ABSTRACT
p53 is regulated by heterodimer E3 ligase Mdm2-Mdm4 via RING domain interaction. Mdm2 transcripts undergo alternative splicing, and Mdm2 splice isoforms are increased in cancer and induced by DNA damage. Although 2 major Mdm2 splice isoforms that do not bind to p53 were reported to impact the p53 pathway, the underlying biochemical mechanisms were not understood. Here, we show that these Mdm2 splice isoforms ubiquitinate Mdm2 and Mdm4 in vivo and regulate the activity of Mdm2-Mdm4 E3 complex in cells. The Mdm2 isoforms are capable of promoting p53 ubiquitination in the absence of Mdm2 or Mdm4. The 2 isoforms stimulate Mdm2 or Mdm4 activity for p53 ubiquitination in vivo and promote degradation of p53 and Mdm4 in cells. However, the Mdm2 isoforms have opposing effects on the steady-state p53 levels depending on the stoichiometric ratios of Mdm2, Mdm4 and the isoforms, causing either decreased or increased p53 levels in cells. Our data indicate that the Mdm2 splice isoforms can act as independent E3 ligases for p53 when Mdm2 and Mdm4 are absent, form potent heterodimer E3 ligases with either Mdm2 or Mdm4 for targeting p53 degradation, or act as inhibitory regulators of Mdm2-Mdm4 E3 ligase activity by downregulating Mdm4. These findings suggest that Mdm2 splice isoforms may play critical roles in the regulatory loop of p53/Mdm2-Mdm4 via a RING domain-mediated biochemical mechanism.
KEYWORDS: degradation, Mdm2, p53, RING domain, splice isoforms, ubiquitination
Introduction
The centerpiece of the regulatory mechanisms for restricting p53 activity is Mdm2-Mdm4 E3 complex.1-3 The N-terminus of Mdm2 and Mdm4 binds to p53 to mask its transactivation domain whereas their C-terminal RING domains bind to each other to form a heterodimer.4-6 Mdm2 is the key E3 ligase for regulating p53 degradation.7-9 We previously reported that the RING domain-mediated heterodimerization of Mdm2 and Mdm4 activates the polyubiquitination activity of the E3 complex that targets p53 for degradation.10 Mouse genetics studies indicated that RING-RING interaction of Mdm2-Mdm4 domain is essential for restricting p53 activity during embryonic development, since mutation of either Mdm2 or Mdm4 RING domain causes p53-dependent embryonic lethality.11-13
Mdm2 alternative splice isoforms were reported years ago.14 Two major splice isoforms Mdm2-A and Mdm2-B (referred thereafter as Mdm2-A/B) do not bind to p53 and induce p53-independent cell growth in vivo and tumorigenesis in vivo.15-17 However, Mdm2-A overexpression in Mdm2-a transgenic mice causes perinatal lethality involving p53 activation and senescence,18 suggesting that Mdm2-A has significant impact on the p53 pathway, even though it does not directly bind to p53. How Mdm2 splice isoforms exert their effects on the p53/Mdm2/Mdm4 regulatory loop remains unexplored. To address this, we show that the RING domain of Mdm2 splice variants mediate ubiquitination of Mdm2, Mdm4 and p53, and regulate their abundance in context- and stoichiometry-dependent manners. Therefore, the findings in this report revealed Mdm2 splice isoforms as critical components of the central regulatory loop of p53/Mdm2-Mdm4.
Results and discussions
Mdm2 isoforms can trans-ubiquitinate Mdm2 in vitro
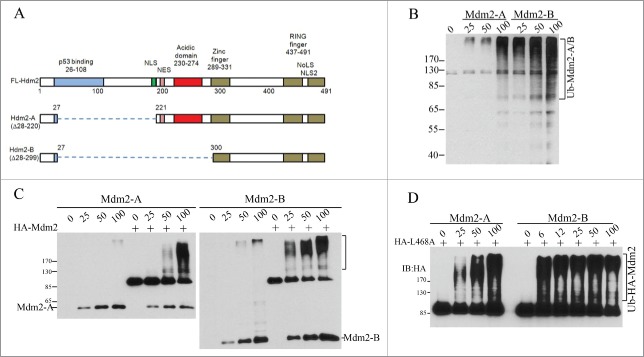
Our previous work has shown that RING domain interaction of Mdm2 and Mdm4 plays a dual role in the interplay of p53 with Mdm2/Mdm4 E3 complex. As an activator, Mdm4 is required for Mdm2-mediated p53 polyubiquitination. As a substrate, Mdm4 competes with p53 and Mdm2 for accepting ubiquitins from E2 enzymes accompanied by Mdm4 degradation. Thus, the relative stoichiometric ratios of Mdm2, Mdm4 and p53 are determinants of the varying effects of Mdm2 on p53 and Mdm4.10 Although Mdm2-A/B isoforms lack the p53-binding region, they both have and intact RING domain (Fig. 1A). We hypothesize that the 2 isoforms can directly regulate Mdm2-Mdm4 E3 complex via RING-RING interaction with Mdm4, in a same manner as Mdm2 interacts with Mdm4. To test this hypothesis, we performed in vitro ubiquitination assays with purified recombinant proteins expressed from insect cells. We found that both the human Mdm2-A/B (Hdm2-A or Hdm2-B, for simplicity, we use Mdm2-A/B) possess E3 ligase activity as indicated by their autoubiquitination (Fig. 1B). Both isoforms can activate the E3 ligase activity of Mdm2 in forms of autoubiquitination of full-length Mdm2 in a concentration-dependent manner (Fig. 1C), indicating that heterodimers Mdm2-Mdm2-A/B E3 ligases can be formed in vitro. We previously reported that human Mdm2 RING domain mutant Mdm2L468A is an E3-dead mutant with an intact RING domain structure. Mdm2L468A is capable of interacting with Mdm4, but it cannot bind to E2, rendering complete loss of its E3 ligase activity.10,20 However, this E3-dead Mdm2L468A can be efficiently ubiquitinated by Mdm2-A/B in vitro, indicating trans-ubiqutination of Mdm2 full length protein by the splice isoforms (Fig. 1D).
Figure 1.
Mdm2-A/B possesses intrinsic E3 ligase activity and can trans-ubiquitinate full length Mdm2 in vitro. (A) A diagram of Mdm2-A/B structure. (B) Mdm2-A/B autoubiquitination. Western blotting of polyubiquitin chain after in vitro ubiquitination reaction with indicated amounts of Mdm2-A/B. (C) Mdm2-A/B ubiquitinates full length Mdm2. (D) Trans-ubiquitination of full length enzyme-dead Mdm2L468A by Mdm2-A/B in vitro.
Mdm2 isoforms are super active E3 ligase for ubiquitination of Mdm4 and p53 in vivo
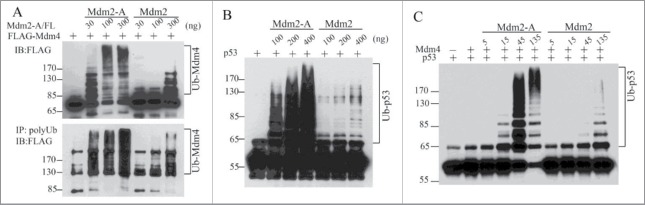
How does Mdm2-A/B affect the ubiquitination of Mdm4 and p53 in vivo? Our results indicated that Mdm2-A is approximately 10-times more active than Mdm2 in promoting ubiquitination of Mdm4 in vitro (Fig. 2A). Similarly, Mdm2-A alone potently promotes p53 ubiquitination, with a several-fold more activity than full-length Mdm2 (Fig. 2B). Strikingly, Mdm2-A-Mdm4 heterodimer E3 is ∼10-fold more active than Mdm2-Mdm4 E3 complex in p53 polyubiquitination (Fig. 2C). Similar results were obtained from Mdm2-B (data not shown). These in vitro E3 ligase assays suggest that Mdm2-A/B by themselves act as more active E3 ligase than Mdm2, and as more active regulators of the RING domain heterodimer E3 activities. It is puzzling at first glimpse that Mdm2 isoforms can ubiquitinate p53 without having p53-binding domain. Two possibilities may explain these results. First, the RING domain itself is involved in p53 substrate recognition. This possiblity was suggested by earlier observations that Mdm2 RING domain when swapped with the RING domain of Praja1 protein resulted in loss of p53 ubiquitination by Mdm2-Praja1-RING hybrid protein, although the N-ternminal p53-binding region of Mdm2 was intact in the hybrid proteins.9 The second possibility is that in in vitro biochemical assays, the requirement for a stable E3-substrate binding is low such that Mdm2-A/B can ubiquitinates p53 without the need of stable protein-protein interactions. Interestingly, Mdm2-A-Mdm4 heterodimers show super E3 ligase activity for p53 polyubiquitination compared with Mdm2-Mdm4. This finding suggests that Mdm2-A promotes processiveness of the heterodimer E3 ligase, possibly owing to a better interaction with RING domain of Mdm4 due to the lack of p53 binding domain. In addition, it demonstrates that Mdm2-A can fully compensate the biochemical role of full-length Mdm2 in Mdm2-Mdm4-mediated p53 ubiquitination.
Figure 2.
Mdm2-A potently ubiquitinates Mdm4, p53 and strongly stimulates p53 ubiquitination by Mdm2-Mdm4 in vitro. (A) Mdm2-A is more active than Mdm2 in ubiquitination of Mdm4. (B) Mdm2-A is more active than Mdm2 in p53 ubiquitination. (C) Mdm2-A-Mdm4 is more active heterodimer E3 ligase than Mdm2-Mdm4 in promoting p53 polyubiquitination in vitro.
Mdm2 isoforms alone can regulate steady-state levels of p53 in cells and acts as modifiers of p53/Mdm2/Mdm4 regulatory circuit in a stoichiometry-dependent manner
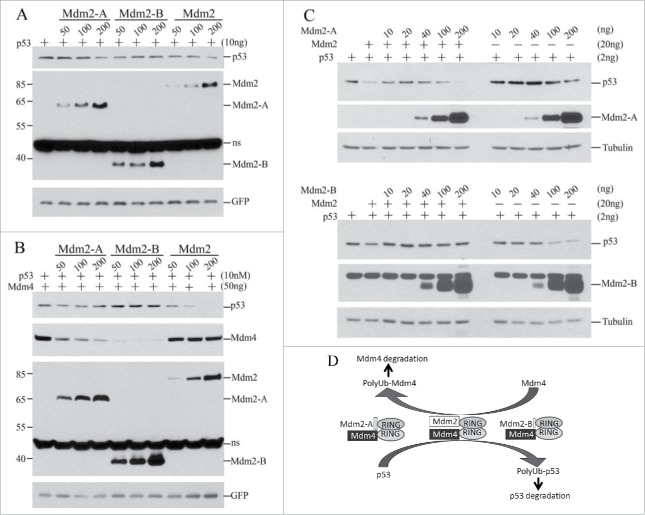
How does Mdm2-A/B affect p53 steady-state levels in cells? We first tested the effects of Mdm2-A/B in p53/mdm2/mdm4 triple knockout mouse embryonic fibroblasts (TKO MEFs) in co-transfection experiments. In the absence of Mdm4, Mdm2 decreased p53 steady-state levels in a concentration-dependent manner, suggesting that Mdm2 may form homodimer E3 complex at high levels to promote p53 degradation in cells through selfoligomerization.21 Mdm2-A alone decreased p53 levels at higher expression levels, while Mdm2-B decreased p53 levels at low concentrations but increased p53 levels at high concentrations (Fig. 3A). However, when Mdm4 was co-transfected, Mdm2 and Mdm2-A more efficiently decreased p53 levels (Fig. 3A versus 3B). Moreover, Mdm2-Mdm4 is more efficient than Mdm2-A-Mdm4 in promoting p53 degradation (Fig. 3B, Mdm2-A vs. Mdm2). This result suggests that Mdm2 and Mdm2-A are functionally replaceable with each other in forming active heterodimer E3 with Mdm4. In contrast to Mdm2-A, Mdm2-B constantly increased p53 levels in the presence of Mdm4 (Fig. 3B). Of note, Mdm2-B does not have the acidic domain as Mdm2-A (Fig. 1A). The deficiency of Mdm2-B in promoting p53 degradation is consistent with previous observations that the acidic domain of Mdm2 plays a critical role in p53 ubiquitination and degradation.22,23 Strikingly, both splice isoforms promoted robust degradation of Mdm4, in contrast to the weaker effect of full-length Mdm2 on Mdm4 levels (Fig. 3B). These data suggest that high expression levels of Mdm2-A promote significant degradation of Mdm4 leading to reduced abundance of the Mdm2-Mdm4 heterodimers that further result in p53 accumulation, when all these molecules coexist. This situation applies to Mdm2-a transgenic mice and explains why these mice suffer from p53 activation and senescence.18 Moreover, Mdm2-B solely decreased p53 levels at low concentrations (Fig. 3A, 50 ng of Mdm2-B) while Mdm2-A at low concentrations worked together with Mdm4 to promote p53 degradation (Fig. 3B, 50, 100 ng of Mdm2-A). These data from TKO MEFs suggest that Mdm2-A/B at low expression levels can be crucial promotors of p53 degradation when Mdm2 and Mdm4 are insufficient, or unavailable.
Figure 3.
Effect of Mdm2-A/B on steady-state levels of p53 and Mdm4 in cells. (A) Co-transfection of p53 with indicated amounts of Mdm2 or isoforms in TKO MEFs followed by WB 24h after transfection. (B) As performed in A but with addition of Mdm4. (C) As performed in B but in PC3 cells. (D) A model of RING-domain heterodimer E3 ligases in regulation of the p53/Mdm2/Mdm4 loop.
To test whether Mdm2-A/B also modulates Mdm2-Mdm4-mediated p53 regulation in human cells, we performed experiments in human prostate cancer cell line PC3 cells under the background of endogenous Mdm4 expression. We found that both Mdm2-A and Mdm2-B at low concentrations (10–40 ng) protected p53 from Mdm2-Mdm4-mediated degradation (Fig. 3C). However at high expression levels, Mdm2-A or Mdm2-B cooperated with endogenous Mdm4 to promote p53 degradation (Fig. 3C, 100, 200ng). These effects are opposed to the stoichiometric effects of Mdm2-A/B in TKO cells (Fig. 3A and B). Therefore, these data suggest that Mdm2-A/B modifies the outputs of Mdm2-Mdm4-mediated p53 regulation in both stoichiometry-dependent and cell type-dependent manners.
Based on our biochemical cellular evidence, we propose the following model for p53 regulation by Mdm2-Mdm4 and Mdm2 splice isoforms (Fig. 3D). Full-length Mdm2-Mdm4 heterodimers are the major E3 complex for p53 regulation. In the absence of Mdm2, Mdm2-A/B at high expression levels can downregulate p53 in complex with either Mdm2 or Mdm4. In this case, Mdm2 splice isoforms act to compensate the absence of either Mdm2 or Mdm4. However, when both full-length Mdm2 and Mdm4 are expressed, Mdm2-A/B promotes Mdm4 degradation to downregulate the abundance of Mdm2-Mdm4 heterodimer E3 complex to maintain basal p53 expression. In this case, Mdm2 isoforms act as a buffering molecule for the activity of Mdm2-Mdm4 in a stoichiometry-dependent manner. This complex regulatory module ensures a delicate regulation of p53 degradation in a time and tissue-specific manner for the destined biologic consequence of p53 during development. It has been reported that Mdm2 expression during development is tissue-specific and independent of p53.24 For example, Mdm2 is not expressed in liver, muscle and thymus on 12.5 dpc and it is not expressed in colon epithelium, intestine, liver, muscle and salivary gland on 14.5 dpc.24 Tissue specific knockout of Mdm2 or Mdm4 in mice revealed differential rquirements of Mdm2 and Mdm4 for p53 regulation in different tissues at different stages.25-27
The significance of RING-domain-mediated regulation of the p53 pathway by Mdm2 isoforms remians to be tested in vivo. However, the biochemical model we proposed here provides an explanation for the puzzling genetic findings from the rescue experiments of Mdm2 and Mdm4 knockout mice in the field. The rescue of Mdm4 knockout and Mdm4 RING domain mutant mice by p53 deficiency concluded that Mdm4 has a non-overlapping function of Mdm4 with Mdm2.12,13,19 However, this conclusion is difficult to reconcile with the finding that an Mdm2 transgene containing Mdm2 gene and its native promotor can completely rescue Mdm4-deficiency-induced p53-dependent lethality, leading to a conclusion that Mdm2 and Mdm4 are functionally overlapping in development.28 Based on our biochemical model, it is higly likely that the Mdm2 splice isoforms expressed from the Mdm2 transgene, rather than the overexpression of full-length Mdm2 per se, may have compensated for the loss of Mdm4 as a RING-domain stimulater of full length Mdm2 activity to fully suppress p53 during development.
Materials and methods
Cell culture, transfections and western blotting
p53/mdm2/mdm4 triple knockout MEFs19 (from Gigi Lozano, MD Anderson Cancer Center, Houston, TX) and p53-null prostate cancer cell line PC3 were maintained in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% fetal calf serum (FCS, Atlanta Biologicals, Inc. GA, USA) and antibiotics. Transfection was performed with Lipofectamine™ 2000 (Invitrogen). Western blotting analysis was performed with following antibodies, DO-1 for p53, 4B11 for human Mdm2 (both were gifts from Moshe Oren, Weizmann Institute of Science, Israel), and Human Mdm4 was detected with a rabbit polyclonal antibody from Proteintech (Cat #17914–1-AP). HA-Mdm2 isoforms and FLAG-Mdm4 were detected with either anti-HA (HA.11, Covance, Princeton, NJ) or anti-FLAG (Sigma, M2, F1804) and polyubiquitin with mouse anti-ubiquitin antibody from BD (cat#550944).
Plasmid and recombinant protein preparation, and in vivo and in vivo ubiquitination assays
FLAG-MdmX and HA-Mdm2 (human) constructs for insect cell expression and protein purification were described previously.10 HA-Mdm2A and HA-Mdm2-B expression plasmids were PCR-cloned with full length human Mdm2 as a template into pcDNA3.1 for mammalian expression or pFastBact vectors for recombinant protein expression in insect cells. Human Mdm2-a cDNA has a deletion of Mdm2 coding region spanning from amino acid 27 to 221 and Mdm2-b cDNA has a deletion of Mdm2 coding region spanning from amino acid 27 to 300. The method for recombinant protein expression and purification in insect cells was described previously.10 In vitro and in vivo ubiquitination assays for p53, Mdm2 and Mdm4 were performed as described previously.10
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgment
The authors sincerely thank Dr. Moshe Oren (Weizmann Institute of Science, Israel) for p53 and Mdm2 antibodies and Dr. Gigi Lozano (MD Anderson Cancer Center) for p53/mdm2/mdm4 triple knockout MEFs and scientific communications on Mdm2 splice isoforms.
Funding
This research was supported, in part, by ACS developmental fund (XW) and by the NCI Cancer Center Support Grant to the Roswell Park Cancer Institute (CA016056).
Notes on contributors
XW conceived the study. XW and CF designed the experiments and CF conducted the experiments. XW and CF analyzed the results. XW wrote the paper.
References
- [1].Marine JC, Francoz S, Maetens M, Wahl G, Toledo F, Lozano G. Keeping p53 in check: essential and synergistic functions of Mdm2 and Mdm4. Cell Death Differ 2006; 13:927-34; PMID:16543935; http://dx.doi.org/ 10.1038/sj.cdd.4401912 [DOI] [PubMed] [Google Scholar]
- [2].Wade M, Wang YV, Wahl GM. The p53 orchestra: Mdm2 and Mdmx set the tone. Trends Cell Biol 2010; 20:299-309; PMID:20172729; http://dx.doi.org/ 10.1016/j.tcb.2010.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Levav-Cohen Y, Goldberg Z, Tan KH, Alsheich-Bartok O, Zuckerman V, Haupt S, Haupt Y. The p53-Mdm2 loop: a critical juncture of stress response. Subcell Biochem 2014; 85:161-86; PMID:25201194; http://dx.doi.org/ 10.1007/978-94-017-9211-0_9 [DOI] [PubMed] [Google Scholar]
- [4].Tanimura S, Ohtsuka S, Mitsui K, Shirouzu K, Yoshimura A, Ohtsubo M. MDM2 interacts with MDMX through their RING finger domains. FEBS Lett 1999; 447:5-9; PMID:10218570; http://dx.doi.org/ 10.1016/S0014-5793(99)00254-9 [DOI] [PubMed] [Google Scholar]
- [5].Vigneron A, Vousden KH. p53 and Mdm2: an auld alliance. Cell Cycle 2010; 9:865-66; PMID:20348845; http://dx.doi.org/ 10.4161/cc.9.5.11040 [DOI] [PubMed] [Google Scholar]
- [6].Karni-Schmidt O, Lokshin M, Prives C. The roles of MDM2 and MDMX in cancer. Annu Rev Pathol 2016; 11:617-44; PMID:27022975; http://dx.doi.org/ 10.1146/annurev-pathol-012414-040349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature 1997; 387:296-99; PMID:9153395; http://dx.doi.org/ 10.1038/387296a0 [DOI] [PubMed] [Google Scholar]
- [8].Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature 1997; 387:299-303; PMID:9153396; http://dx.doi.org/ 10.1038/387299a0 [DOI] [PubMed] [Google Scholar]
- [9].Fang S, Jensen JP, Ludwig RL, Vousden KH, Weissman AM. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J Biol Chem 2000; 275:8945-51; PMID:10722742; http://dx.doi.org/ 10.1074/jbc.275.12.8945 [DOI] [PubMed] [Google Scholar]
- [10].Wang X, Wang J, Jiang X. MdmX protein is essential for Mdm2 protein-mediated p53 polyubiquitination. J Biol Chem 2011; 286:23725-34; PMID:21572037; http://dx.doi.org/ 10.1074/jbc.M110.213868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Itahana K, Mao H, Jin A, Itahana Y, Clegg HV, Lindstrom MS, Bhat KP, Godfrey VL, Evan GI, Zhang Y. Targeted inactivation of Mdm2 RING finger E3 ubiquitin ligase activity in the mouse reveals mechanistic insights into p53 regulation. Cancer Cell 2007; 12:355-66; PMID:17936560; http://dx.doi.org/ 10.1016/j.ccr.2007.09.007 [DOI] [PubMed] [Google Scholar]
- [12].Pant V, Xiong S, Iwakuma T, Quintas-Cardama A, Lozano G. Heterodimerization of Mdm2 and Mdm4 is critical for regulating p53 activity during embryogenesis but dispensable for p53 and Mdm2 stability. Proc Natl Acad Sci U S A 2011; 108:11995-12000; PMID:21730132; http://dx.doi.org/ 10.1073/pnas.1102241108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Huang L, Yan Z, Liao X, Li Y, Yang J, Wang ZG, Zuo Y, Kawai H, Shadfan M, Ganapathy S, Yuan ZM. The p53 inhibitors MDM2/MDMX complex is required for control of p53 activity in vivo. Proc Natl Acad Sci U S A 2011; 108:12001-06; PMID:21730163; http://dx.doi.org/ 10.1073/pnas.1102309108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Matsumoto R, Tada M, Nozaki M, Zhang CL, Sawamura Y, Abe H. Short alternative splice transcripts of the mdm2 oncogene correlate to malignancy in human astrocytic neoplasms. Cancer Res 1998; 58:609-13; PMID:9485008 [PubMed] [Google Scholar]
- [15].Fridman JS, Hernando E, Hemann MT, de Stanchina E, Cordon-Cardo C, Lowe SW. Tumor promotion by Mdm2 splice variants unable to bind p53. Cancer Res 2003; 63:5703-06; PMID:14522887 [PubMed] [Google Scholar]
- [16].Steinman HA, Burstein E, Lengner C, Gosselin J, Pihan G, Duckett CS, Jones SN. An alternative splice form of Mdm2 induces p53-independent cell growth and tumorigenesis. J Biol Chem 2004; 279:4877-86; PMID:14612455; http://dx.doi.org/ 10.1074/jbc.M305966200 [DOI] [PubMed] [Google Scholar]
- [17].Volk EL, Fan L, Schuster K, Rehg JE, Harris LC. The MDM2-a splice variant of MDM2 alters transformation in vivo and the tumor spectrum in both Arf- and p53-null models of tumorigenesis. Mol Cancer Res: MCR 2009; 7:863-69; PMID:19491200; http://dx.doi.org/ 10.1158/1541-7786.MCR-08-0418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Volk EL, Schuster K, Nemeth KM, Fan L, Harris LC. MDM2-A, a common Mdm2 splice variant, causes perinatal lethality, reduced longevity and enhanced senescence. Dis Models Mech 2009; 2:47-55; PMID:19132120; http://dx.doi.org/ 10.1242/dmm.000992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Parant J, Chavez-Reyes A, Little NA, Yan W, Reinke V, Jochemsen AG, Lozano G. Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nat Genet 2001; 29:92-95; PMID:11528400; http://dx.doi.org/ 10.1038/ng714 [DOI] [PubMed] [Google Scholar]
- [20].Linke K, Mace PD, Smith CA, Vaux DL, Silke J, Day CL. Structure of the MDM2/MDMX RING domain heterodimer reveals dimerization is required for their ubiquitylation in trans. Cell Death Differ 2008; 15:841-48; PMID:18219319; http://dx.doi.org/ 10.1038/sj.cdd.4402309 [DOI] [PubMed] [Google Scholar]
- [21].Poyurovsky MV, Priest C, Kentsis A, Borden KL, Pan ZQ, Pavletich N, Prives C. The Mdm2 RING domain C-terminus is required for supramolecular assembly and ubiquitin ligase activity. EMBO J 2007; 26:90-101; PMID:17170710; http://dx.doi.org/ 10.1038/sj.emboj.7601465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kawai H, Wiederschain D, Yuan ZM. Critical contribution of the MDM2 acidic domain to p53 ubiquitination. Mol Cell Biol 2003; 23:4939-47; PMID:12832479; http://dx.doi.org/ 10.1128/MCB.23.14.4939-4947.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Meulmeester E, Frenk R, Stad R, de Graaf P, Marine JC, Vousden KH, Jochemsen AG. Critical role for a central part of Mdm2 in the ubiquitylation of p53. Mol Cell Biol 2003; 23:4929-38; PMID:12832478; http://dx.doi.org/ 10.1128/MCB.23.14.4929-4938.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Leveillard T, Gorry P, Niederreither K, Wasylyk B. MDM2 expression during mouse embryogenesis and the requirement of p53. Mech Dev 1998; 74:189-93; PMID:9651526; http://dx.doi.org/ 10.1016/S0925-4773(98)00074-4 [DOI] [PubMed] [Google Scholar]
- [25].Grier JD, Xiong S, Elizondo-Fraire AC, Parant JM, Lozano G. Tissue-specific differences of p53 inhibition by Mdm2 and Mdm4. Mol Cell Biol 2006; 26:192-98; PMID:16354690; http://dx.doi.org/ 10.1128/MCB.26.1.192-198.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Maetens M, Doumont G, Clercq SD, Francoz S, Froment P, Bellefroid E, Klingmuller U, Lozano G, Marine JC. Distinct roles of Mdm2 and Mdm4 in red cell production. Blood 2007; 109:2630-33; PMID:17105817; http://dx.doi.org/ 10.1182/blood-2006-03-013656 [DOI] [PubMed] [Google Scholar]
- [27].Valentin-Vega YA, Box N, Terzian T, Lozano G. Mdm4 loss in the intestinal epithelium leads to compartmentalized cell death but no tissue abnormalities. Differentiation 2009; 77:442-49; PMID:19371999; http://dx.doi.org/ 10.1016/j.diff.2009.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Steinman HA, Hoover KM, Keeler ML, Sands AT, Jones SN. Rescue of Mdm4-deficient mice by Mdm2 reveals functional overlap of Mdm2 and Mdm4 in development. Oncogene 2005; 24:7935-40; PMID:16027727; http://dx.doi.org/ 10.1038/sj.onc.1208930 [DOI] [PubMed] [Google Scholar]