting stronger binding (lower KM) of ATP to Prp43 in the presence of Pfa1GP (Figure 3C). Thus, Pfa1FL reduced the catalytic efficiency of Prp43 (kcat/KM) ∼3-fold. Yet, these data indicate that Pfa1FL can bind productively to Prp43 via its GP motif and stimulate Prp43΄s ATPase activity in the absence of spliceosomes, but it cannot do the same in the ILS. Thus, the N-terminal domain of Pfa1FL (NTD-Pfa1, see Figure 1B) impedes Prp43-ATP-dependent dissociation of ILS by preventing Pfa1΄s binding to the spliceosome and/or productive Prp43΄s ATP hydrolysis (see below).
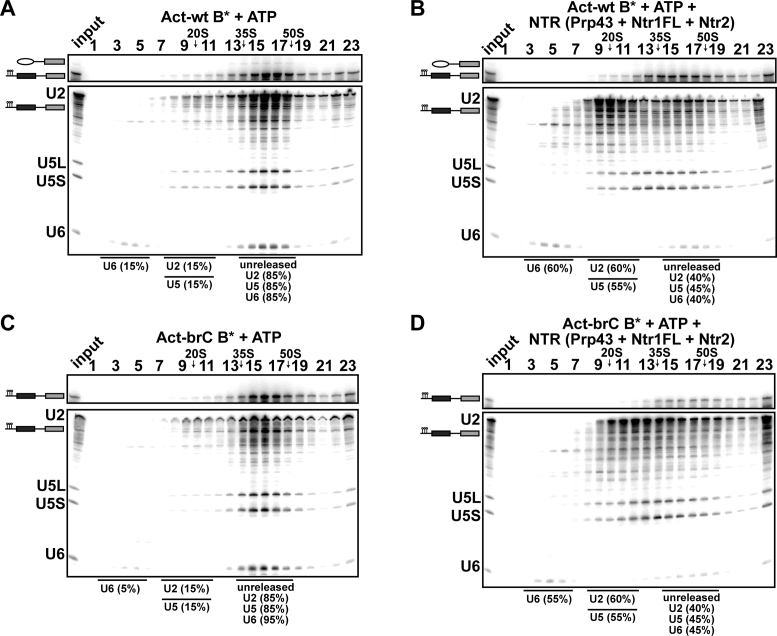
Ntr1-CTD prevents Prp43 from disassembling Bact spliceosomes assembled on an optimal/wild-type pre-mRNA substrate
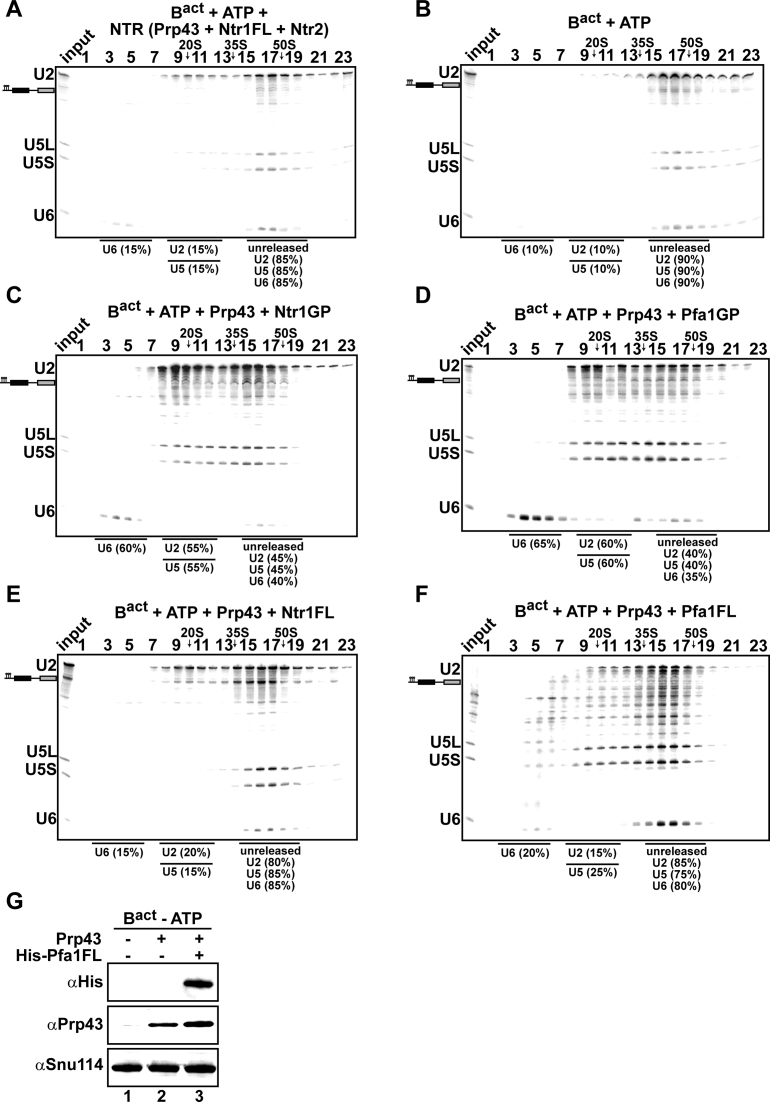
Earlier data showed that Prp43_Ntr1GP can dismantle purified wild-type Bact complexes efficiently, whereas in the presence of Ntr1 and Ntr2 the Bact complex is not disassembled by Prp43 (42), indicating that Ntr2 and Ntr1-CTD may impede a productive interaction of the Ntr1GP motif with Prp43. To determine whether the Ntr1-CTD plays a role in preventing Prp43 from disrupting properly assembled Bact spliceosomes that are not disassembled by the NTR (41), we next investigated whether Prp43 can dissociate Bact spliceosomes together with full-length Ntr1. Consistent with previous results (41), purified Bact ΔPrp2 complexes (lacking Prp2) that were assembled on a wild-type Actin7 (Act7-wt) pre-mRNA, remained stable during incubation with the NTR and ATP, (Figure 4A, compare to the negative control with ATP alone in Figure 4B). On the contrary, the addition of Prp43, ATP and Ntr1GP, led to efficient dissociation of Act7-wt Bact ΔPrp2 complexes (∼60% of the U6 snRNA and 55% of the U2 and U5 snRNPs were dissociated), yielding free U6 snRNA that migrates on top of the gradient, a 20S U2 snRNP, and a U5 snRNP that exhibits a broad migration behavior (see (42) for a detailed description); this indicates that all snRNPs were separated from each other (Figure 4C). Addition of Pfa1GP also led to efficient dissociation of the Bact complex (∼60% of U6 snRNA and the U2 and U5 snRNPs were dissociated) (Figure 4D); thus Pfa1GP can activate Prp43 and promote disassembly also in the context of Bact. In contrast, the majority (∼80%) of U2, U5 and U6 were not dissociated from the Bact ΔPrp2 complexes when incubated with Ntr1FL, ATP and Prp43 (Figure 4E). Likewise, only minimal disassembly was observed when Bact ΔPrp2 complexes were incubated with Pfa1FL, Prp43 and ATP (Figure 4F), although western blotting showed that Pfa1FL binds efficiently to Bact ΔPrp2 spliceosomes (Figure 4G, lane 3). In sum, in contrast to the bona fide ILS substrate, Ntr1FL impedes Prp43΄s disassembly of Bact spliceosomes, indicating that the Ntr1-CTD plays a role in preventing Prp43 from disassembling early/wild-type spliceosomes that should not be disassembled.
Figure 4.
Prp43 dissociates Bact spliceosomes in co-operation with the GP motif of Ntr1 or Pfa1 but not with full-length cofactors. 10–30% glycerol gradient sedimentation of purified Bact ΔPrp2 complexes incubated with ATP, and (A) NTR, (B) no recombinant protein, (C) Prp43 and Ntr1GP, (D) Prp43 and Pfa1GP, (E) Prp43 and Ntr1FL, (F) Prp43 and Pfa1FL. Samples were analyzed as described in Figure 2. Quantifications were performed with ImageQuant software (Molecular Dynamics). U2 and U5: numbers represent the percentage of U2 or U5 snRNAs released in the peak fractions (sum of fractions 8–12) or associated with the Bact ΔPrp2 complex (unreleased, sum of fractions 15–19) relative to the U2 or U5 snRNAs distributed in fractions 8–12 and 15–19, the sum of which was set to 100%. U6: numbers represent the percentage of U6 snRNA released in the peak fractions (sum of fractions 2–6) or associated with the Bact ΔPrp2 complex (unreleased, sum of fractions 15–19) relative to the U6 snRNA distributed in the fractions 2–6 and 13–17, the sum of which was set to 100%. (G) Purified Bact ΔPrp2 complexes formed on radiolabeled Act7-wt pre-mRNA were incubated without recombinant proteins (lane 1) or with 2-fold molar excess of either Prp43 (lane 2) or Prp43 plus His-Pfa1FL (lane 3) in the absence of ATP. Samples were loaded on distinct glycerol gradients. Proteins from the peak fraction were recovered and separated by SDS-PAGE on a 4–12% Bis-TrisNuPAGE polyacrylamide gel (Invitrogen) and visualized by Western blotting using anti-Snu114, anti-Prp43 or anti-polyhistidine-tag antibodies (Quiagen).
In conclusion, the finding that Prp43 in cooperation with the GP motifs of Ntr1 or Pfa1, efficiently dismantles the Bact ΔPrp2 complex, indicates that the isolated GP motifs can bind efficiently to the CTD domain of Prp43, leading to activation of Prp43 and thus dissociation of the Bact ΔPrp2 complex. On the contrary, with full-length Pfa1, Ntr1 or the NTR complex dissociation was not observed, indicating that in the Bact ΔPrp2 complex the Ntr1-CTD (or the NTD-Pfa1) impedes a productive interaction between Ntr1GP (or Pfa1GP) and Prp43.
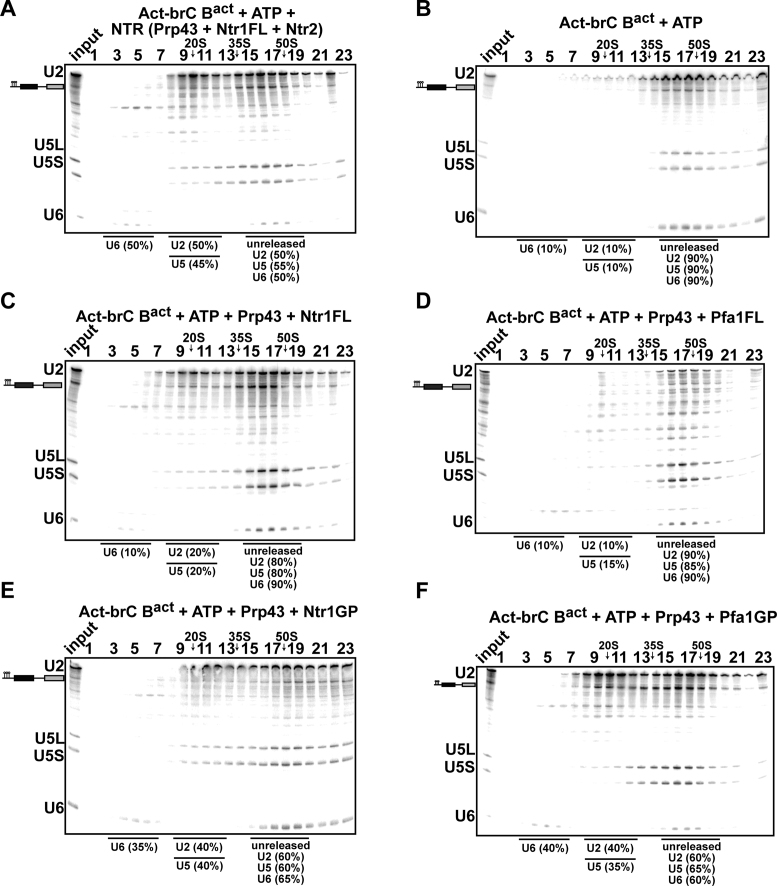
NTR dismantles Bact complexes assembled on a suboptimal/mutated pre-mRNA substrate
Our data so far indicate that the Ntr1-CTD prevents Prp43-mediated disassembly of early spliceosomal complexes that are engaged with an optimal/wild-type substrate. As the NTR is also involved in discarding spliceosomes assembled on suboptimal substrates (39), we next tested whether the NTR can discriminate between Bact spliceosomes that are engaged with an optimal or suboptimal substrate. As a suboptimal substrate we chose an Actin pre-mRNA in which the branch-site (BS) adenosine (A) was mutated to C (henceforth termed Act-brC), because Act-brC pre-mRNA allows the formation of activated spliceosomes but is a poor substrate for step 1 catalysis (46). In contrast to Act7-wt Bact ΔPrp2 complexes (Figure 4A), a substantial fraction of Act-brC Bact ΔPrp2 complexes (∼50%) was disassembled when incubated with the NTR, but not with ATP alone (∼10%) (Figure 5A and B). These results suggest that the structure of the Bact ΔPrp2 complex differs to some extent depending on whether it is engaged with Act7-wt or Act-brC pre-mRNA, such that Ntr1-CTD (and Ntr2) can productively interact with the spliceosome and with Prp43 only within Act-brC Bact ΔPrp2 complexes and thus lead to their dismantling. Surprisingly, in contrast to the NTR, Ntr1FL together with Prp43 and ATP stimulated disassembly of Act-brC Bact spliceosomes less efficiently (∼20%) (Figure 5C). As expected, background disassembly (∼10%) was observed for Pfa1FL (Figure 5D). Addition of the GP motif of Ntr1 or Pfa1 (plus Prp43 and ATP), lead to significant disassembly, as ∼40% of the U2 and U5 snRNPs were dissociated from Act-brC Bact spliceosomes (Figure 5E and F). All together our data indicate that the non-GP domains of Ntr1 (or Pfa1) impedes activation of Prp43. Ntr2 might have a role in discriminating between spliceosomes assembled on suboptimal substrates versus those assembled on optimal/wild-type substrates.
Figure 5.
The NTR complex disassembles Bact spliceosomes formed on a suboptimal substrate. 10–30% glycerol gradient sedimentation of purified Bact ΔPrp2 complexes assembled on Act-brC pre-mRNA incubated with ATP and (A) NTR, (B) no recombinant protein, (C) Prp43 and Ntr1FL, (D) Prp43 and Pfa1FL, (E) Prp43 and Ntr1GP, (F) Prp43 and Pfa1GP. Samples were analyzed as described in Figure 2. Quantification of snRNAs released or associated with the Act-brC Bact ΔPrp2 complex (unreleased) was performed as described in Figure 4.
NTR dismantles B* complexes assembled on optimal or suboptimal pre-mRNA substrates
It was previously reported that the NTR complex can disassemble also affinity-purified spliceosomes associated with optimal/wild-type pre-mRNA substrates but only when stalled at specific stages in the splicing pathway (41). It was suggested that the susceptibility of the spliceosome to be disassembled by the NTR might require the removal of certain proteins from the catalytic center as for example the SF3a/b complex, which would lead to the formation of a B* complex (post-Prp2 stage) (19,41). NTR would be recruited to the spliceosome only at defined stages because Ntr2 binds with high affinity only at these defined stages. Moreover, competitive exclusion of Ntr2 from the spliceosome under normal splicing conditions was suggested (41). Consistent with these data we observed that Bact ΔPrp2 complexes transformed into B* complexes after the action of Prp2 (Spp2 and ATP), are indeed more susceptible to the NTR-mediated disassembly (Figure 6A and B). As anticipated, the NTR complex can also disassemble Act-brC B* complexes as efficiently as Act-brC Bact (Figure 6C and D). These experiments indicate that Ntr2 and Ntr1-CTD have an available binding site in the B* and Act-brC B* complexes, thus assuring a productive interaction of Prp43 with the Ntr1GP.
Figure 6.
The NTR complex disassembles B* spliceosomes assembled on optimal or suboptimal substrates. 10–30% glycerol gradient sedimentation of reconstituted and purified B* complexes incubated with (A) ATP, (B) NTR. 10–30% glycerol gradient sedimentation of purified B* complexes assembled on Act-brC pre-mRNA incubated with (C) ATP, (D) NTR. Quantification of snRNAs released or associated with the Act-wt or Act-brC B* complexes (unreleased) was performed as described in Figure 4.
DISCUSSION
The Ntr1-CTD prevents Prp43-mediated disassembly of early/wild-type Bact spliceosomes
Prp43 fused to Ntr1΄s G-patch motif is as efficient as the NTR in ILS disassembly, yielding identical dissociation products and recognizing its natural ILS target even in the absence of Ntr1΄s C-terminal-domain (CTD) and Ntr2 (42). However, the question whether Ntr2 and the Ntr1-CTD play a role in controlling directly or indirectly the productive interaction of Ntr1΄s GP motif with Prp43 in the spliceosome was never addressed. Here, we show that Ntr1΄s G-patch motif alone (i.e. not fused to Prp43), is sufficient for Prp43-mediated ILS disassembly, thus its primary function is to couple Prp43 ATP hydrolysis to ILS disassembly, while the Ntr1-CTD and Ntr2 do not play a major role in regulating the productive interaction of Ntr1GP with Prp43 in ILS disassembly (Figures 2 and 7A). Furthermore, we show that the GP motif of Pfa1, one of Prp43΄s cofactors in ribosome biogenesis, can replace the GP motif of the spliceosomal cofactor Ntr1 and can activate Prp43-mediated disassembly of ILSs in a similar manner (Figures 2, 3 and 7A). This is consistent with previous results indicating that the specificity for interaction with the spliceosome resides in Prp43 and is independent of the G-patch motif bound by Prp43 (42). To the best of our knowledge, the present work is the first ‘direct’ evidence of Prp43-mediated dissociation of a molecular machine promoted by isolated G-patches.
Figure 7.
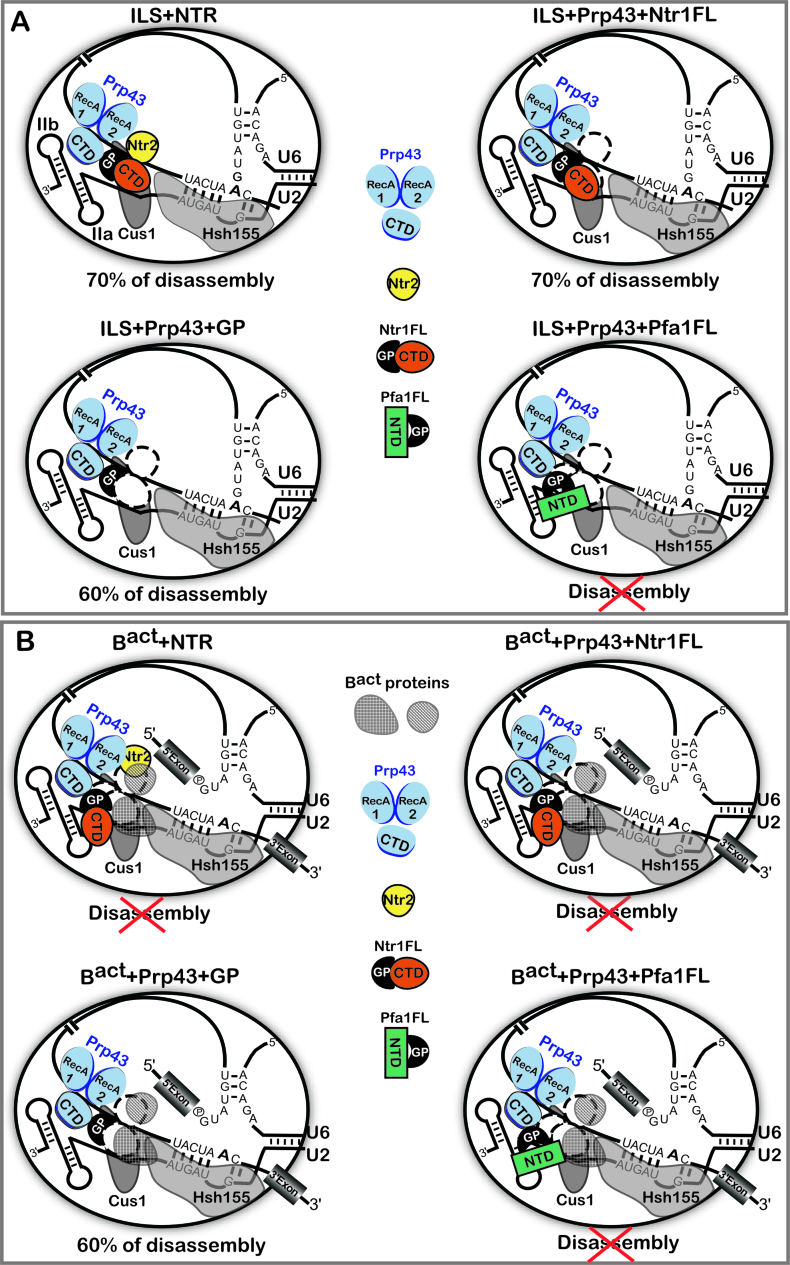
Schematic representation of the regulation of Prp43 by its cofactors in the disassembly of spliceosomes. (A) The ILS is schematically shown with bound Prp43, Ntr1 and Ntr2. The U2 snRNA is shown with the stem IIa and b and two of the U2 proteins, Cus1 and Hsh155, bound on the pre-mRNA anchoring site, according to (6). The DEAH-box helicase Prp43 is depicted schematically as proposed previously (49,52), with the canonical helicase core comprising the RecA1 and RecA2 domains. The conserved C-terminal domain (CTD) is also shown. Prp43 is bound to its binding site on the pre-mRNA intron (42). Ntr1FL and Pfa1FL indicate Ntr1 and Pfa1 full-length, respectively. GP indicates the G-patch motif. In the presence of Pfa1FL the ILS is not disassembled, indicated by a red cross. Efficiency of Prp43-mediated disassembly is indicated and represents the percentage of intron lariat RNA dissociated from the ILS. (B) The Bact complex is schematically shown as in (A) with proteins of the Bact complex obstructing the binding sites for Ntr2 and Ntr1-CTD. In the presence of the NTR, Ntr1FL or Pfa1FL, the Bact complex is not disassembled, indicated by red crosses. Efficiency of Prp43-mediated disassembly promoted by the GP-motif is indicated and represents the percentage of U2, U5 and U6 dissociated from the Bact complex.
How is Prp43 controlled by its cofactors to discard selectively only ILS but not earlier complexes that should not to be discarded? Our data provide evidence that the Ntr1-CTD allows a productive interaction of Prp43 with the GP motif of Ntr1 only in spliceosomes that have to be disassembled (ILS) (Figures 2 and 7A). Indeed, the Ntr1-CTD does not impede the adjacent GP motif to enter into productive binding with the CTD region of Prp43 on the ILS. On the contrary, the Bact spliceosome is not disassembled by Prp43 in the presence of Ntr1FL or Ntr1FL and Ntr2, (Figures 4 and 7B), while Prp43 together with the isolated Ntr1GP can dissociate purified wild-type Bact complexes efficiently. Our data suggest that the availability of a Ntr1-CTD/Ntr2 binding site might be a major contributing factor to determine whether a given spliceosomal complex is disrupted by Prp43. This may depend on the structural/compositional difference between the Bact complex and the ILS (11,12,26,47,48), which likely affects the ability of Ntr1-CTD/Ntr2 to stably/productively dock to the Bact complex and/or to the Bact-bound Prp43, while the GP motif alone can bind productively to the CTD of Prp43 in both spliceosomal complexes (Figure 7). Ntr1 thus regulates the activity of Prp43 at multiple levels; the Ntr1GP activates Prp43 and is also sufficient for coupling Prp43΄s ATPase activity to the mechanical work required for spliceosome disassembly. The Ntr1-CTD appears to play the role of a ‘doorkeeper’, guaranteeing a productive interaction of the Ntr1GP motif with Prp43 only when a binding site for Ntr1-CTD exists or is available next to the Prp43-binding site on the spliceosome - as is the case for the ILS (Figure 7).
Ntr1 cannot be replaced by the ribosomal cofactor Pfa1 in spliceosome disassembly
Here we tested whether the ribosomal cofactor of Prp43, Pfa1 might replace the spliceosomal cofactor Ntr1 using our spliceosome disassembly assay. Both cofactors are able to interact via their GP motif with Prp43 in vivo (28), and in vitro this interaction is sufficient to stimulate Prp43΄s ATPase and unwinding activities (36,43) (and Figure 3). In this study, the extent of stimulation of Prp43 by Pfa1FL is very similar to that previously observed by Lebaron and co-workers (36). Yet, the RNA-dependent ATPase activity of Prp43 in the presence or absence of Pfa1FL is lower here as compared to previous studies (36). These differences in RNA-stimulated ATPase activity are likely due to higher concentrations of the RNA cofactor used in previous studies [1500-fold excess of total yeast RNA over Prp43 (36,49)], as compared to a 5-fold excess of poly-A20 RNA over Prp43 used here. Alternatively, total yeast RNA may contain RNA of different length, and longer RNAs may influence stimulation of ATP hydrolysis by Prp43, as previously shown (50).
Our data showed that Ntr1FL together with Prp43 is sufficient to promote significant ILS disassembly (∼70%) (Figures 2 and 7A). In contrast, Pfa1FL, does not activate Prp43 for ILS disassembly, even though it can activate Prp43 in the absence of spliceosomes (Figure 3 and 7A). Although the isolated GP motif of Pfa1 can promote disassembly, the entire protein (which is not a spliceosomal protein) cannot. As we showed that Pfa1FL binds efficiently to Bact (Figure 4), this indicates that the non-GP domain of Pfa1 (NTD-Pfa1) precludes a productive interaction of the Pfa1GP with Prp43 in any spliceosomal complex including the ILS (Figure 7), despite the fact that Pfa1FL can productively dock to pre-ribosomal complexes and activate Prp43 in ribosome biogenesis.
A biochemical/structural proofreading function for the NTR
To ensure the fidelity of splicing, several spliceosomal ATPases proofread various steps of the splicing process by discriminating against mutated suboptimal substrates (38,40). Spliceosomes formed on suboptimal substrates are subsequently rejected and subjected to a discard pathway involving Prp43 (Ntr1 and Ntr2), which disassembles the discarded complexes. Thus, in addition to the ILS, discarded spliceosomes formed on mutated pre-mRNAs are also substrates for the NTR complex. The NTR normally functions first after the release of the mRNA from the spliceosome or when defective spliceosomes are earmarked for the discard pathway by a RNA helicase/ATPase. Here we demonstrated that the NTR can discriminate in vitro between Bact complexes formed on an optimal substrate (wild-type Act7 pre-mRNA), as opposed to those assembled on a suboptimal substrate (Act-brC pre-mRNA), in which the branch site adenosine of the Actin pre-mRNA was mutated to cytosine; the NTR disassembled the latter but not the wild-type Bact complex (Figures 4 and 5). As both purified Bact complexes lack the ATPase Prp2 and are incubated solely with the NTR, disassembly is uncoupled from a potential kinetic proofreading step, involving Prp2. During the latter, the Bact to B* transition mediated by Prp2 in vivo or in cell extracts, would potentially be slower with Bact complexes formed on the mutated Actin pre-mRNA, hindering their progression and presumably allowing the productive binding of the NTR. Our data suggest that this kinetic difference is not needed for the NTR to discriminate, and that instead (or additionally) structural/compositional changes in spliceosomes formed on suboptimal substrates likely play a major role in enabling/triggering NTR-mediated disassembly. These structural/compositional differences likely affect the ability of the NTR or Ntr1 and Ntr2 to stably/productively dock to the structurally compromised spliceosomes. For instance, immunoprecipitation experiments indicated that Prp5 binds with increased affinity to brC spliceosomes (51), whereas Prp5 is not bound to a brA/wild-type Bact complex (13). Prp5 remains bound in pre-catalytic spliceosomes when the configuration of the U2–branch site interaction is impaired (51). Thus, a compromised conformation of the U2–branch site interaction may create conditions or change molecular interactions that make the spliceosome susceptible to disassembly by NTR. Thus, a function of the NTR may be to ‘proofread’ or inspect the RNP structure of spliceosomal complexes. Moreover, our data indicate that the major contribution for the prevention of disassembly is driven by Ntr1 via its CTD domain. Indeed, in both spliceosomes, Act7-wt Bact and Act-brC Bact, the presence of Ntr1FL and Prp43, impedes the disassembly; while the presence of the GP motif and Prp43 allows the disassembly (Figures 4 and 5). Our data suggest that Ntr2 has a major role in recognition of defective spliceosomes, likely by stabilizing Ntr1 and allowing Prp43 to enter a productive interaction with the Ntr1GP in defective spliceosomes (Figure 5A and C and Figure 7B).
To date, several 3D structures of Prp43 in the ADP-bound (post-catalytic) state were solved (49,52,53), but no structure of Prp43 with a bound GP motif could be determined so far. The 3D structure of Prp43 bound to one of its G-patch cofactors or within the spliceosome (11,12,47) will give new insights into the molecular mechanism of the regulation of Prp43.
ACKNOWLEDGEMENTS
We thank T. Conrad for excellent technical assistance.
FUNDING
Deutsche Forschungsgemeinschaft [SFB 860 to R.F. and R.L.]. Funding for open access charge: Deutsche Forschungsgemeinschaft [SFB 860 to R.F. and R.L.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Wahl M.C., Will C.L., Lührmann R.. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009; 136:701–718. [DOI] [PubMed] [Google Scholar]
- 2. Cordin O., Beggs J.D.. RNA helicases in splicing. RNA Biol. 2013; 10:83–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cordin O., Hahn D., Beggs J.D.. Structure, function and regulation of spliceosomal RNA helicases. Curr. Opin. Cell Biol. 2012; 24:431–438. [DOI] [PubMed] [Google Scholar]
- 4. O’Day C.L., Dalbadie-McFarland G., Abelson J.. The Saccharomyces cerevisiae Prp5 protein has RNA-dependent ATPase activity with specificity for U2 small nuclear RNA. J. Biol. Chem. 1996; 271:33261–33267. [DOI] [PubMed] [Google Scholar]
- 5. Gozani O., Feld R., Reed R.. Evidence that sequence-independent binding of highly conserved U2 snRNP proteins upstream of the branch site is required for assembly of spliceosomal complex A. Genes Dev. 1996; 10:233–243. [DOI] [PubMed] [Google Scholar]
- 6. Schneider C., Agafonov D.E., Schmitzová J., Hartmuth K., Fabrizio P., Lührmann R.. Dynamic contacts of U2, RES, Cwc25, Prp8 and Prp45 proteins with the pre-mRNA branch-site and 3΄ splice site during catalytic activation and step 1 catalysis in yeast spliceosomes. PLoS Genet. 2015; 11:e1005539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. McPheeters D.S., Muhlenkamp P.. Spatial organization of protein-RNA interactions in the branch site-3΄ splice site region during pre-mRNA splicing in yeast. Mol. Cell. Biol. 2003; 23:4174–4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wu J.A., Manley J.L.. Base pairing between U2 and U6 snRNAs is necessary for splicing of a mammalian pre-mRNA. Nature. 1991; 352:818–821. [DOI] [PubMed] [Google Scholar]
- 9. Datta B., Weiner A.M.. Genetic evidence for base pairing between U2 and U6 snRNA in mammalian mRNA splicing. Nature. 1991; 352:821–824. [DOI] [PubMed] [Google Scholar]
- 10. Madhani H.D., Guthrie C.. A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell. 1992; 71:803–817. [DOI] [PubMed] [Google Scholar]
- 11. Rauhut R., Fabrizio P., Dybkov O., Hartmuth K., Pena V., Chari A., Kumar V., Lee C.T., Urlaub H., Kastner B. et al. Molecular architecture of the Saccharomyces cerevisiae activated spliceosome. Science. 2016; 353:1399–1405. [DOI] [PubMed] [Google Scholar]
- 12. Yan C., Wan R., Bai R., Huang G., Shi Y.. Structure of a yeast activated spliceosome at 3.5 A resolution. Science. 2016; 353:904–911. [DOI] [PubMed] [Google Scholar]
- 13. Fabrizio P., Dannenberg J., Dube P., Kastner B., Stark H., Urlaub H., Lührmann R.. The evolutionarily conserved core design of the catalytic activation step of the yeast spliceosome. Mol. Cell. 2009; 36:593–608. [DOI] [PubMed] [Google Scholar]
- 14. Warkocki Z., Schneider C., Mozaffari-Jovin S., Schmitzová J., Hobartner C., Fabrizio P., Lührmann R.. The G-patch protein Spp2 couples the spliceosome-stimulated ATPase activity of the DEAH-box protein Prp2 to catalytic activation of the spliceosome. Genes Dev. 2015; 29:94–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ohrt T., Prior M., Dannenberg J., Odenwälder P., Dybkov O., Rasche N., Schmitzová J., Gregor I., Fabrizio P., Enderlein J. et al. Prp2-mediated protein rearrangements at the catalytic core of the spliceosome as revealed by dcFCCS. RNA. 2012; 18:1244–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim S.H., Lin R.J.. Spliceosome activation by PRP2 ATPase prior to the first transesterification reaction of pre-mRNA splicing. Mol. Cell. Biol. 1996; 16:6810–6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lardelli R.M., Thompson J.X., Yates J.R. 3rd, Stevens S.W.. Release of SF3 from the intron branchpoint activates the first step of pre-mRNA splicing. RNA. 2010; 16:516–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chiu Y.F., Liu Y.C., Chiang T.W., Yeh T.C., Tseng C.K., Wu N.Y., Cheng S.C.. Cwc25 is a novel splicing factor required after Prp2 and Yju2 to facilitate the first catalytic reaction. Mol. Cell. Biol. 2009; 29:5671–5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Warkocki Z., Odenwälder P., Schmitzová J., Platzmann F., Stark H., Urlaub H., Ficner R., Fabrizio P., Lührmann R.. Reconstitution of both steps of Saccharomyces cerevisiae splicing with purified spliceosomal components. Nat. Struct. Mol. Biol. 2009; 16:1237–1243. [DOI] [PubMed] [Google Scholar]
- 20. Schwer B., Guthrie C.. PRP16 is an RNA-dependent ATPase that interacts transiently with the spliceosome. Nature. 1991; 349:494–499. [DOI] [PubMed] [Google Scholar]
- 21. Horowitz D.S. The mechanism of the second step of pre-mRNA splicing. Wiley Interdiscip. Rev. RNA. 2012; 3:331–350. [DOI] [PubMed] [Google Scholar]
- 22. Company M., Arenas J., Abelson J.. Requirement of the RNA helicase-like protein PRP22 for release of messenger RNA from spliceosomes. Nature. 1991; 349:487–493. [DOI] [PubMed] [Google Scholar]
- 23. Arenas J.E., Abelson J.N.. Prp43: An RNA helicase-like factor involved in spliceosome disassembly. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:11798–11802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Martin A., Schneider S., Schwer B.. Prp43 is an essential RNA-dependent ATPase required for release of lariat-intron from the spliceosome. J. Biol. Chem. 2002; 277:17743–17750. [DOI] [PubMed] [Google Scholar]
- 25. Tsai R.T., Fu R.H., Yeh F.L., Tseng C.K., Lin Y.C., Huang Y.H., Cheng S.C.. Spliceosome disassembly catalyzed by Prp43 and its associated components Ntr1 and Ntr2. Genes Dev. 2005; 19:2991–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fourmann J.B., Schmitzová J., Christian H., Urlaub H., Ficner R., Boon K.L., Fabrizio P., Lührmann R.. Dissection of the factor requirements for spliceosome disassembly and the elucidation of its dissociation products using a purified splicing system. Genes Dev. 2013; 27:413–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pandit S., Lynn B., Rymond B.C.. Inhibition of a spliceosome turnover pathway suppresses splicing defects. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:13700–13705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Banerjee D., McDaniel P.M., Rymond B.C.. Limited portability of G-patch domains in regulators of the Prp43 RNA helicase required for pre-mRNA splicing and ribosomal RNA maturation in Saccharomyces cerevisiae. Genetics. 2015; 200:135–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Aravind L., Koonin E.V.. G-patch: a new conserved domain in eukaryotic RNA-processing proteins and type D retroviral polyproteins. Trends Biochem. Sci. 1999; 24:342–344. [DOI] [PubMed] [Google Scholar]
- 30. Tsai R.T., Tseng C.K., Lee P.J., Chen H.C., Fu R.H., Chang K.J., Yeh F.L., Cheng S.C.. Dynamic interactions of Ntr1-Ntr2 with Prp43 and with U5 govern the recruitment of Prp43 to mediate spliceosome disassembly. Mol. Cell. Biol. 2007; 27:8027–8037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Boon K.L., Auchynnikava T., Edwalds-Gilbert G., Barrass J.D., Droop A.P., Dez C., Beggs J.D.. Yeast ntr1/spp382 mediates prp43 function in postspliceosomes. Mol. Cell. Biol. 2006; 26:6016–6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Christian H., Hofele R.V., Urlaub H., Ficner R.. Insights into the activation of the helicase Prp43 by biochemical studies and structural mass spectrometry. Nucleic Acids Res. 2014; 42:1162–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lebaron S., Froment C., Fromont-Racine M., Rain J.C., Monsarrat B., Caizergues-Ferrer M., Henry Y.. The splicing ATPase prp43p is a component of multiple preribosomal particles. Mol. Cell. Biol. 2005; 25:9269–9282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Leeds N.B., Small E.C., Hiley S.L., Hughes T.R., Staley J.P.. The splicing factor Prp43p, a DEAH box ATPase, functions in ribosome biogenesis. Mol. Cell. Biol. 2006; 26:513–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Combs D.J., Nagel R.J., Ares M. Jr, Stevens S.W.. Prp43p is a DEAH-box spliceosome disassembly factor essential for ribosome biogenesis. Mol. Cell. Biol. 2006; 26:523–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lebaron S., Papin C., Capeyrou R., Chen Y.L., Froment C., Monsarrat B., Caizergues-Ferrer M., Grigoriev M., Henry Y.. The ATPase and helicase activities of Prp43p are stimulated by the G-patch protein Pfa1p during yeast ribosome biogenesis. EMBO J. 2009; 28:3808–3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Guglielmi B., Werner M.. The yeast homolog of human PinX1 is involved in rRNA and small nucleolar RNA maturation, not in telomere elongation inhibition. J. Biol. Chem. 2002; 277:35712–35719. [DOI] [PubMed] [Google Scholar]
- 38. Semlow D.R., Staley J.P.. Staying on message: ensuring fidelity in pre-mRNA splicing. Trends Biochem. Sci. 2012; 37:263–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mayas R.M., Maita H., Semlow D.R., Staley J.P.. Spliceosome discards intermediates via the DEAH box ATPase Prp43p. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:10020–10025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Koodathingal P., Staley J.P.. Splicing fidelity: DEAD/H-box ATPases as molecular clocks. RNA Biol. 2013; 10:1073–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen H.C., Tseng C.K., Tsai R.T., Chung C.S., Cheng S.C.. Link of NTR-mediated spliceosome disassembly with DEAH-box ATPases Prp2, Prp16, and Prp22. Mol. Cell. Biol. 2013; 33:514–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fourmann J.B., Dybkov O., Agafonov D.E., Tauchert M.J., Urlaub H., Ficner R., Fabrizio P., Lührmann R.. The target of the DEAH-box NTP triphosphatase Prp43 in Saccharomyces cerevisiae spliceosomes is the U2 snRNP-intron interaction. eLife. 2016; 5:e15564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tanaka N., Aronova A., Schwer B.. Ntr1 activates the Prp43 helicase to trigger release of lariat-intron from the spliceosome. Genes Dev. 2007; 21:2312–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yean S.L., Lin R.J.. U4 small nuclear RNA dissociates from a yeast spliceosome and does not participate in the subsequent splicing reaction. Mol. Cell. Biol. 1991; 11:5571–5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Agarwal K.C., Miech R.P., Parks R.E. Jr. Guanylate kinases from human erythrocytes, hog brain, and rat liver. Methods Enzymol. 1978; 51:483–490. [DOI] [PubMed] [Google Scholar]
- 46. Tseng C.K., Liu H.L., Cheng S.C.. DEAH-box ATPase Prp16 has dual roles in remodeling of the spliceosome in catalytic steps. RNA. 2011; 17:145–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yan C., Hang J., Wan R., Huang M., Wong C.C., Shi Y.. Structure of a yeast spliceosome at 3.6-angstrom resolution. Science. 2015; 349:1182–1191. [DOI] [PubMed] [Google Scholar]
- 48. Sun C., Rigo N., Fabrizio P., Kastner B., Luhrmann R.. A protein map of the yeast activated spliceosome as obtained by electron microscopy. RNA. 2016; 22:1427–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Walbott H., Mouffok S., Capeyrou R., Lebaron S., Humbert O., van Tilbeurgh H., Henry Y., Leulliot N.. Prp43p contains a processive helicase structural architecture with a specific regulatory domain. EMBO J. 2010; 29:2194–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tanaka N., Schwer B.. Mutations in PRP43 that uncouple RNA-dependent NTPase activity and pre-mRNA splicing function. Biochemistry. 2006; 45:6510–6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Liang W.W., Cheng S.C.. A novel mechanism for Prp5 function in prespliceosome formation and proofreading the branch site sequence. Genes Dev. 2015; 29:81–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. He Y., Andersen G.R., Nielsen K.H.. Structural basis for the function of DEAH helicases. EMBO Rep. 2010; 11:180–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tauchert M.J., Fourmann J.B., Christian H., Lührmann R., Ficner R.. Structural and functional analysis of the RNA helicase Prp43 from the thermophilic eukaryote Chaetomium thermophilum. Acta Crystallogr. F Struct. Biol. Commun. 2016; 72:112–120. [DOI] [PMC free article] [PubMed] [Google Scholar]