Abstract
UV radiation is the most common risk factor for skin cancer. Patients with the autosomal recessive DNA repair disorder xeroderma pigmentosum (XP) suffer high incidence of skin cancer after sunlight exposure. XP-mutant mice are attractive models to study this syndrome, as they, too, develop UV radiation-induced skin tumors, mimicking the human phenotype. Recombinant adenovirus carrying the human XPA gene was used for in vivo gene therapy in UVB-irradiated skin of such mice. Virus s.c. injection led to the expression of the XPA protein in basal keratinocytes and prevented deleterious effects in the skin, including late development of squamous cell carcinoma. Thus, efficient adenovirus gene delivery to the skin is a promising tool for reconstitution of specific DNA repair defects in XP patients.
Keywords: DNA repair, gene therapy, ultraviolet, keratinocytes
Curing hereditary human diseases by providing functional copies of relevant genes is an active area of ongoing research (1). However, to date, so-called gene therapy has met with relatively limited success. Among the many complexities that limit progress in this general field is the relatively paucity of human hereditary diseases that obviously lend themselves to this therapeutic approach.
Xeroderma pigmentosum (XP) is a human autosomal recessive disease characterized by hypersensitivity to sunlight and a marked predisposition to skin cancer (2). Indeed, this condition is the most sun-sensitive human disorder known. XP individuals have an ≈1,000-fold increased risk of developing cancer at sun-exposed areas, primarily the skin (3-5). Most cases of XP arise from inactivating mutations in any of seven genes designated XPA-XPG that are required for nucleotide excision repair (NER) of DNA damage caused by exposure to sunlight (6). The mean age of diagnosis is 3 years, and the mean age of the onset of skin cancers is 8 years (7). Despite the fact that skin cancer is one of the more surgically tractable forms of cancer, XP individuals suffer multiple skin cancers, including malignant melanoma. Hence, the disease is typically associated with a 30- to 40-year reduction in life span (3). Aside from surgical removal of individual skin cancers, sometimes accompanied by reconstructive surgery using unexposed tissue from the same patient, current therapies only involve isotretinoin application. The most effective “treatment” of XP is stringent avoidance of all sources of UVB radiation from very early childhood (8-11).
The XPA genetic complementation group (defective in the XPA gene) comprises one of the largest groups among XP patients (12). This gene encodes a protein involved in the initial damage-recognizing steps of NER and the stabilization of the multiprotein repair complex assembled at sites of DNA damage (13). Mice defective in the highly conserved Xpa gene (as well as multiple other XP-mutant mice) have been generated by conventional gene targeting and represent attractive models for the human disease. In particular, exposure of the shaved dorsal skin of mice to UV light results in multiple skin lesions that typically progress to tumors, mainly squamous cell carcinoma (SCC) (14, 15).
Skin is a highly accessible organ for gene therapy. In this study, we report the use of a recombinant adenovirus as a vehicle for delivery of human XPA cDNA to Xpa-mutant mice. We observed complete recovery of defective DNA repair in mouse embryo fibroblasts (MEF) in culture after infection with the virus. Additionally, Xpa mice infected with adenovirus by s.c. injection were protected from the phenotypic consequences of UVB radiation, including tumor development. These results offer further perspectives for the therapeutic use of recombinant adenovirus in the complementation of specific molecular defects in DNA repair, with the prospect of devising tools for the prevention of skin tumors in XP patients.
Methods
MEF. Cell studies were performed with Xpa-knockout (Xpa-/-) and wild-type (WT) MEF isolated from embryonic day 13.5 embryos. All MEF were routinely grown at 37°C in a 5% CO2 humidified atmosphere in DMEM (Invitrogen), supplemented with 15% FCS (Invitrogen), and antibiotics at 1 μg/ml each of penicillin and streptomycin and 2.5 μg/ml fungizone.
Adenovirus Purification and MEF Infection. Production and purification of adenovirus vector encoding human XPA gene and the EGFP (AdyXPA) was described in ref. 16. Recombinant adenovirus infection for all cells tested was performed as described in ref. 17. In summary, ≈104 cells in 3-cm-diameter dishes were infected with 0.5 ml of the virus suspension in DMEM for 1 h at 37°C. Virus titration was attained by gene transfer unit methodology. The gene transfer unit determines the number of cells that express a reporter gene after contact with the virus (18). The titer of AdyXPA was determined by the proportion of infected cells expressing the EGFP detected by fluorescence microscopy.
Cell Survival. MEF cells were irradiated with a germicidal lamp emitting UV light, predominantly at 254 nm (UVC). Cell survival was measured 1 week later by the addition of the tetrazolium salt XTT (final concentration, 0.12 mg/ml) to the culture medium. Surviving cells, with active mitochondria, cleave the XTT substrate into an orange formazan dye. The amount of formazan dye formed after 2-h incubation was measured by using a spectrophotometer (Genesys 5, Spectronic, Westbury, NY) measuring OD at 450 and 650 nm. Cell survival was calculated as the percentage of absorbance of UV-irradiated cells in relation to the absorbance of untreated cells.
Unscheduled DNA Synthesis (UDS). Analysis of DNA repair synthesis was carried out as described in ref. 19, with modifications. Briefly, 104 cells were grown on glass coverslips for 24 h. After 24 h of culture in a serum-deprived medium (0.5% FCS), 10 μCi/ml [methyl-3H]thymidine (86.0 Ci/mmol, Amersham; 1 Ci = 37 GBq) was added to the medium for 1 h. The cells were washed with PBS and then UV-irradiated with 10 J/m2. After 3 h in thepresence of [methyl-3H]thymidine, followed by a chase of 1 h with unlabeled thymidine (100 μM), the cells were fixed with methanol/acetic acid (3:1). The cells mounted onto glass slides were washed three times with 5% trichloroacetic acid for 15 min each, then rinsed twice with 70% ethanol and once with absolute ethanol. The slides were dipped into an emulsion (EM-1, Amersham) and exposed for 1 week at 4°C. After development, the mean number of grains per nucleus was obtained by counting at least 30 non-S-phase nuclei.
Western Blot Analysis. The proteins from whole cellular extracts were analyzed by using SDS/PAGE. Thirty micrograms of total protein was loaded per lane and subsequently transferred to Hybond-C membrane (Amersham) and probed with the specific antibodies anti-XPA polyclonal (Santa Cruz Biotechnology), anti-EGFP (Clontech), and anti-β-actin. Band intensity was determined by using a GS700 densitometer and molecular analyst software, both from Bio-Rad.
In Vivo Analysis. At the beginning of the experiment, all mice were 6-9 weeks old. The mice used were all in a CBA and C57BL/6 hybrid genetic background (14). Twenty-six Xpa-/- mice were infected with AdyXPA, 24 Xpa-/- mice were infected with an empty virus (mock-treated mice), and 24 WT mice were injected with empty virus also. The mice were checked on a daily basis for skin ulceration or tumor development and for any side effect caused by the adenovirus infection. All mice were back-shaved and infected s.c. with 200 μl of a viral solution containing 2 × 109 gene transfer units (AdyXPA or mock virus) in buffer (10 mM Tris, pH 7.4/1 mM MgCl2/10% glycerol). EGFP fluorescence in the skin was detected by direct exposure of the shaved mouse to a UVC source 24 h after AdyXPA infection.
Mouse Skin Irradiation. American Philips F40 sunlamps, positioned above the cages, were used as a UVB source (280-320 nm). Twenty-four hours after infection, the mice previously shaved were irradiated for 30 min with an acute UVB dose of 3.4 kJ/m2 per day for 4 days. Mice were killed for histology analysis at 24 h, 3 weeks, and 2 and 5 months after the last UVB irradiation.
RNA Isolation and RT-PCR. Mouse skin RNA was isolated from 40- to 60-mm2 skin sections after back-shaving. After addition of 2-3 ml of an ice-cold 3 M LiCl/6 M urea solution, the skin sample was immediately homogenized on ice by using a Diax 600 homogenizer (Heidolph, Kelheim, Germany) set at 22,000 rpm. The homogenate was left at 4°C overnight, and after centrifugation at 113,000 × g for 20 min, the pellet was resuspended in 0.5 ml of 10 mM Tris·HCl/0.1 mM EDTA/0.5% SDS/0.1 mg/ml proteinase K and incubated at 37°C for 30 min. Total RNA was obtained by subsequent phenol extraction, centrifugation, and ethanol precipitation of the supernatant. The pellet was dissolved in 10 mM Tris·HCl/0.1 mM EDTA. cDNA synthesis was performed by using reverse transcriptase (Superscript II RNase H, Invitrogen). A standard PCR was performed on the cDNA by using the following pair of primers for XPA cDNA detection: (forward) 5′-GGC-TTC-TTA-GAA-GAGGAA-GAG-A-3′ and (reverse) 5′-TGC-TCG-CCG-CAA-TTCTTT-TAC-TTT-TTT-3′. As a control, the cDNA of 18S ribosome subunit was used: (forward) 5′-GTA-ACC-CGT-TGA-ACC-CCATT-3′ and (reverse) 5′-CCA-TCC-AAT-CGG-TAG-TAG-CG-3′. As a positive control for the RT-PCR, a plasmid containing the XPA cDNA was used.
Histology Analysis. The mice were killed by CO2 asphyxiation. The UVB-irradiated skin was excised, fixed by immersion in 10% paraformaldehyde, and embedded in paraffin. The paraffin blocks were sectioned, and sections were stained with hematoxylin/eosin (H&E) for histopathological examination or immunostained with anti-XPA antibody for in situ analysis. Briefly, the slides were blocked with PBS containing 5% goat serum and incubated with polyclonal rabbit anti-XPA (Santa Cruz Biotechnology) in blocking solution overnight. After the incubation, the slides were washed and incubated for 2 h with biotin-conjugated goat anti-rabbit Ab (Vector Laboratories). The slides were then washed and incubated with alkaline phosphatase-conjugated ABC kit (Vectastain Elite, Vector Laboratories) 1:100 for 45 min at room temperature, stained with new fuchsin, and counterstained with Mayer's hematoxylin (WWR International).
Results
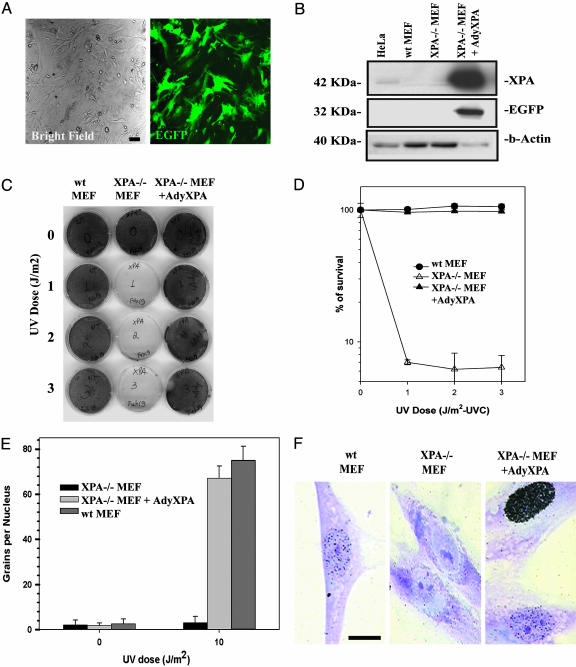
Xpa-/- MEF were infected with an adenovirus vector carrying both human XPA and EGFP cDNA (AdyXPA, ref. 16). After 24 h, cells were examined by fluorescence microscopy for EGFP expression and viability and for DNA repair capacity after exposure to UV radiation. All infected cells expressed EGFP (Fig. 1A) with no evidence of altered morphology or cytotoxicity. Western analysis of whole-cell extracts from MEF confirmed expression of both proteins (Fig. 1B). Quantitative densitometric analysis revealed about a 50-fold increased expression of XPA protein in infected Xpa-/- MEF, compared with uninfected HeLa cells. Xpa-/- MEF showed markedly reduced survival levels, compared with WT controls, after exposure to UV radiation (Fig. 1 C and D). However, after adenovirus infection, the cells exhibited levels of survival indistinguishable from WT cells. When Xpa-/- MEF were infected with recombinant adenovirus carrying a different DNA repair gene (Xpc) (16), no correction of UV-radiation sensitivity was observed (data not shown), indicating that the AdyXPA-mediated correction is gene-specific. To directly demonstrate correction of defective NER, UDS was measured before and after UV irradiation. Xpa-/- MEF exhibited reduced UDS levels after UV radiation exposure (3.3 ± 2.7 grains per nucleus) (Fig. 1 E and F). In contrast, after infection of Xpa-/- MEF with AdyXPA, the level of UDS increased to 67.2 ± 5.3 grains per nucleus, comparable with that in NER-proficient cells (75.1 ± 6.1) (Fig. 1 E and F). These results demonstrate complete correction of defective NER in Xpa-/- cells infected with the recombinant adenovirus.
Fig. 1.
Adenovirus-mediated gene transfer and complementation of Xpa-/- MEF. The experiments were performed 24 h after infection with the recombinant virus. (A) Fluorescence microscope analysis showing EGFP expression in Xpa-/- MEF. (B) Western blot analysis of infected cells. The cells were prepared as described in Methods, and anti-XPA or anti-EGFP antibodies were used as probes and anti-β-actin antibody as a control. (C) Survival curves after UV irradiation. Cells were irradiated at the indicated doses, and cell survival was measured 1 week later by the addition of XTT to the culture medium. (D) Quantification of the survival data in C. Cell lines and treatments are indicated in the inset. (E) UDS. Cells were irradiated with the indicated doses, and UDS is expressed by grains of [methyl-3H]thymidine per nucleus. Cell lines are indicated in the inset. (F) Aspect of the nuclei in UV-irradiated cells. Heavily marked nuclei indicate S-phase cells. (Scale bar, 10 μm.)
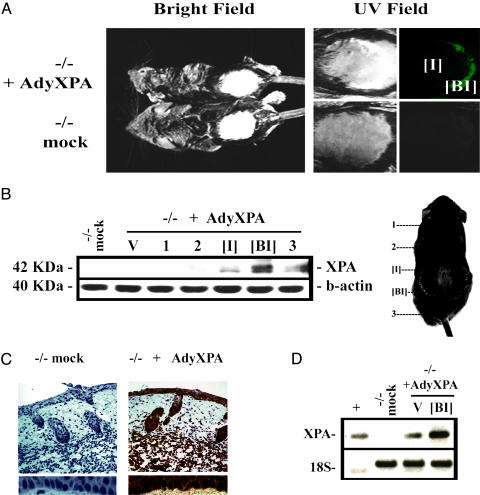
To extend the in vitro experiments described above to live mice, Xpa-mutant mice were infected by s.c. injection of the shaved skin with either AdyXPA or an empty adenovirus vector. The mice were killed 24 h later and examined for expression of EGFP and XPA proteins. A radial pattern of EGFP expression extending from the injection site was consistently observed, with highest levels posterior to the injection site (Fig. 2A). XPA expression, detected both by Western blot analysis and immunohistochemistry using an XPA-specific antibody, showed the same pattern of expression (Fig. 2 B and C). Skin sections (Fig. 2C) indicated the presence of XPA protein in cells of the epidermis, including keratinocytes in the basal layer and also in fibroblasts of the dermis. Similarly, RT-PCR of total RNA extracted from skin demonstrated the presence of human XPA mRNA in Xpa-/--infected mice at the site of injection. However, with this more sensitive technique, lower levels of XPA mRNA were even detected in the ventral skin of the animal, indicating extensive spreading of the virus via the s.c. space (Fig. 2D).
Fig. 2.
Expression of XPA and EGFP genes in Xpa-/- mice after infection with AdyXPA virus. Mice were killed 24 h after infection and exposed to UVC-light for EGFP detection. The skin was than removed for XPA protein detection and RNA extraction. (A) EGFP fluorescence showing a radial pattern of the adenovirus spreading, indicating two distinct regions: the injection site [I] and the region below the injection site [BI]. (B) XPA protein expression measured by Western blotting in the positions indicated in the mouse image (1 and 2 are the most anterior regions, 3 is the most posterior, and V represents the skin extracted from ventral portion). (C) (Upper) Immunohistochemistry using anti-XPA human antibody in cross sections of the dorsal skin of Xpa-/- mock-treated and AdyXPA-infected mouse skin. (×100.) (Lower) Basal layer of keratinocytes from both mock and AdyXPA-infected mice. Positive cells are stained brown. (×1,000.) (D) RT-PCR indicating the presence of XPA mRNA in AdyXPA-infected mouse at the site of injection [I] and in the ventral portion of the animal (V). A plasmid containing the human XPA cDNA was used as positive control (+), a mock-treated skin sample was used as negative control (-), and primers designed for 18S ribosomal RNA detection were used as loading control.
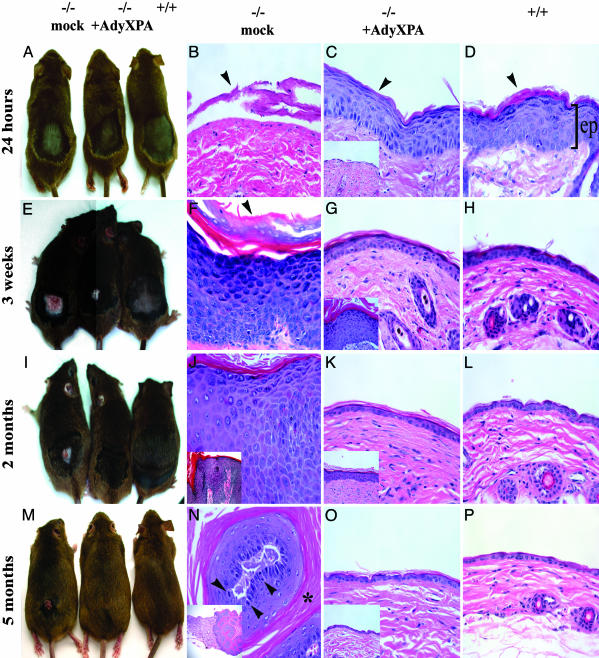
Xpa mice infected with AdyXPA, mock-infected Xpa mice, and WT mice were irradiated on the shaved dorsal skin for 4 days with a daily UVB dose of 3.4 kJ/m2. The mice were killed 24 h, 3 weeks, and 2 and 5 months later, and skin was processed for histological examination. Mice that were killed 24 h after treatment showed no obvious abnormalities visible to the naked eye (Fig. 3A). However, histological examination showed extensive epidermal loss in Xpa-/- mock-infected mice (Fig. 3B). In contrast, both AdyXPA-infected (Fig. 3C) and WT mice (Fig. 3D) showed epidermal hyperplasia and keratoses. In the former group, these features were most prominent posterior to the injection site where expression of XPA and of EGFP was the highest (Fig. 2).
Fig. 3.
Effects of AdyXPA infection in Xpa-/- mice skin after the UVB treatment. Back-shaved mice were injected with either mock virus or AdyXPA, 24 h before 4 days of UVB exposure. (A, E, I, and M) Overviews of the dorsal skin of the mock-infected Xpa-/- (Left), AdyXPA-infected (Center), and WT (Right) mice at different times after irradiation. (B-D, F-H, J-L, and N-P) Representative examples of hematoxylin/eosin-stained cross sections of the dorsal skin of the mice shown in A, E, I, and M. (B) Xpa-/- mock-infected showing substantial loss of epidermis (arrowhead). (C and D) Moderate epidermal hyperplasia and keratoses (arrowheads) in AdyXPA-infected (C) and WT skin (D). (F) Xpa-/- mock-infected mouse skin showing acute epidermal hyperplasia and hyperkeratoses (arrowhead). (G and H) Slight epidermal hyperplasia in AdyXPA-infected (G) and WT (H) mouse skin. (J) In situ SCC in Xpa-/- mock-infected skin. (Inset, lower magnification: ×100.) (K and L) Normal appearance of the skin in AdyXPA-infected (K) and WT (L). (N) Differentiated SCC in Xpa-/- mock-infected skin showing large extent of abnormal keratinization (asterisk) invading the adjacent dermis. (Inset, lower magnification: ×40.) Cells were markedly irregular in size and shape, and an enhanced number of mitotic figures (arrowheads) was observed. (O and P) Normal appearance of skin from AdyXPA-infected (O) and WT mice (P). The Insets in C, G, K, and O represent the effect observed in the [I] site of AdyXPA-infected mice, showing a delayed skin recovery from UVB injury. [I], injection site; [BI], region below injection site; ep, epidermis. (×400 for all sections unless otherwise indicated.)
Three weeks later, mock-treated Xpa mice presented acute epidermal hyperplasia and hyperkeratoses that could be visually detected (Fig. 3E). Microscopically these mice showed atypical basal keratinocytic layer and actinic keratoses (a dysplastic epidermal preneoplastic state) (Fig. 3F). WT and AdyXPA-infected mice, at the [BI] site, showed only slight hyperplasia with no hyperkeratoses or inflammation (Fig. 3 G and H).
Two months after treatment, the great majority (90%) of the mock-treated Xpa mice presented at least one SCC in situ in which keratinocytic atypia spanned the full thickness of the epidermis (Fig. 3 I and J). However, the [BI] site in Xpa-/- mice infected with recombinant adenovirus was histologically normal and indistinguishable from the skin of WT mice (Fig. 3 K and L).
Five months after the UVB treatment, all of the Xpa mice infected with AdyXPA showed no detectable abnormalities either by visual (Fig. 3M) or microscopic (Fig. 3 O and P) examination. On the other hand, all of the mock-treated mice presented either SCC in situ (55%) or well differentiated SCC (45%) invading the adjacent dermis (Fig. 3 M and N). Additionally, the number of cells in mitosis was increased [≈2-3 per ×400 field (Fig. 3N)], compared with WT and Xpa-/--infected skin (<0.001 mitosis per ×400 field).
The histology of the skin at the injection site [I] is shown in the insets in Fig. 3 C, G, K, and O. Initially, features resembling the skin of mock-treated Xpa-/- mice were observed, with loss of the epidermal layer followed by robust epidermal hyperplasia and hyperkeratoses (Fig. 3 C and G Insets). However, these lesions began to regress at 2 and 5 months (Fig. 3 K and O Insets), and by the end of the experiment, no differences were observed between WT and infected mice. The data shown in Fig. 3 are recapitulated quantitatively in Tables 1 and 2.
Table 1. UVB-induced hyperplasia in WT, XPA-mock-infected, and XPA-infected mice.
| Hyperplasia, no. of cell layers*
|
|||
|---|---|---|---|
| Time after UV treatment | XPA mock | XPA + AdyXPA | WT |
| 24 h | 0.1 ± 0.2 | [I] 0.2 ± 0.21 | 5.2 ± 1.1 |
| [BI] 8.3 ± 0.7 | |||
| 3 weeks | 20.2 ± 3.4 | [I] 12.1 ± 1.9 | 2.9 ± 0.5 |
| [BI] 3.9 ± 1.4 | |||
| 2 months | 22.3 ± 2.3 | [I] 5.3 ± 1.1 | 2.1 ± 2.8 |
| [BI] 2.5 ± 1.5 | |||
Hyperplasia is given as the number of epidermal cell layers at the indicated times after in vivo exposure to UVB. Data are means ± SEM (n ≥ 3) or variation (n = 2).
Table 2. UVB-induced SCC in WT, XPA-mock-infected, and XPA-infected mice.
| Skin neoplastic transformation as a result of UVB exposure, no. of mice affected*
|
|||
|---|---|---|---|
| Time after UV treatment | XPA mock | XPA + AdyXPA | WT |
| 2 months | 18/20 | 0/22 | 0/20 |
| 5 months | 18/18 | 0/20 | 0/18 |
Number of mice that presented persistent SCC (in situ or well differentiated) after 2 or 5 months of UV treatment per total mice UV-irradiated.
Discussion
In the present study, we have investigated the ability to correct defective DNA repair and prevent skin abnormalities (including skin cancer) consequent to exposure to UVB radiation, by injection of recombinant adenovirus in an XP mouse model. Adenovirus vectors are a common tool for gene therapy and have provided promising results (20). Despite well characterized immunologic responses when administered in vivo, advanced generations of vectors show reduced toxicity and improved persistence of transgene expression (21, 22).
The adenovirus used in this study encodes the human XPA nucleotide excision repair protein and has been previously tested in cultured human cells (16). The concentration of virus used in the previous and present studies did not reveal toxicity either in cell culture (human or mouse cells) or in living mice. The XPA-encoding adenovirus efficiently infected Xpa-/- MEF (100% transduction, Fig. 1A) and manifested high levels of transgene expression, as well as complete recovery of DNA repair synthesis and cell survival after UV-radiation exposure. Collectively, these results demonstrate that human XPA protein is able to fully restore defective nucleotide excision repair in Xpa-mutant mouse cells.
Subcutaneous application of adenovirus has been previously reported (23). Expression of a reporter transgene (Escherichia coli lacZ) can be observed macroscopically by direct observation of the skin and microscopically in various cell types in the epidermis, including keratinocytes, the progenitor cells for squamous and basal cell carcinomas (23). The direct observation of transgene expression (EGFP) in the present study confirms the potential of these vectors for infection of skin cells. The pattern of EGFP expression observed indicates radial spreading of the virus from injection site in the s.c. space. This finding is corroborated by the pattern of expression of XPA protein. In skin sections of the region posterior to the injection site, cells of the epidermis and of the lower layers of the dermis expressed the human protein, including basal stem cell keratinocytes, which are believed to be the cell types responsible for tumor formation. Protein samples extracted from the dorsal-anterior portion of the skin (anterior the site of injection) revealed considerably reduced XPA expression, compared with the dorsal-posterior portion (below the site of injection). The higher level of XPA expression observed at the dorsal-posterior portion is likely a reflection of physiological distribution of body fluids along with a gravitational effect. Reduced amounts of XPA protein also were detected at the actual sites of injection. This gradient of expression correlates well with the phenotypes observed by histological analysis of the skin in UVB-irradiated Xpa-/- mice infected with AdyXPA. Nonetheless, human XPA mRNA was detected in skin from the ventral nonirradiated region of infected mice, indicating that the virus can spread widely through the s.c. space.
Genetic correction of defective nucleotide excision repair has been achieved in cell culture experiments (16, 24) and in vitro in XPC-deficient keratinocytes derived from skin (25). Enzymatic therapy with liposomes containing bacteriophage T4 endonuclease V has been reported to reduce the frequency of skin cancer in addition to actinic keratoses in XP patients after topical treatment application, possibly because of enhanced DNA repair in the skin (10). This enzyme nicks DNA exclusively at cyclobutane pyrimidine dimers by concerted DNA glycosylase/apyrimidinic lyase activities, generating repair intermediates that can be resolved by other DNA repair systems in XP cells. However, T4 endonuclease V does not attack the other major mutagenic photoproduct in DNA [(6-4) photoproducts], which is addressed by nucleotide excision repair. Because all seven XP genes representing different complementation groups for the disease have already been cloned, the adenovirus vector approach offers the potential of specific correction of defective nucleotide excision repair in all XP individuals.
The present study provides encouraging perspectives toward in vivo skin therapy for XP. However, techniques for topical application of adenovirus vectors must be improved. Alternatives are particle bombardment with a gene gun (26) and direct topical application of adenovirus to the skin (27, 28). The use of epicutaneous patches has been shown to provide effective absorption of an adenovirus-vaccine vector (29). Alternatively, the use of liposomecomplexed adenovirus (30) also may improve the ability of gene transfer by such vectors in skin.
The most important limitation of the use of adenovirus vectors for gene correction protocols is that it elicits strong immune responses in the host. This feature reduces transgene expression in vivo and the efficiency of repeated vector administration. Although cell culture experiments with AdyXPA indicate that XPA protein is still functional 2 months after virus infection (16), treatment of XP patients would certainly require periodic vector applications. Recent efforts have been dedicated to the development of high-capacity (gutless) adenovirus vectors that lead to reduced host immunological responses and long-term gene expression, with encouraging results (21, 22). Additionally, a binary adenovirus vector carrying the DNA replication system, derived from Epstein-Barr virus, has been shown to be useful for sustained gene expression in replicating cells (31).
The success of gene delivery to skin cells and genetic correction in Xpa-mutant mice provides an exciting technology to investigate DNA repair functions in vivo. These results also motivate continued searches for improved methodologies to optimize gene therapy protocols with the aim of offering an improved quality of life for XP patients.
Acknowledgments
We thank Dr. J. Cleaver for helpful discussions and encouragement; Dr. K. Tanaka (Osaka University, Japan) for providing the XPA mice; P. L. Fischhaber for critically reviewing the manuscript; and A. Doughty, R. Daniel, and J. Marshburn for valuable technical assistance. This work was supported by the Xeroderma Pigmentosum Society (Craryville, NY) and the Department of Pathology at the University of Texas Southwestern Medical Center, Dallas. Additional funding was provided by Fundaçao de Amparo a Pesquisa do Estado de São Paolo (Brazil) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (Distrito Federal, Brazil). A.R.M. was a Pew Latin American Fellow in biomedical sciences.
Author contributions: M.C.N.M., A.R.M., E.C.F., and C.F.M. designed research; M.C.N.M., A.R.M., and D.K.B. performed research; M.C.N.M., A.R.M., D.K.B., E.C.F., and C.F.M. analyzed data; and M.C.N.M., A.R.M., E.C.F., and C.F.M. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: XP, xeroderma pigmentosum; AdyXPA, recombinant adenovirus carrying the human XPA gene; SCC, squamous cell carcinoma; NER, nucleotide excision repair; MEF, mouse embryo fibroblasts; UDS, unscheduled DNA synthesis.
References
- 1.Jacoby, J. (2004) Gene Ther. 11, 427-428. [DOI] [PubMed] [Google Scholar]
- 2.Cleaver, J. E. (1968) Nature 218, 652-656. [DOI] [PubMed] [Google Scholar]
- 3.Kraemer, K. H., Lee, M. M. & Scotto, J. (1987) Arch. Dermatol. 123, 241-250. [DOI] [PubMed] [Google Scholar]
- 4.Hoeijmakers, J. H. (2001) Nature 411, 366-374. [DOI] [PubMed] [Google Scholar]
- 5.Berneburg, M. & Lehmann, A. R. (2001) Adv. Genet. 43, 71-102. [DOI] [PubMed] [Google Scholar]
- 6.Costa, R. M. A., Chiganças, V., Galhardo, R. S., Carvalho, H. & Menck, C. F. M. (2003) Biochimie 85, 1083-1099. [DOI] [PubMed] [Google Scholar]
- 7.Kraemer, K. H., Lee, M. M., Andrews, A. D. & Lambert, W. C. (1994) Arch. Dermatol. 130, 1018-1021. [PubMed] [Google Scholar]
- 8.Horiki, S., Miyauchi-Hashimoto, H., Tanaka, K., Nikaido, O. & Horio, T. (2000) Arch. Dermatol. Res. 292, 511-518. [DOI] [PubMed] [Google Scholar]
- 9.Kraemer, K. H., DiGiovanna, J. J., Moshell, A. N., Tarone, R. E. & Peck, G. L. (1988) New Engl. J. Med. 318, 1633-1637. [DOI] [PubMed] [Google Scholar]
- 10.Yarosh, D., Klein, J., O'Connor, A., Hawk, J., Rafal, E. & Wolf, P. (2001) Lancet 357, 926-929. [DOI] [PubMed] [Google Scholar]
- 11.Atabay, K., Celebi, C., Cenetoglu, S., Baran, N. K. & Kiymaz, Z. (1991) Plast. Reconstr. Surg. 87, 1121-1125. [DOI] [PubMed] [Google Scholar]
- 12.Friedberg, E. C., Walker, G. C. & Siede, W. (1995) DNA Repair and Mutagenesis (Am. Soc. Microbiol., Washington, DC), pp. 633-672.
- 13.Volker, M., Moné, M. J., Karmakar, P., van Hoffen, A., Schul, W., Vermeulen, W., Hoeijmakers, J. H. J., van Driel, R., van Zeeland, A. A. & Mullenders, L. H. F. (2001) Mol. Cell 8, 213-224. [DOI] [PubMed] [Google Scholar]
- 14.Nakane, H., Takeuchi, S., Yuba, S., Saijo, M., Nakatsu, Y., Murai, H., Nakatsuru, Y., Ishikawa, T., Hirota, S., Kitamura, Y., et al. (1995) Nature 377, 165-168. [DOI] [PubMed] [Google Scholar]
- 15.de Vries, A., van Oostrom, C. T. M., Hofhuis, F. M. A., Dortant, P. M., Berg, R. J. W., de Gruijl, F. R., Wester, P. W., van Kreijl, C. F., Capel, P. J. A., van Steeg, H., et al. (1995) Nature 377, 169-173. [DOI] [PubMed] [Google Scholar]
- 16.Muotri, A. R., Marchetto, M. C. N., Zerbini, L. F. C., Libermann, T. A., Ventura, A., Sarasin, A. & Menck, C. F. M. (2002) Hum. Gene Ther. 13, 1833-1844. [DOI] [PubMed] [Google Scholar]
- 17.Graham, F. L. & Prevec, L. (1992) Biotechnology 20, 363-390. [DOI] [PubMed] [Google Scholar]
- 18.Nyberg-Hoffman, C., Shabram, C. P., Li, W., Giroux, D. & Aguilar-Cordova, E. (1997) Nat. Med. 3, 808-811. [DOI] [PubMed] [Google Scholar]
- 19.Cleaver, J. E. & Thomas, G. H. (1981) in DNA Repair: A Laboratory Manual of Research Procedures, eds. Friedberg, E. C. & Hanawalt, P. C. (Dekker, New York), pp. 227-287.
- 20.Graham, F. L. (2000) Immunol. Today 21, 426-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.St. George, J. A. (2003) Gene Ther. 10, 1135-1141. [DOI] [PubMed] [Google Scholar]
- 22.Volpers, C. & Kochanek, S. (2004) J. Gene Med. 6, S164-S171. [DOI] [PubMed] [Google Scholar]
- 23.Setoguchi, Y., Jaffe, H. A., Danel, C. & Crystal, R. G. (1994) J. Invest. Dermatol. 102, 415-421. [DOI] [PubMed] [Google Scholar]
- 24.Zeng, L., Sarasin, A. & Mezzina, M. (1998) Cell Biol. Toxicol. 14, 105-110. [DOI] [PubMed] [Google Scholar]
- 25.Arnaudeau-Begard, C., Brellier, F., Chevallier-Lagente, O., Hoeijmakers, J., Bernerd, F., Sarasin, A. & Magnaldo, T. (2003) Hum. Gene Ther. 14, 983-996. [DOI] [PubMed] [Google Scholar]
- 26.Lu, B., Scott, G. & Goldsmith, L. A. (1996) Proc. Assoc. Am. Physicians 108, 165-172. [PubMed] [Google Scholar]
- 27.Lu, B., Federoff, H. J., Wang, W., Goldsmith, L. A. & Scott, G. (1997) J. Invest. Dermatol. 108, 803-808. [DOI] [PubMed] [Google Scholar]
- 28.Tang, D. C., Shi, Z. & Curiel, D. T. (1997) Nature 388, 729-730. [DOI] [PubMed] [Google Scholar]
- 29.Shi, Z., Zeng, M., Yang, G., Siegel, F., Cain, L. J., van Kampen, K. R., Elmets, C. A. & Tang, D. C. (2001) J. Virol. 75, 11474-11482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, E. M., Hong, S. H., Lee, Y. J., Kang, Y. H., Choi, K. C., Kim, I. H. & Lim, S. J. (2004) J. Cancer Res. Clin. Oncol. 130, 169-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kreppel, F. & Kochanek, S. (2004) J. Virol. 78, 9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]