Abstract
Domain V of 23S rRNA, gyrA and gyrB Quinolones Resistance-Determining Region (QRDR), and pbp-1A gene point mutations were investigated in Helicobacter pylori-resistant isolates from three centres of Buenos Aires. Minimal inhibitory concentrations (MICs) were performed in 197 isolates from 52 H. pylori-positive naive patients by agar dilution method. Point mutations were achieved by amplification and sequencing of the target genes, and their association with resistance was determined by natural transformation assays. Resistance rates were as follows: metronidazole 28.8%, clarithromycin (CLA) 26.9%, levofloxacin (LEV) 32.7%, and amoxicillin (AMX) 7.6%. Nearly one-third of patients carried multidrug-resistant isolates. A2143G or A2142G in domain V of 23S-rRNA was found in all isolates showing high level of resistance to CLA (MIC >2 mg/L), accounting for 76.0% (38/50) of those with the resistant phenotype. The mutations A2267G or T1861C carried by 8/12 isolates with MIC 1–2 mg/L (low level) did not confer resistance by transformation. Substitutions at GyrA position 87 or 91, mainly N87K and D91G, were found in 92.8% (52/56) of the LEV-resistant isolates: 48 isolates with MIC 4–64 mg/L and 4/8 isolates with MIC 2 mg/L. The remaining four harboured K133N, also present in susceptible isolates. None of the substitutions in GyrB demonstrated to confer resistance. Transformation proved that PBP-1A N562Y and/or T556S substitutions confer the AMX resistance in our isolates, showing an additive effect. In conclusion, the usually reported mutations related to CLA, LEV, and AMX resistance were found in our isolates. However, low-level CLA resistance seems not to be due to mutations in Domain V of 23S rRNA gene.
Keywords: : 23S rRNA, gyrA, pbp-1A, mutations, H. pylori
Introduction
The effectiveness of conventional treatment regimens aimed to Helicobacter pylori eradication has declined due to the growing resistance to several antimicrobial agents and the high emergence of multidrug-resistant strains in several geographic areas.1–3 The progressive reduction of primary susceptibility patterns seems to parallel with the community antimicrobial consumption.4
In general, bismuth salts, amoxicillin (AMX), and tetracycline (TET) show a slighter emergence of resistance than other antimicrobial agents.1 The difference in the emergence of primary resistance among antimicrobial agents might be explained by the mechanism(s) involved in its evolution. Resistance to AMX requires mainly amino acid substitutions in the acyl transpeptidase domain of PBP-1A,5–7 although substitutions in PBP-2 and PBP-3 could elevate the minimal inhibitory concentration (MIC) value.8 However, other mechanisms could be involved in the high levels of AMX-resistant strains.6,9
H. pylori resistance to clarithromycin (CLA) has been frequently associated with single point mutations in the peptidyl transferase region of domain V of 23S rRNA. The most common mutations are A2143G and A2142G, while A2142C is associated with CLA resistance to a lesser extent. However, resistant isolates with other mutations seem to be emerging.10,11 Mutations outside domain V of 23S rRNA or in other genes seem to contribute to CLA resistance.12,13
Single point mutations in Quinolones Resistance Determining Region (QRDR) of gyrA appear to be the main event leading to fluoroquinolone resistance.14 Amino acid substitutions at positions 91 (D91G, N, A, Y or H) and 87 (N87L, I, A or K) of GyrA were most frequently associated with levofloxacin (LEV) resistance, although substitutions at position 88 (A88V or P) or at position 86 (D86N) have been described.15 Rimbara et al. suggested that amino acid substitution at position 463 in GyrB could be a novel mechanism of fluoroquinolone resistance in H. pylori.16
Several reports suggested that overexpression of the efflux pump systems in H. pylori were associated with multidrug resistance.17–19
For geographic regions with high rates of resistance to CLA and LEV, different empiric regimens have been considered, including bismuth and nonbismuth-based quadruple therapies (sequential or concomitant), as well as triple therapies where AMX is administered several times a day for an optimal concentration at the gastric mucosa level.1 However, all these options also included an ecological cost for the human microbiome.1 In this era of increasing prevalence of antimicrobial resistance, tailored treatment based on the individual characterization of H. pylori susceptibility appears to be a reasonable future alternative.1 At this point, the genotypic detection of H. pylori resistance to antimicrobial agents, mainly to CLA and LEV, using whole sample of DNA isolated directly from gastric biopsy tissues would avoid performing H. pylori culture systematically.20,21 For this purpose, there is an ongoing surveillance of mutations in the main antimicrobial target genes that could confer resistance. However, phenotypic methods will continue to have an advantage when resistance to the same antimicrobial agent is caused by several mechanisms and are still reliable for surveying antimicrobial susceptibility in population-based studies.
This study was aimed to investigate the point mutations in gyrA and gyrB QRDR, domain V of 23S rRNA, and pbp-1A genes in H. pylori primary-resistant isolates recovered in three centers of Buenos Aires City as a way to contribute to the assessment of probable unknown mutations related to resistance as well as to find the occurrence of those described previously.
Materials and Methods
Patients and biopsies
From November 2011 to March 2013, 52 H. pylori-positive patients without previous eradication therapy were recruited from one hospital and two community-based endoscopy centers. Written consents were provided by all patients and this study was approved by a local Human Research Ethics Committee. Four simultaneous biopsies (two from antrum: A1, mid-greater curvature; A2, antral lesser curvature within 2 cm of the pylorus; and two from corpus: C1, middle portion of the greater curvature of the corpus; C2, lesser curvature within 3 cm of the Z line) were obtained by upper gastrointestinal endoscopy from each patient. All biopsies were cultured as described previously.22
Antimicrobial susceptibility test
A screening of resistant isolates was performed by subculturing swabs of bacteria from isolation plates of each biopsy to Mueller–Hinton agar plates (Oxoid) supplemented with 5% aged sheep blood with and without metronidazole (MTZ) (8 mg/L), CLA (0.5 mg/L), AMX (0.125 mg/L), LEV (1 mg/L), and TET (1 mg/L), respectively, as described previously.23 When growth on plates with antimicrobials was observed, the expansion of single colonies from the entire population present on the plate without antimicrobials was performed to obtain confluent cultures for MIC determination and DNA extraction, as described previously.24 MIC to MTZ, CLA, AMX, LEV, TET, rifampicin, and rifabutin of all isolates recovered from the 52 patients was determined by agar dilution method.24 H. pylori ATCC 43504 was used as a quality control strain. The susceptibility of isolates was categorized according to EUCAST breakpoints (http://eucast.org/clinical_breakpoints/).
Strain delineation and molecular detection of gyrA and gyrB QRDR, domain V of 23S rRNA, and pbp-1A point mutation
DNA was extracted from confluent cultures with fewer than three “in vitro” passages as described previously.23 For strain delineation, the Random Amplified Polymorphic DNA (RAPD)-PCR was performed in all isolates with the commonly used primer pMAV 17B (5′-CACTCGTCGGGAATGCCCT-3′), after the optimization of this technique, according to Berg et al.25,26 After PCR, amplification products were electrophoretically separated in 1.2% agarose gels. For 01 matrix construction, banding patterns were analyzed by GelQuest sofware (SequentiX—Digital DNA Processing). A total of 89 bands were scored for their presence or absence. Banding pattern similarities were achieved by the Dice coefficient using Free-Tree–freeware program.27,28
For amplification and sequencing gyrA and gyrB QRDR, the following sets of primers were used: gyrAQRDR-F: 5′-TTTRGCYTATTCMATGAGCGT-3′ and gyrAQRDR-R: 5′-GCAGACRCTTGGTARAATA-3; gyrBQRDR-F: 5′-YGCAAAAGCCAGAGAAG CCA-3 and gyrBQRDR-R: 5′-ACATGCCCTTGTTCAATCAGC-3, respectively. The domain V of 23S-rRNA was amplified with CLA-F 5′-AAAGAG TCCCTCCCGACTGT-3 and CLA-R 5′-CCCCAGTCAAACTACCCAC-3. The whole pbp-1A gene was amplified using the following sets of primers: (1) PBP-1F: 5′-TAGCCATTCTTATCGCTC-3 and PBP-1R: 5′-CGACTAGCATGGTGATTT-3; (2) PBP-2F: 5′-AACCGCAAGTTTAGGGTA-3 and PBP-2R: 5′-GATCATGCT AGCGTTTAAGT-3; (3) PBP-3F: 5′-ACGCGTCTAATGAAGATG-3′ and PBP-3R: 5′-GTGATGCTTTCAATGAGC-3; and (4) PBP-4F: 5′-GGGAGCTTTGCTATCTCA-3 and PBP-4R: 5′-GTTCCTCGCTATCGTCTG-3; yielding overlapping fragments. PCR products were purified with the NucleoSpin purification kit (Macherey–Nagel) according to the manufacturer's instructions. Nucleotide sequences of both chains were obtained using an ABI 3500xL Genetic Analyzer sequencer (Applied Biosystems) or by submitting purified PCR products to Macrogen (Macrogen, Inc.).
The sequences studied in this study were assigned with the following GenBank accession numbers: gyrA, KT958622-KT958662; gyrB, KT958664-KT958703; domain V 23S-rRNA, KT958589-KT958604 and KX148071; and pbp-1A, KT958605, KT958606, KT958609, KT958610, KT958613, and KT958615-KT958620.
Natural transformation
Natural transformation of DNA was carried out in line with Lin et al.29 using fragments containing gyrA, gyrB, and domain V of 23S rRNA mutations not previously described and those amplified with primers PBP-3F and PBP-3R, PBP-4F and PBP-4R, and PBP-3F and PBP-4R for pbp-1A. Susceptible isolates showing MICs 0.064, 0.125, and 0.032 mg/L to CLA, LEV, and AMX, respectively, were used as recipient strains. The sequences of the antimicrobial target genes studied of recipient strains were assigned with the following GenBank accession numbers: domain V 23S-rRNA, KT958588, gyrA, KT958621; gyrB, KT958663; and pbp-1A, KT966878. As controls, bacteria were transformed with TE(10 mM Tris–HCl, 1 mM EDTA) without DNA. All transformants obtained from each experiment were analyzed by DNA sequencing and MIC determination. The corresponding GenBank accession numbers of transformants are KT958607, KT958608, KT958611, KT958612, and KT958614.
Results
One-hundred ninety-seven isolates were recovered from the 52 patients studied. The 197 isolates were susceptible to TET. The MIC90 of rifampicin and rifabutin were 1 mg/L (range 0.25–8 mg/L) and 0.5 mg/L (range 0.125–8 mg/L), respectively.
Isolates resistant to MTZ, CLA, LEV, or AMX in one or more biopsies were found in 15 (28.8%), 14 (26.9%), 17 (34.6%), and 4 (7.6%) of the 52 patients, respectively (Tables 1 and 2). The rate of resistance to MTZ is consistent with that reported in Argentina by a systematic review of resistance in Latin America.30 In contrast, CLA and LEV showed a higher rate of resistance.30
Table 1.
Levofloxacin, Clarithromycin, or Metronidazole MICs of the 197 Isolates from the 52 Patients
| Patients (No. of isolates)a | GyrA substitution | GyrB substitution | LEVbMIC | 23S-RNA mutations | CLAbMIC | MTZbMIC |
|---|---|---|---|---|---|---|
| Susceptible isolates | ||||||
| 21 (77) | NA | NA | 0.125–0.5 | NA | 0.064–0.125 | 2–8 |
| 2 (6) | NA | NA | 0.5 | G1939A | 0.5c | 4 |
| 1 (4) | K133N | None | 0.25 | NA | 0.064 | 4 |
| 1 (4) | N87T | None | 0.25 | NA | 0.064 | 2 |
| Resistant isolates to one or more antimicrobial agents | ||||||
| 2 (8) | NA | NA | 0.5 | NA | 0.064 | 64 |
| 1 (3/1)d | NA | NA | 0.5 | NA | 0.064 | 16/0.5 |
| 1 (4) | NA | NA | 0.25 | A2143G | 128 | 4 |
| 1 (4) | NA | NA | 0.5 | A2143G, T2182C | 32 | 0.5 |
| 1 (1/1/2) | N87K/N87K/N87Id | None | 2/2/32d | G1939A, A2306G | 0.064 | 128/4/128 |
| 1 (1/3) | N87Tf/N87I | I418N, K461E/None | 0.5/16 | NA | 0.064 | 4 |
| 2 (8)e | D91G | None | 32–64 | None | 0.064 | 2–8 |
| 1 (4)e | D91N | None | 32 | NA | 0.064 | 2 |
| 1 (2/2) | None/D91G | NA | 0.5/4 | NA | 0.064 | 1 |
| 1 (4) | None | None | 1 | A2143G | 16 | 2 |
| 1 (4) | D91G | None | 8 | A2142G | 32 | 0.125 |
| 1 (4) | N87K | None | 16 | A2267Gf | 1 | 4 |
| 1 (2)e | None | None | 0.125 | A2143G | 32 | 64 |
| 1 (3/1) | None | R484K, D481E/None | 1/0.125 | None | 0.5 | 32/16 |
| 1 (4) | N87K | None | 16 | NA | 0.064 | 128 |
| 1 (4) | E105G-D145Gf | NA | 1 | A2143G | 16 | 128 |
| 1 (4) | N87K | None | 16 | A2143G | 32 | 2 |
| 1 (2/2) | None/D91N | None | 1/8 | A2143G | 8 | 32 |
| 1 (2/2) | N87K/None | None | 2/0.5 | NA | 0.064 | 64 |
| 1 (4) | K133Ne | None | 2 | A2143G | 128 | 128 |
| 1 (3/1) | N87K/None | None | 4/0.5 | A2306G | 2/0.5 | 64 |
| 1 (2/1/1) | N87Tf, F149S/E105G, D145G, N152Ie/N87I | None | 0.5/1/32 | A2143G/A2143G, C1872A, C1882T/A2143G | 128 | 1 |
| 1 (3/1) | D91A/D91N | None/E381Ge | 16/4 | C1944T, G2212A/NA | 0.25/0.064 | 64/2 |
| 1 (1/3) | NA | NA | 0.5 | T1861Cf | 2 | 8/64 |
| 1 (3/1) | D91G/K133Nf | R484Kf/none | 64/0.5 | NA | 0.064/2 | 16/4 |
GyrA and GyrB amino acid substitutions and Domain V 23S-rRNA mutations.
Four biopsies/patient were studied. In brackets is shown the number of isolates recovered from the indicated number of patients.
LEV, levofloxacin; CLA, clarithromycin; MTZ, metronidazole.
MIC 0.5 mg/L: intermediate susceptibility to CLA.
Patients harboring isolates with different MIC values or mutations recovered from different biopsies.
Patients harbored isolates with resistance to amoxicillin: 23U, 27U 29U, 78E (Table 2).
Amino acid substitutions in GyrA or GyB or mutations in Domain V 23S-r-RNA gene not related to resistance. GyrA N87T was described by Tankovic et al. in the susceptible population.14
MIC, minimal inhibitory concentration.
Table 2.
Amino Acid Substitutions in the Acyl Transpeptidase Domain of PBP-1A
| Position of amino acid substitutions in the acyl transpeptidase domain of PBP-1A | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strains | 406 | 417 | 473 | 480 | 515 | 535 | 543 | 552 | 556 | 560 | 562 | 596+ | 604 | MIC |
| 26695 | E | S | F | A | M | D | S | I | T | N | N | Y | 0.032 | |
| 63F (recipient) | T | T | F | V | M | N | S | I | T | N | N | G | Y | 0.032 |
| 29UA1 | E | T | F | A | M | N | H | I | T | N | N | Y | 0.064 | |
| 29UC2 (donor) | A | T | F | A | M | N | S | I | S | N | Y | Y | 2 | |
| T29UC2Frag 3 + 4a,b | A | T | F | A | M | N | S | I | S | N | Y | G | Y | 2 |
| T29UC2Frag 4b | T | T | F | A | M | N | S | I | S | N | Y | G | Y | 1–2 |
| 23UC2 (donor) | A | T | F | A | M | N | S | I | S | N | N | Y | 1 | |
| T23UC2Frag 4 | T | T | F | A | M | N | S | I | S | N | N | G | Y | 0.5–1 |
| T23UC2Frag 3 + 4 | A | T | F | A | M | N | S | I | S | N | N | G | Y | 1 |
| 27UC1 (donor) | A | S | F | A | M | N | S | I | S | T | N | Y | 1 | |
| T27UC1F4 Frag 3 + 4 | T | T | F | A | M | N | S | I | S | T | N | G | Y | 1 |
| Low susceptible strains | ||||||||||||||
| 78EA2 | A | T | F | A | I | N | R | V | T | N | N | H | 0.25 | |
| 61EA2 | A | S | F | A | I | N | R | I | T | N | N | Y | 0.125 | |
| Susceptible strains | ||||||||||||||
| 55RC1 | A | S | F | A | M | N | S | I | T | N | N | Y | 0.032 | |
| 148EA1, A2 | T | S | F | A | M | N | H | I | T | N | N | Y | 0.032 | |
| 34UA2 | E | S | F | A | I | N | S | I | T | N | N | S | Y | 0.016 |
T: Transformant.
Frag 3 + 4: pbp-1A DNA fragment amplified with primer PBP-3F (nucleotide positions 956–973 of Helicobacter pylori 26695) and PBP-4R (nucleotide positions 1490–1507). Frag 4: pbp-1A DNA fragment 4 amplified with primers PBP-4F (nucleotide positions 1411–1428) and PBP-4R (nucleotide positions 1949–1966).
Isolates with different susceptibility patterns among biopsies of a single individual were detected in 11/52 (21.2%) patients (MTZ: four patients, LEV: five patients, one for AMX, and one for LEV-MTZ-CLA) (Tables 1 and 2). This result suggested that the analysis of more than one biopsy increased the possibility to find resistant isolates in a single individual.
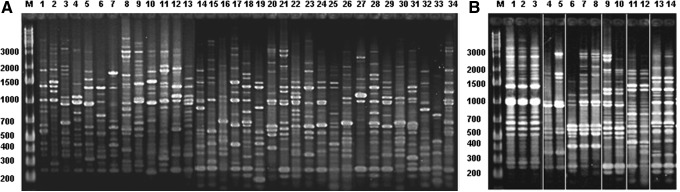
The RAPD-PCR of the 197 isolates showed 52 distinguishable banding patterns. Fingerprint similarity of isolates interpatients' was 15–75% (Dice: 0.15–0.75), and all isolates from a single individual showed an identical banding pattern, except for six of them. These six patients harbored isolates with slight differences in the RAPD-PCR profiles (Dice: 0.83–0.96). Taking into account the cutoff at the 80% of similarity level for strain differentiation, the isolates from each of these patients can be considered to be closely related.26 Figure 1A shows the banding patterns found in 34 of the 52 patients studied and Fig. 1B shows those found in the six patients harboring isolates with slightly different profiles.
FIG. 1.
RAPD-PCR. (A) Banding patterns of isolates from 34 out of the 52 patients studied. (B) Banding patterns found in the six patients that harbor isolates with slight differences in their profile. M: Molecular marker, 1 kb ladder, Fermentas, Thermo Fisher Scientific. RAPD-PCR, Random Amplified Polymorphic DNA-PCR.
The RAPD-PCR and AFLP (Amplified Fragment Length Polymorphism)-PCR that include the analysis of variations in the whole genome are very suitable to define H. pylori mixed infections as well as microevolution in a single host “per se” or the relationship among isolates showing microevolution by other genomic changes.31–34 The high discriminatory power of RAPD-PCR to determine inter- and intrapatient variation of H. pylori strains was extensively proven, as each independent strain typically yields a banding pattern that is reproducibly different from those of other strains.31–37 Thus, by this genotyping technique, our results suggest that antimicrobial-resistant H. pylori typically develops from preexisting susceptible strains rather than coinfections with different strains.
The patients included in this study represent two different populations in terms of socioeconomic status and prevalence of H. pylori colonization/infection (34.6% in patients from the hospital-based endoscopy center vs. 15% and 12.8% in patients from the two community-based endoscopy centers, respectively). AMX-resistant isolates were mainly recovered from patients assisted in the hospital-based endoscopy unit.
The rate of patients' harboring isolates with multidrug-resistant phenotypes was 18/52 (34. 6%) (LEV-CLA: four patients, LEV-MTZ: four patients; LEV-CLA-MTZ: four patients; LEV-AMX: three patients, CLA-MTZ in two patients, and CLA-MTZ-AMX in one), Table 1.
In this study, 38/50% (76.0%) of isolates phenotypically resistant to CLA showed MICs in the range 8–128 mg/L, and the A2143G mutation in the domain V of 23S-rRNA gene was carried by 34/38 of them (Table 1). The other 4/38 isolates harbored the mutation A2142G (Table 1). The remaining 12 isolates exhibited MIC 1–2 mg/L and showed A2267G or T1861C mutations, not related to CLA resistance by transformation (8/12), A2306G, found also in the susceptible ones (3/12), or “wild type” genotype (1/12). The mutation G1939A alone or associated with A2306G and the double mutation C1944T-G2212A were found in isolates with MIC 0.064–0.5 mg/L (Table 1).
As shown in Table 1, 48/56 (85.7%) isolates with phenotypic resistance to LEV exhibited a MIC range of 4–64 mg/L. These 48 isolates harbored GyrA amino acid substitutions at positions 87 or 91, prevailing N87K and D91G. The substitution N87K was also found in 4/8 isolates with low-level resistance (MIC 2 mg/L) (Table 1). The remaining four low-level-resistant isolates, recovered from one patient, harbored K133N, also carried by susceptible ones. In two patients, the finding of resistant isolates with different GyrA amino acid substitutions located at different biopsies suggests the independent evolution toward resistance (Table 1). Four isolates with the double E105G-D145G substitution and one with the triple E105G-D145G-N152I showed MIC 1 mg/L (Table 1). However, the transformation did not relate these amino acid variations with increasing MIC.
GyrB amino acid substitutions were most frequently found in susceptible isolates showing I418N or double I418N-K461E substitutions, as well as the R484K or D481E described previously (Table 1).14 One isolate carrying the GyrA D91N showed E381G substitution in GyrB, and transformation indicated that the last one did not contribute to resistance (Table 1).
Table 2 shows PBP-1A amino acid variations in the AMX-resistant isolates and in several susceptible ones. The isolate with AMX MIC 2 mg/L harbored almost all the amino acid changes described by Kwon et al.6 in strains with β-lactam resistance (E406A, S417T, T556S, and N562Y) (29UC2, Table 2). In contrast, the isogenic susceptible isolate 29UA1 lacked substitutions at position 406, 556, and 562, but showed S543H observed also in other susceptible ones (Table 2). Two extra AMX-resistant isolates showed amino acid variations at positions 406, 417, and 556 (23U), or at 406 and 556 only (27U) (Table 2).
DNA fragment amplified by primers 3F-3R (PBB-1A fragment 3) included the amino acid substitutions at positions 406 and 417 and the DNA fragment amplified primers 4F-4R (PBB-1A fragment 4), those at positions 556 and 562. These fragments and the one amplified with primer 3F-4R (PBP-1A fragment 3 + 4) were used for transformation. With a selecting concentration of 0.25 mg/L of AMX, transformants were obtained with 29UC2 and 23UC1 fragments 4 and 3 + 4, and with 27UC1 fragment 3 + 4 only (Table 2). Hence, no transformants were obtained with the fragment containing E406A and S417T alone. The MIC of randomly selected transformants indicated that substitutions at fragments 4 had the highest impact on the MIC value of the clinical resistant isolates (Table 2). The substitution S543R was observed in isolates with MIC 0.125–0.25 mg/L (Table 2).
Discussion
In this study, we found a high rate of resistance to CLA and LEV. Multidrug- resistant isolates were recovered from 1/3 patients supporting that testing these resistances is essential to tailor the therapy. Point mutations A2143G and A2142G in domain V of 23S-rRNA were found in all isolates with CLA MIC >2 mg/L, with a wide prevalence of A2143G. In contrast, the CLA low-level-resistant isolates lacked mutations at positions 2142 and 2143. This finding suggests the presence of an alternative mechanism of resistance or mutations in 23S-rRNA gene outside domain V, in these isolates.
Shen et al., demostrated “in vitro” that A2143G mutation was detectable in isolates with MIC 32 mg/L, but not in the low-level-resistant ones, selecting CLA-resistant isolates from the wild-type H. pylori 25695 after serial passages into growing concentrations of this antimicrobial.38
Binh et al., analyzed the low-level- and high-level-resistant isolates obtained from H. pylori 25695 after passages through serial concentrations of CLA by whole-genome sequencing.13 The authors also reported the presence of A2143G in the high-level-resistant isolates only.13 However, the selected low-level-resistant isolates showed mutations in genes outside the 23S rRNA.13 Transformation indicated that mutations in hp1048 (infB) and hp1314 (rpl22) seem to be linked to the CLA low-level-resistant phenotype.13 Transformants containing a single mutation in infB (G160A), 9 bp insertion in rpl22, 3 bp deletion in rpl22, or mutation in domain V of 23S-rRNA showed MICs of 0.5, 2, 4, and 32 mg/L, respectively, while transformants containing double mutations (in the 23S rRNA and infB or rpl22) showed MICs >256 mg/L.13
Fontana et al. found that the transition T2717C linked to the CLA low-level resistance in seven clinical isolates. This mutation is located in a highly conserved region of domain VI of the 23S rRNA.12 However, several studies reported the presence of A2143G and in a lesser extent A2142G in isolates with low-level resistance as well as in intermediately susceptible ones.39–41 In addition, different levels of CLA resistance without mutations in the 23S rRNA have also been reported.12,39,40
In this study, the transition G1939A, previously suggested by Garrido and Toledo42 as related to CLA resistance, was found in isolates with MIC range of 0.064–0.5 mg/L.
Resistance to LEV was due to substitution at position 87 (K, I) and 91 (G, N, A) of GyrA QRDR in the 92.8% of the resistant isolates. N87K and D91G were prevalent. N87K was found in isolates with low level and high level of LEV resistance and N87I, D91G, D91N, and D91A in the last ones only. It is well known that the distribution of amino acid variations conferring resistance differed according to the geographical origins of strains. However, D91G and N87K seemed to be the major mutations described around the world.15 The amino acid changes at positions 88 or 86 were not found in our isolates.15 None of the substitutions found in GyrB proved conferring resistance. Hence, the four resistant isolates recovered from one patient who lacked GyrA amino acid variations could perhaps carry a gyrA or gyrB mutation in a region distinct from the QRDRs.14 In line with Tankovic et al., we also found several amino acid substitutions in the QRDRs of both GyrA and GyrB in susceptible isolates.14
Substitutions at PBP-1A were reported in several studies to likely alter AMX binding, including S414R, T438M, F473L, S543R, T556S, and N562T.5–10 Kwon et al.6 suggested that the acquisition of β-lactam resistance was consistently associated with the transfer of a mosaic block of amino acid substitutions contained in the C-terminal portion of PBP-1A. According to Gerrits et al., several amino acid variations in or adjacent to the second (SKN402-404) and third (KTG555-557) conserved penicillin-binding protein motifs (PBM) can mediate AMX resistance in H. pylori. Hence, E406A, S417T, S414R (located in or adjacent to the second PBM), T555S, and N561Y (located in or adjacent to the third PBM) substitutions represent the major factors in resistance.
By homology modeling analyses, Qureshi et al. defined the importance of specific amino acids (including those mentioned above and S543R, among others) in the AMX resistance.43 The authors reported that over the proximity of amino acid changes to PBMs “per se” it is the impact of the specific amino acid change within the putative binding cleft of the PBP-1A transpeptidase region, which appears to define those substitutions deemed most important in AMX resistance. Combinations of specific substitutions can additively increase the level of AMX resistance.43
The isolate 29UC2 harboring T556S and N562T showed a higher MIC than 23UC1 and 27UC1 with T556S only. The MICs of their transformants obtained with fragment 4 were close to the clinical isolates. The lack of transformants with fragment 3 and the similarity of MIC values of the transformants obtained with fragment 3 + 4 and 4 suggested that E406A and/or S471T played a minor role in the AMX resistance in our isolates. At this point, Qureshi et al. also found that E406T, which is very close to the SKN402-404 PBM, does not affect AMX susceptibility.43
Substitution S543R was found in two isolates with MIC 0.125–0.25 mg/L lacking the additional amino acid variations described in the other resistant isolates. Qureshi et al., reported MIC 0.25 mg/L when the single substitution S543R was created in H. pylori 26695.43 S543R was described jointly with T556S and N562T and with V469M and F473L in high-level-resistant strains, supporting the additive effect of several amino acid changes to increase the AMX MIC.6,43 Of note, in this study, the S543H substitution was found in isolates with MIC 0.032–0.064. S414R substitution, found in AMX-resistant isolates in different geographic areas, was absent in the isolates of this study and in those reported previously.7,44
Conclusion
In this study, mutations A2143G and A2142 in Domain V of the 23S-rRNA gene were the only ones found in high-level CLA-resistant isolates. In the low-level-resistant isolates, the lack of mutations at positions 2142 and 2143 and the finding of mutations at other positions that did not confer resistance suggested nucleotide variations outside the Domain V of 23S-rRNA or alternative mechanisms of resistance. GyrA QRDR substitution at positions 87 and 91 was found in 92.8% of LEV-resistant isolates, prevailing N87K and D91G. GyrA QRDR was found at low- and high-level-resistant isolates. None of the substitutions found in GyrB QRDR could be related to resistance. PBP-1A substitutions T556S and N562T proved to confer AMX resistance in isolates with MIC 1–2 mg/L. S543R substitution was found in two isolates with MIC 0.125 and 0.5 mg/L, lacking other PBP-1A amino acid variations related to the increase of AMX MIC.
Acknowledgments
The authors thank Lucas E. Zapata, CONICET supporting staff, for his contribution with the sequencing of DNAs amplified from transformants and Sergio Mazzini for providing language assistance. This study was supported by grants BID 1728 OC-AR PICT 2010-1492 provided by Agencia Nacional de Promoción Científica y Tecnológica and 20020120100171BA from Universidad de Buenos Aires (UBACyT 2013–2016).
Disclosure Statement
No competing financial interests exist.
References
- 1.Mégraud F. 2013. Current recommendations for Helicobacter pylori therapies in a world of evolving resistance. Gut Microbes. 4:541–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almeida N., Romãozinho J.M., Donato M.M., Luxo C., Cardoso O., Cipriano M.A., Marinho C., Fernandes A., Calhau C., and Sofia C. 2014. Helicobacter pylori antimicrobial resistance rates in the central region of Portugal. Clin. Microbiol. Infect. 20:1127–1133 [DOI] [PubMed] [Google Scholar]
- 3.Shokrzadeh L., Alebouyeh M., Mirzaei T., Farzi N., and Zali M.R. 2015. Prevalence of multiple drug-resistant Helicobacter pylori strains among patients with different gastric disorders in Iran. Microb. Drug Resist. 21:105–110 [DOI] [PubMed] [Google Scholar]
- 4.Liou J.M., Chang C.Y., Chen M.J., Chen C.C., Fang Y.J., Lee J.Y., Wu J.Y., Luo J.C., Liou T.C., Chang W.H., Tseng C.H., Wu C.Y., Yang T.H., Chang C.C., Wang H.P., Sheu B.S., Lin J.T., Bair M.J., and Wu M.S. 2015. The Primary Resistance of Helicobacter pylori in Taiwan after the National Policy to Restrict Antibiotic Consumption and Its Relation to Virulence Factors-A Nationwide Study. PLoS One. 10:e0124199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerrits M.M., Godoy A.P., Kuipers E.J., Ribero M.L., Stoof J., Mendonça S., van Vliet A.H., Pedrazzoli J., Jr., and Kusters J.G. 2002. Multiple mutations in or adjacent to the conserved penicillin-binding protein motifs of the penicillin-binding protein 1A confer amoxicillin resistance to Helicobacter pylori. Helicobacter. 11:181–187 [DOI] [PubMed] [Google Scholar]
- 6.Kwon D.H., Dore M.P., Kim J.J., Kato M., Lee M., Wu J.Y., and Graham D.Y. 2003. High-level beta-lactam resistance associated with acquired multidrug resistance in Helicobacter pylori. Antimicrob. Agents Chemother. 47:2169–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matteo M.J., Granados G., Olmos M., Wonaga A., and Catalano M. 2008. Helicobacter pylori amoxicillin heteroresistance due to point mutations in PBP-1A in isogenic isolates. J. Antimicrob. Chemother. 61:474–477 [DOI] [PubMed] [Google Scholar]
- 8.Rimbara E., Noguchi N., Kawai T., and Sasatsu M. 2008. Mutations in penicillin-binding proteins 1, 2 and 3 are responsible for amoxicillin resistance in Helicobacter pylori. J. Antimicrob. Chemother. 61:995–998 [DOI] [PubMed] [Google Scholar]
- 9.Qureshi N.N., Gallaher B., and Schiller N.L. 2014. Evolution of amoxicillin resistance of Helicobacter pylori in vitro: characterization of resistance mechanisms. Microb. Drug Resist. 20:509–516 [DOI] [PubMed] [Google Scholar]
- 10.De Francesco V., Zullo A., Ierardi E., Giorgio F., Perna F., Hassan C., Morini S., Panella C., and Vaira D. 2010. Phenotypic and genotypic Helicobacter pylori clarithromycin resistance and therapeutic outcome: benefits and limits. J. Antimicrob. Chemother. 65:327–332 [DOI] [PubMed] [Google Scholar]
- 11.De Francesco V., Zullo A., Giorgio F., Saracino I., Zaccaro C.C., Hassan C., Ierardi E., Di Leo A., Fiorini G., Castelli V., Lo Re G., and Vaira D. 2014. Change of point mutations in Helicobacter pylori rRNA associated with clarithromycin resistance in Italy. J. Med. Microbiol. 63:453–457 [DOI] [PubMed] [Google Scholar]
- 12.Fontana C., Favaro M., Minelli S., Criscuolo A.A., Pietroiusti A., Galante A., and Favalli C. 2002. New site of modification of 23S rRNA associated with clarithromycin resistance of Helicobacter pylori clinical isolates. Antimicrob. Agents Chemother. 46:3765–3769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Binh T.T., Shiota S., Suzuki R., Matsuda M., Trang T.T., Kwon D.H., Iwatani S., and Yamaoka Y. 2014. Discovery of novel mutations for clarithromycin resistance in Helicobacter pylori by using next-generation sequencing. J. Antimicrob. Chemother. 69:1796–1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tankovic J., Lascols C., Sculo Q., Petit J.C., and Soussy C.J. 2003. Single and double mutations in gyrA but not in gyrB are associated with low- and high-level fluoroquinolone resistance in Helicobacter pylori. Antimicrob. Agents Chemother. 47:3942–3944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia M., Raymond J., Garnier M., Cremniter J., and Burucoa C. 2012. Distribution of spontaneous gyrA mutations in 97 fluoroquinolone-resistant Helicobacter pylori isolates collected in France. Antimicrob. Agents Chemother. 56:550–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rimbara E., Noguchi N., Kawai T., and Sasatsu M. 2012. Fluoroquinolone resistance in Helicobacter pylori: role of mutations at position 87 and 91 of GyrA on the level of resistance and identification of a resistance conferring mutation in GyrB. Helicobacter. 17:36–42 [DOI] [PubMed] [Google Scholar]
- 17.Liu Z.Q., Zheng P.Y., and Yang P.C. 2008. Efflux pump gene hefA of Helicobacter pylori plays an important role in multidrug resistance. World J. Gastroenterol. 14:5217–5222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Amsterdam K., Bart A., and van der Ende A. 2005. Helicobacter pylori TolC efflux pump confers resistance to metronidazole. Antimicrob. Agents Chemother. 49:1477–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Z., Liu Z.Q., Zheng P.Y., Tang F.A., and Yang P.C. 2010. Influence of efflux pump inhibitors on the multidrug resistance of Helicobacter pylori. World J. Gastroenterol. 16:1279–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y.K., Kuo F.C., Liu C.J., Wu M.C., Shih H.Y., Wang S.S., Wu J.Y., Kuo C.H., Huang Y.K., and Wu D.C. 2015. Diagnosis of Helicobacter pylori infection: current options and developments. World J. Gastroenterol. 21:11221–11235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mégraud F., Bénéjat L., Ontsira Ngoyi E.N., and Lehours P. 2015. Molecular approaches to identify Helicobacter pylori antimicrobial resistance. Gastroenterol. Clin. North Am. 44:577–596 [DOI] [PubMed] [Google Scholar]
- 22.Matteo M.J., Granados G., Pérez C.V., Olmos M., Sanchez C., and Catalano M. 2007. Helicobacter pylori cag pathogenicity island genotype diversity within the gastric niche of a single host. J. Med. Microbiol. 56:664–669 [DOI] [PubMed] [Google Scholar]
- 23.Matteo M.J., Pérez C.V., Domingo M.R., Olmos M., Sanchez C., and Catalano M. 2006. DNA sequence analysis of rdxA and frxA from paired metronidazole-sensitive and -resistant Helicobacter pylori isolates obtained from patients with heteroresistance. Int. J. Antimicrob. Agents. 27:152–158 [DOI] [PubMed] [Google Scholar]
- 24.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Seventeenth Informational Supplement M100-S17. CLSI, Wayne, PA, USA, 2007 [Google Scholar]
- 25.Kim J.J., Kim J.G., and Kwon D.H. 2003. Mixed-infection of antibiotic susceptible and resistant Helicobacter pylori isolates in a single patient and underestimation of antimicrobial susceptibility testing. Helicobacter. 8:202–206 [DOI] [PubMed] [Google Scholar]
- 26.Berg D.E., Lelwala-Guruge J., Incecik E.T., Srivastava K., and Akopyants N.S. 1997. H. pylori DNA fingerprinting using the Arbitrarily Primed PCR (AP-PCR) or Random Amplified Polymorphic DNA (RAPD) Method. Methods Mol. Med. 8:117–132 [DOI] [PubMed] [Google Scholar]
- 27.Struelens M.J. Members of the European Study Group on Epidemiological Markers (ESGEM), of the European Society for Clinical Microbiology and Infectious Diseases (ESCMID). 1996. Consensus guidelines for appropriate use and evaluation of microbial epidemiologic typing systems. Clin. Microbiol. Infect. 2:2–11 [DOI] [PubMed] [Google Scholar]
- 28.Pavlícek A., Hrdá S., and Flegr J. 1999. Free-Tree—freeware program for construction of phylogenetic trees on the basis of distance data and bootstrap/jackknife analysis of the tree robustness. Application in the RAPD analysis of genus Frenkelia. Folia Biol. (Praha). 45:97–99 [PubMed] [Google Scholar]
- 29.Lin E.A., Zhang X.S., Levine S.M., Gill S.R., Falush D., and Blaser M.J. 2009. Natural transformation of Helicobacter pylori involves the integration of short DNA fragments interrupted by gaps of variable size. PLoS Pathog. 5:e1000337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Camargo M.C., García A., Riquelme A., Otero W., Camargo C.A., Hernandez-García T., Candia R., Bruce M.G., and Rabkin C.S. 2014. The problem of Helicobacter pylori resistance to antibiotics: a systematic review in Latin America. Am. J. Gastroenterol. 109:485–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuipers E.J., Israel D.A., Kusters J.G., Gerrits M.M., Weel J., Van Der E., Van Der H., Wirth H.P., Höök-Nikanne J., Thompson S.A., and Blaser M.J. 2000. Quasispecies development of Helicobacter pylori observed in paired isolates obtained years apart from the same host. J. Infect. Dis. 181:273–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carroll I.M., Ahmed N., Beesley S.M., Khan A.A., Ghousunnissa S., Morain C.A., Habibullah C.M., and Smyth C.J. 2004. Microevolution between paired antral and paired antrum and corpus Helicobacter pylori isolates recovered from individual patients. J. Med. Microbiol. 53:669–677 [DOI] [PubMed] [Google Scholar]
- 33.Lundin A., Björkholm B., Kupershmidt I., Unemo M., Nilsson P., Andersson D.I., and Engstrand L. 2005. Slow genetic divergence of Helicobacter pylori strains during long-term colonization. Infect. Immun. 73:4818–4822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Linz B., Windsor H.M., Gajewski J.P., Hake C.M., Drautz D.I., Schuster S.C., and Marshall B.J. 2013. Helicobacter pylori genomic microevolution during naturally occurring transmission between adults. PLoS One. 8:e82187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akopyanz N., Bukanov N.O., Westblom T.U., Kresovich S., Berg D.E. 1992. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 20:5137–5142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu H., Fero J.B., Mendez M., Carpenter B.M., Servetas S.L., Rahman A., Goldman M.D., Boren T., Salama N.R., Merrell D.S., and Dubois A. 2015. Analysis of a single Helicobacter pylori strain over a 10-year period in a primate model. Int. J. Med. Microbiol. 305:392–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ben Mansour K., Fendri C., Battikh H., Garnier M., Zribi M., Jlizi A., and Burucoa C. 2016. Multiple and mixed Helicobacter pylori infections: comparison of two epidemiological situations in Tunisia and France. Infect. Genet. Evol. 37:43–48 [DOI] [PubMed] [Google Scholar]
- 38.Shen J., Zhang J.Z., Ke Y., and Deng D. 2005. Formation of A2143G mutation of 23S rRNA in progression of clarithromycin resistance in Helicobacter pylori 26695. Microb. Drug. Resist. 11:100–106 [DOI] [PubMed] [Google Scholar]
- 39.Versalovic J., Shortridge D., Kimbler K., Griffy M.V., Beyer J., Flamm R.K., Tanaka S.K., Graham D.Y., and Go M.F. 1996. Mutations in 23S rRNA are associated with clarithromycin resistance in Helicobacter pylori. Antimicrob. Agents Chemother. 40:477–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Versalovic J., Osato M.S., Spakovsky K., Dore M.P., Reddy R., Stone G.G., Shortridge D., Flamm R.K., Tanaka S.K., and Graham D.Y. 1997. Point mutations in the 23S rRNA gene of Helicobacter pylori associated with different levels of clarithromycin resistance. Antimicrob. Agents Chemother. 40:283–286 [DOI] [PubMed] [Google Scholar]
- 41.Boyanova L., Markovska R., Yordanov D., Gergova G., and Mitov I. 2016. Clarithromycin resistance mutations in Helicobacter pylori in association with virulence factors and antibiotic susceptibility of the strains. Microb. Drug Resist. 22:227–232 [DOI] [PubMed] [Google Scholar]
- 42.Garrido L., and Toledo H. 2007. Novel genotypes in Helicobacter pylori involving Domain V of the 23S rRNA Gene. Helicobacter. 12:505–509 [DOI] [PubMed] [Google Scholar]
- 43.Qureshi N.N., Morikis D., and Schiller N.L. 2011. Contribution of specific amino acid changes in penicillin binding protein 1 to amoxicillin resistance in clinical Helicobacter pylori isolates. Antimicrob. Agents Chemother. 55:101–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim B.J., and Kim J.G. 2013. Substitutions in penicillin-binding protein 1 in amoxicillin-resistant Helicobacter pylori strains isolated from Korean patients. Gut Liver. 7:655–660 [DOI] [PMC free article] [PubMed] [Google Scholar]