The sex-determining protein FEM1, which contains a VHL-box, B/C-box, cullin 2-box, and several ANK repeat domains, belongs to the substrate-recognizing adaptor proteins of Cullin 2/Rbx1/Elongin B/C E3 ligases (also known as Cullin-RING ligase 2, CRL2). In C. elegans, FEM1, together with 2 cofactors, FEM2 and FEM3, promotes the poly-ubiquitination and subsequent degradation of the Gli-family transcription factor sex-determining transformer protein 1 (TRA-1) to control the development of male phenotype.1 Notably, 3 distinct FEM1 isoforms (FEM1A, 1B, and 1C) are encoded in H. sapiens genome with multiple biologic functions, including anti-inflammation, apoptosis, insulin secretion, as well as being implicated in the pathogenesis of the polycystic ovary syndrome (PCOS). However, besides Gli1, the mammalian homolog of TRA-1,2 the downstream ubiquitin substrates of FEM1 in mammals remain largely unknown, which prevents our full understanding of the molecular mechanism through which FEM1 exerts its biologic functions in numerous important processes such as sex determination in mammals. To this end, in this volume of Cell Cycle, an elegant study from the Pagano group identified the stem-loop-binding protein (SLBP) as a bona fide ubiquitin substrate for the CRL2FEM1 E3 ligase complex. This important study has shed light into our fundamental understanding of the biologic features of FEM1 and its possible connection to various human diseases, including PCOS.
SLBP is a well-characterized master regulator of histone mRNA. Due to lack of introns and the polyadenylated tail, the stem-loop structure at the 3′ end of histone pre-mRNAs (H1, H2A, H2B, H3, and H4) is critical for the histone pre-mRNAs processing by endonucleolytic cleavage and controls the transport, translation and stability of histone mRNAs. Functionally, SLBP binds the stem-loop structure of histone pre-mRNAs to regulate their processing, translation, and stability in a highly regulated manner.3 SLBP is predominantly retained in the nucleus during the G1 and G2 phases of the cell cycle, while during S phase it localizes in the cytoplasm, exerting its pivotal role in regulating virtually every aspects of histone mRNA processing. In keeping with this key biological function, cells in S phase witness the highest levels of SLBP protein, accompanied by high levels of histone mRNA and DNA replication. At the end of S phase, SLBP is phosphorylated at Thr-61 and Thr-62 by CK2 and cyclin A2-CDK1,4,5 respectively, facilitating its degradation by SCFCyclin F in the following G2 phase, which is largely dependent on an atypical RxL motif present in SLBP.6 Interestingly, there is evidence that SLBP may also be degraded during the G1 phase when Cyclin F is not abundantly expressed. However, the E3 ligase responsible for its decay in G1 phase remained elusive until this report.7
In a recent paper published in Cell Cycle, Dankert and colleagues report that SLBP is degraded by CRL2FEM1A/B/C during the G1 phase,7 in addition to its controlled degradation by SCFCyclin F in G2 phase.6 Through a series of rigorous mutagenesis analysis, the authors identified that FEM1A, FEM1B, and FEM1C bound to distinct motifs within the N-terminus of SLBP. As a result, depletion of individual FEM1A, FEM1B, or FEM1C led a modest effect, while depleting all 3 FEM1 isoforms led to a more dramatic accumulation, of SLBP protein abundance during the G1 phase with minimal effects to SLBP abundance during the G2 phase, where SCFCyclin F is largely controls its stability. Furthermore, mutating all degron motifs responsible for SLBP binding with both FEM1 and Cyclin F resulted in a non-degradable version of SLBP across the whole cell cycle.
Hence, these elegant studies from the Pagano group together revealed a synergy between CRL2FEM1A/B/C and SCFCyclin F to orchestrate the timely degradation of SLBP during the G1 and G2 phases,6,7 respectively, thereby synchronizing histone and nucleosome metabolism with cell cycle progression (Fig. 1). This finding advocates for a tight and interwoven network of regulatory mechanisms typically involving the synergistic action from multiple E3 ligases acting during different cell cycle phases to regulate cell cycle-dependent oscillations of key proteins controlling DNA replication, protein synthesis, and other cellular processes. In keeping with this notion, recent reports have revealed that the RING type of E3 ubiquitin ligase such as Skp1-cullin 1-F-box protein complex (SCF) and anaphase promoting complex/cyclosome complex (APC/C) E3 ligases coordinately promote the degradation of several cellular factors in a cell cycle-dependent manner (Fig. 1).
Figure 1.
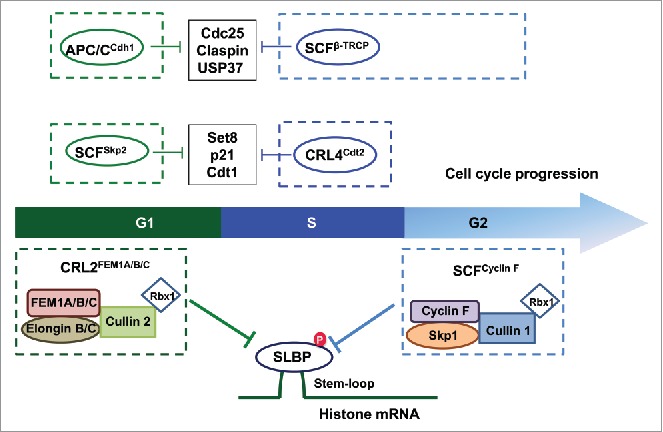
SLBP protein stability is strictly regulated by CRL2FEM1A/B/C in G1 and by SCFCyclin F in G2 phase, respectively. Of note, the stability of many key cellular regulators, including Set8, p21, Cdt1, Cdc25, Claspin and USP37, are also governed by distinct E3 ligases, such as SCF and APC/C, in different cell cycle phases to orchestrate proper cell replication and division events.
Interestingly, given that Cyclin F is not conserved in metazoans and absent in lower eukaryotes, including C. elegans and D. melanogaster, it appears that FEM1 family of proteins, which are conserved in metazoans and lower eukaryotes, might serve as the ancient regulators for SLBP to impact histone metabolism during the cell cycle. This is possibly in part due to the fact that in vertebrates but not invertebrates, in addition to regulating canonical histone mRNAs, SLBP also binds the stem loop of H2AFX mRNAs (encoding H2A.X) and promotes their translation during S phase. Hence, to avoid SLBP-mediated synthesis of H2A.X outside of S phase, vertebrates evolved Cyclin F to promote SLBP degradation during the G2 phase. In this regard, Cyclin F-mediated proteolytic regulation of SLBP may be a relatively late, evolutionarily acquired event. Indeed, a stable SLBP mutant that is insensitive to Cyclin F-mediated degradation led to constant translation of H2A.X throughout the cell cycle, subsequently conferring increased cellular sensitivity to apoptosis upon DNA damage.6 However, it warrants further in-depth studies including generation of conditional knockout (KO) and knockin (KI) mice to explore to what extent the aberrancy in histone processing due to elevated SLBP protein abundance contributes to human pathological conditions such as sex determining or polycystic ovary defects associated with FEM1 deficiency.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- [1].Starostina NG, Lim JM, Schvarzstein M, Wells L, Spence AM, Kipreos ET. A CUL-2 ubiquitin ligase containing three FEM proteins degrades TRA-1 to regulate C. elegans sex determination. Dev Cell 2007; 13:127-39; PMID:17609115; http://dx.doi.org/ 10.1016/j.devcel.2007.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Gilder AS, Chen YB, Jackson RJ 3rd, Jiang J, Maher JF. Fem1b promotes ubiquitylation and suppresses transcriptional activity of Gli1. Biochem Biophys Res Commun 2013; 440:431-6; PMID:24076122; http://dx.doi.org/ 10.1016/j.bbrc.2013.09.090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet 2008; 9:843-54; PMID:18927579; http://dx.doi.org/ 10.1038/nrg2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Koseoglu MM, Graves LM, Marzluff WF. Phosphorylation of threonine 61 by cyclin a/Cdk1 triggers degradation of stem-loop binding protein at the end of S phase. Mol Cell Biol 2008; 28:4469-79;PMID:18490441;http://dx.doi.org/ 10.1128/MCB.01416-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Zheng L, Dominski Z, Yang XC, Elms P, Raska CS, Borchers CH, Marzluff WF. Phosphorylation of stem-loop binding protein (SLBP) on two threonines triggers degradation of SLBP, the sole cell cycle-regulated factor required for regulation of histone mRNA processing, at the end of S phase. Mol Cell Biol 2003; 23:1590-601;PMID:12588979;http://dx.doi.org/ 10.1128/MCB.23.5.1590-1601.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Dankert JF, Rona G, Clijsters L, Geter P, Skaar JR, Bermudez-Hernandez K, Sassani E, Fenyö D, Ueberheide B, Schneider R, Pagano M. Cyclin F-Mediated Degradation of SLBP Limits H2A. X Accumulation and Apoptosis upon Genotoxic Stress in G2. Mol Cell 2016; 64:507-19; PMID:27773672; http://dx.doi.org/ 10.1016/j.molcel.2016.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Dankert JF, Pagan JK, Starostina NG, Kipreos ET. Pagano FEM1 proteins are ancient regulators of SLBP degradation. Cell Cycle 2017; PMID:28118078; http://dx.doi.org/ 10.1080/15384101.2017.1284715 [DOI] [PMC free article] [PubMed] [Google Scholar]


