ABSTRACT
The ability of some transcription factors to remain bound to specific genes on condensed mitotic chromosomes has been suggested to play a role in their rapid transcriptional reactivation upon mitotic exit. We have recently shown that SOX2 and OCT4 remain associated to mitotic chromosomes, and that depletion of SOX2 at the mitosis-G1 (M-G1) transition impairs its ability to maintain pluripotency and drive neuroectodermal commitment. Here we report on the role of SOX2 at the M-G1 transition in regulating transcriptional activity of embryonic stem cells. Using single cell time-lapse analysis of reporter constructs for STAT3 and SOX2/OCT4 activity, we show that SOX2/OCT4 do not lead to more rapid transcriptional reactivation in G1 than STAT3, a transcription factor that is excluded from mitotic chromosomes. We also report that only few endogenous target genes show decreased pre-mRNA levels after mitotic exit or in other cell cycle phases in the absence of SOX2 at the M-G1 transition. This suggests that bookmarked SOX2 target genes are not differently regulated than non-bookmarked target genes, and we discuss an alternative hypothesis on how mitotic bookmarking by SOX2 and other sequence-specific transcription factors could be involved in transcriptional regulation.
KEYWORDS: Embryonic stem cells, mitotic bookmarking, pluripotency, Sox2, transcription
Introduction
The regulation of cellular differentiation is intimately connected to the cell cycle, the M and G1 phases being particularly critical for cells to choose between alternative fates. M phase is a privileged window for reprogramming and rewiring of transcriptional programs,1,2 and components of the cell cycle machinery are involved in the initial decision of pluripotent stem cells to commit to neuroectodermal versus mesendodermal fate,3 a decision that is made in G1.4 Although underlying mechanisms are not fully understood, it has been speculated that the arrest of transcriptional activity occurring during mitosis may provide newly born daughter cells with an enhanced flexibility in their choice between restoring the mother cell gene expression program or redirecting their transcriptional activity toward a different fate. This would also imply that cells maintaining their identity over multiple rounds of divisions, such as stem cells, may require dedicated mechanisms acting in these windows of lower phenotypic stability to shield them from the influence of differentiation signals.
The marking of specific loci on condensed chromosomes, dubbed mitotic bookmarking, was proposed to transmit epigenetic marks to daughter cells for them to “remember” the gene expression program of the mother cell. Such marks were reported across a broad range of transcriptional regulation mechanisms, consisting in the retention of histone modifications,5 chromatin modifiers,6,7 general transcription factors8 and sequence-specific transcription factors9-15 on specific loci of mitotic chromosomes. Two seminal studies have shown that the chromatin writer MLL and reader Brd4 remain bound to specific genes and allow their rapid transcriptional reactivation upon mitotic exit.6,7 This prompted the idea that bookmarked genes have a kinetic advantage in their transcriptional regulation in G1, and that bookmarking of key cell fate determinants plays a role in maintaining cell identity during division. However, and in contrast to these chromatin modifier/reader, robust evidence supporting a role in rapid transcriptional reactivation for sequence-specific bookmarking transcription factors is lacking. While one study on GATA1 reported that genes bound during mitosis are enriched in cell fate regulators,9 there is no clear evidence that genes bookmarked by GATA116 or other sequence-specific transcription factors10,12,14 undergo faster transcriptional reactivation than non-bookmarked genes. Perhaps more importantly, these reports did not provide direct evidence of the involvement of mitotic bookmarking in cell fate regulation.
We recently screened for mitotic chromosome binding of pluripotency transcription factors and reported that SOX2 and OCT4 remain bound to mitotic chromosomes of embryonic stem (ES) cells through their DNA-binding domains.15 Using single molecule imaging, we found that both SOX2 and OCT4 display long-lived DNA binding events compatible with their association to specific sites on mitotic chromatin. Furthermore, we showed that SOX2 is enriched on a small number of genes during mitosis,15 thus qualifying SOX2 as a mitotic bookmarking transcription factor. We then investigated the functional relevance of SOX2 mitotic bookmarking by fusing it to a degron sequence from Cyclin B, allowing its degradation at the M-G1 transition.9 This allowed us to show that the absence of SOX2 at the M-G1 transition results in decreased pluripotency maintenance and abolishes its capacity to enhance neuroectodermal commitment, thus providing the first experimental link between mitotic bookmarking of a sequence-specific transcription factor and regulation of cell fate decisions. However, the molecular mechanisms by which SOX2 acts at the M-G1 transition to regulate cell fate remain unclear. Here we provide additional data on transcriptional regulation of SOX2 target genes in the presence or absence of SOX2 at the M-G1 transition and discuss how mitotic bookmarking could regulate gene expression.
Results
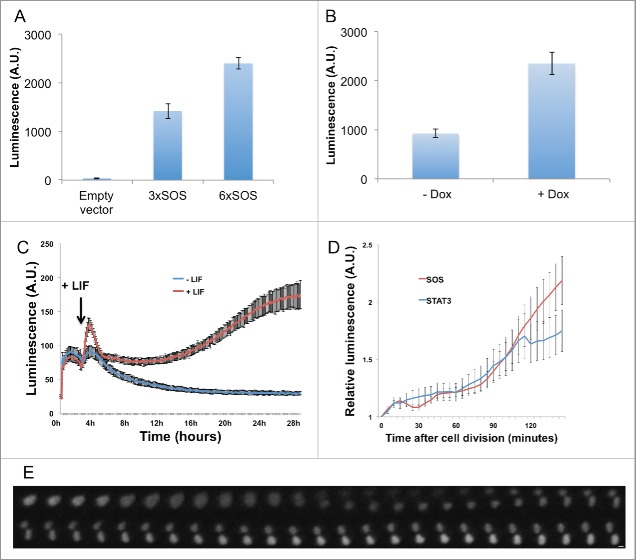
We first asked whether SOX2 has the intrinsic property to mediate particularly fast transcriptional reactivation upon mitotic exit. In pluripotent stem cells, SOX2 activates thousands of target genes by heterodimerizing with OCT4,17,18 which also binds to mitotic chromosomes. We engineered a construct containing repeats of a SOX2/OCT4 binding sequence upstream of a short-lived luciferase reporter to investigate the kinetics of transcriptional reactivation after mitotic exit mediated by SOX2. Since in this construct, both the luciferase protein and encoding mRNA are destabilized, single cell luminescence kinetics is a good proxy for transcriptional activity. We compared the transcriptional activity of constructs containing 0, 3 or 6 tandem repeats of SOX2/OCT4 binding sites (hereafter referred as to 6xSOS) after their transient transfection in ES cells, and as expected luminescence levels scaled with the number of SOX2/OCT4 binding sites (Fig. 1A). To verify that the reporter could be directly transactivated by SOX2 and OCT4, we generated NIH-3T3 cells (that do not endogenously express SOX2 or OCT4) for (dox)-inducible expression of YPet-SOX2 and Halo-OCT4 (see Methods). We transfected the 6xSOS reporter in this cell line and observed an increase in luminescence after dox induction, indicating that SOX2 and OCT4 transactivated the reporter (Fig. 1B). To compare transcriptional kinetics of the SOX2/OCT4 reporter with a reporter driven by STAT3, a transcription factor that does not bind mitotic chromosomes,15 we generated an ES cell line reporting for STAT3 activity using a reporter construct containing 7 repeats of a STAT3 binding sequence upstream of the short-lived luciferase reporter. Since STAT3 is activated by the LIF/JAK/STAT3 pathway in ES cells,19 we validated the reporter by monitoring its activity before and after addition of LIF to previously LIF-starved ES cells, which resulted in a rapid and sustained increase in luminescence levels (Fig. 1C). We then generated two ES cell lines by lentiviral transduction of the 6xSOS or 7xSTAT3 reporter and performed time-lapse luminescence microscopy in the pluripotent state. We measured single-cell luminescence levels by manual tracking just after division for at least 30 frames (2.5 h) to assess differences in post-mitotic transcriptional reactivation of both reporters. We did not observe any difference in the kinetics of post-mitotic increase in luminescence signals between the two reporters, suggesting that SOX2/OCT4 do not lead to particularly rapid transcriptional reactivation after mitotic exit as compared to STAT3 (Fig. 1D-E).
Figure 1.
(A) Luciferase activity of plasmids containing 0, 3 or 6 repeats of the SOS sequence upstream of a destabilized nuclear luciferase. (B) Activity of the 6xSOS sequence in NIH-3T3 with or without dox induction of YPet-Sox2 and Halo-Oct4 expression. (C) Luciferase activity driven by the 7xSTAT3 reporter in ES cells with or without addition of LIF in LIF-starved stable 7xSTAT3 ES cells. (D) Averaged single cell luminescence intensities normalized on values of the first frame after division for 6xSOS and 7xSTAT3 cell lines. N = 56 (6xSOS); N = 45 (7xSTAT3). Error bars: SE. (E) Time series of a dividing ES cell expressing a destabilized nuclear luciferase under the control of the 6xSOS sequence. Time resolution: 5 minutes. Scale bar: 5μM. Red: MD* cell line. Blue: MD cell line.
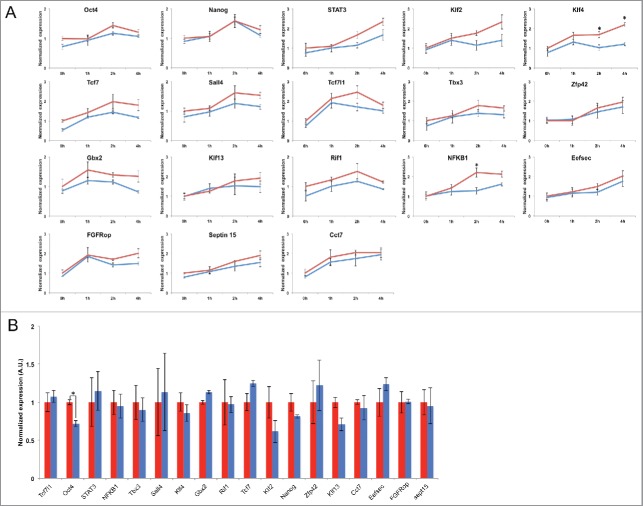
To investigate the transcriptional reactivation of SOX2 target genes after mitosis, we took advantage of the cell lines in which SOX2 is fused to a mitotic degron (MD), allowing to degrade SOX2 at the M-G1 transition, or a mutant thereof (MD*) that does not lead to mitotic degradation of SOX2. SNAP-MD-Sox2 and SNAP-MD*-Sox2 cell lines were generated in the 2TS22C cell line background, allowing dox-inducible loss of endogenous SOX2. Importantly, the two cell lines express the same amount of SOX2 on average, even though SOX2 is degraded at the M-G1 transition in the SNAP-MD-Sox2 but not in the SNAP-MD*-Sox2 cell line.15 Therefore, this allows investigating how degradation of SOX2 at the M-G1 transition affects transcriptional reactivation upon mitotic exit. Both cell lines were treated with dox to stop expressing endogenous SOX2 and synchronized using a thymidine block for 14 h followed by a 7 h nocodazole block, as previously reported.20 Cells were then released from mitotic block and RNA was collected at 0h, 1h, 2h and 4h post-release. We also collected RNA from non-synchronized cells in parallel to assess for global dysregulation of gene expression over the whole cell cycle in both cell lines. We performed Quantitative RT-PCR on nascent transcripts using primers spanning the exon-intron junctions of different pluripotency genes and other genes that were bookmarked during mitosis, as well as two control genes that are not known to be regulated by Sox2 and did not have a Sox2 ChIP-seq peak in either asynchronous or mitotic cells within 20kb of their 5′ and 3′ end. Strikingly, most Sox2 target genes were not differentially transcribed between the two cell lines after mitotic exit, and the only gene that was bookmarked during mitosis and exhibited significantly higher pre-mRNA level during G1 in the MD* cell line was Nf-Kb1, while other bookmarked genes such as Nanog, Oct4, Rif1, Eefsec and FGFRop had similar pre-mRNA levels during mitotic exit in both cell lines (Fig. 2A). Some pluripotency genes that are known targets of SOX2 but were not bookmarked during mitosis, such as Klf4 and Klf2, were also upregulated more efficiently in G1 in the presence of SOX2 during mitosis (Fig. 2A). Thus, bookmarked genes did not display more dependence on the presence of SOX2 at the M-G1 transition than non-bookmarked genes for their transcriptional reactivation in G1. Interestingly, Oct4 was more markedly downregulated in the asynchronous cell population (Fig. 2B) than early after mitotic exit (Fig. 2A), raising the possibility that SOX2 at the M-G1 transition may prime Oct4 for robust transcription during other phases of the cell cycle. In summary, the absence of SOX2 at the M-G1 transition did not result in a differential regulation of bookmarked vs. non-bookmarked genes, but led to a modest downregulation of several key pluripotency genes.
Figure 2.
(A) pre-mRNA levels of selected genes at 0, 1, 3 and 6 h post-nocodazole release. (B) pre-mRNA levels of the same genes in unsynchronized cells. Error bars: SE. *p<.05. Scale bar: 5μm.
Discussion
Here we analyzed the transcriptional reactivation of SOX2 target genes after mitotic exit and show that a SOX2/OCT4 reporter is not more rapidly reactivated than a reporter for STAT3, which does not bind to mitotic chromosomes. We also showed that genes bookmarked by SOX2 during mitosis are not subject to particularly rapid transcriptional reactivation after mitotic exit. Therefore, and similarly to GATA1 and FOXA1, there is no evidence that SOX2 mediates fast transcriptional reactivation of bookmarked genes in early G1. Consequently, it remains unclear whether gene bookmarking during mitosis by sequence-specific transcription factors is functionally relevant or simply the consequence of local mitotic chromatin architecture. Furthermore, since the residence time of most transcription factors on specific DNA-binding sites, including SOX2 and OCT4, is in the second range,15,21,22 bookmarked genes are only transiently bound during mitosis. Finally there is no evidence that bookmarking transcription factors functionally contribute in modulating the local mitotic chromatin environment. Thus, this questions the idea that bookmarked genes have a competitive advantage against non-bookmarked genes for their reactivation in G1 once chromatin starts decondensing. Interestingly, the Zaret laboratory reported that FOXA1 target genes were more rapidly reactivated than non-target genes upon mitotic exit, irrespectively of their bookmarking status.10 While the scope of the present work does not allow thoroughly addressing this question for SOX2, we did not find strong evidence for particularly rapid transcriptional reactivation of SOX2 targets. One limitation of our study is the modest increase in pre-mRNA levels we observed after mitotic exit. We also performed experiments in which nocodazole treatment was extended to 18 h, but this did not result in lower pre-mRNA amounts in synchronized cells or stronger increase in gene expression after mitotic exit (data not shown). This suggests that the detection of pre-mRNA in mitotically-synchronized cells resulted either from a fraction of cells reaching G1 phase in the presence of nocodazole, or carry-over of unprocessed pre-mRNAs from cells still in the G2 phase. Nevertheless, based on earlier studies in the field,16,10,12 and on the present work, the heavy focus made on the identification and characterization of specific sites bound during mitosis did not reveal clear functional insights. Interestingly, mitotic bookmarking transcription factors are mostly identified by their co-localization with mitotic chromosomes as assessed by microscopy, which is arguably observable because of the large fraction of non-specifically bound molecules.10,15 While most of such transcription factors have been confirmed by ChIP-seq to be also retained on specific loci, there is no evidence that those that are apparently largely evicted from mitotic chromosomes do not remain bound to specific sites. Furthermore, whether either of these two parameters is associated with the function of transcription factors in gene regulation during G1 phase or the regulation of cell fate decisions remains to be elucidated.
Interestingly, we found that several SOX2 target genes were dependent on the presence of SOX2 during the M-G1 transition for their full transcriptional activity in G1 as late as 4 h after nocodazole block release, and that Oct4 transcription was significantly downregulated in asynchronous cells in the absence of SOX2 at the M-G1 transition. This suggests the possibility that SOX2 needs to reach some of its target sites in early G1 to ensure their robust transcription, irrespectively of when each target is maximally transcribed. How could mitotic bookmarking shorten the search time of SOX2 or other mitotically-bound transcription factors for their targets ? Since mitotic chromosomes occupy a relatively small volume of the mitotic cell,23 the sequestration of transcription factor molecules in this compartment substantially increases their local concentration. This may reduce their search time for target sites upon mitotic chromosome decondensation, potentially providing them with a competitive advantage over transcription factors that do not remain bound to mitotic chromosomes. So then why does SOX2 not lead to more rapid transcriptional reactivation of target genes? It could be that SOX2 is generally not rate-limiting for the speed of target gene reactivation, but its timely target occupation may enhance the robustness of SOX2 control over their activity after mitotic exit, the kinetics of their transcriptional reactivation depending on other activators. Since G1 is particularly short in ES cells and is a privileged time window for cell fate decisions, mitotic bookmarking by central pluripotency transcription factors may be particularly important to control the stability of gene regulatory networks through cell divisions. In the future, it will be important to determine how mitotic chromosome binding by other transcription factors such as OCT4 and ESRRB collectively contribute to maintain the pluripotency network.
Methods
The culture conditions of ES cells, generation of cell lines expressing SNAP-MD-Sox2 and SNAP-MD*-Sox2 in the 2TS22C background, and luminescence microscopy are described in our previous study.15
Generation of SOX2/OCT4 and STAT3 sensor constructs and stable cell lines
To generate the SOX2/OCT4 sensor, top and bottom oligos encoding 3 repeats of the CTTTGTTATGCAAAT SOX2/OCT4 consensus sequences spaced by 42 nucleotides were phosphorylated using T4 Polynucleotide kinase (NEB), annealed, and ligated into the NheI site upstream of TATA box and of the coding sequence for destabilized nuclear firefly luciferase in a previously described lentiviral vector backbone.24 E. Coli transformants were then screened for insertion number and orientation, and two constructs with either 1 or 2 insertions in the same orientation (thus containing 3 or 6 repeats) were selected for further experiments. The STAT3 sensor construct was kindly provided by Dr. Ka Yi Hui and Prof. Ueli Schibler, Geneva University, and contains 7 repeats of the CTTCCCGGAA sequence spaced by 84 nucleotides. The 6xSOS and 7xSTAT3 E14 cell lines were generated by lentiviral transduction with the corresponding vectors described above and as previously described.15 To generate a NIH-3T3 cell line expressing Halo-Oct4 and YPet-Sox2 upon dox induction (hereafter referred as to 3T3-hOS), NIH-3T3 were transduced with three lentiviral vectors encoding rtTA3G, TRE3G-Halo-Oct4 and TRE3G-YPet-Sox2. The construction of these vectors and lentiviral transduction of NIH-3T3 is described in reference.15
Transfections and luminescence assays
All transfections were performed using the Xtreme Gene 9 reagent (Roche) according to the manufacturers instructions. To assay the activity of the 6xSOS reporter in ES cells, E14 ES cells were plated in a white gelatinated 96-well plate and transfected the next day with a lentivector containing either 0, 3 or 6 repeats of the SOX2/OCT4 consensus sequence upstream of the short-lived luciferase, or a plasmid driving the expression of eGFP under the control of the PGK promoter (negative control). 48 h after transfection, a luciferase assay was performed using the Steady-Glo® Luciferase Assay System (Promega) on a luminescence plate reader (Infinite F200 Pro, Tecan). To assay the activity of the 6xSOS reporter in NIH-3T3 cells, 3T3hOS cells were plated in 8 wells of a white 96-well plate and transfected the next day with the lentivector containing 6 repeats of the SOX2/OCT4 consensus sequence. 24 h later, 4 of the wells were treated with 0.5μg/ml of dox, and cells from all wells were lysed and assayed for luciferase activity in a luminescence plate reader. To assay the activity of the 7xSTAT3 reporter, 7xSTAT3 ES cells were plated on 6 gelatinated 3.5cm dishes in medium devoid of LIF and 2i. The next day, luciferin was added to the cells at the final concentration of 100μM and luminescence recordings were started using a Lumicycle apparatus. 2 h later, 2μl of home-made LIF were added to the cell culture medium of 3 of the dishes.
ES cell synchronization and quantitative RT-PCR
To synchronize ES cells in M-phase, we used a previously described approach based on dual synchronization at the G1-S transition and at the beginning of M-phase.20 Briefly, ES cells were plated at 300,000 cells per gelatinated 3.5cm dish in complete cell culture medium supplemented with 1μg/ml of dox. The next day, the medium was supplemented with 1.25mM thymidine, and 14 h later, cells were washed and further maintained in medium containing 50ng/ml of nocodazole and 1μg/ml of dox. 7 h after nocodazole addition, cells were washed and further maintained in complete medium and 1μg/ml of dox. Cell cultures were then flash-frozen in liquid nitrogen at 0h, 1h, 2h and 4h post-nocodazole block release, and RNA was extracted using an a GenElute™ Mammalian Total RNA Miniprep Kit Q-PCR (Sigma-Aldrich). Reverse transcription was performed using random hexamers primer using superscript II (Life Technologies). QPCR was performed on a 7900HT Fast Real-Time PCR System (Thermofisher) with SYBR green reagent (Roche). The house-keeping gene Rps9 was used for data normalization. Primers used for RT-QPCR are listed in Table 1.
Table 1.
QPCR primers spanning intron-exon junctions of selected genes.
| Sequence (5′ to 3′) | Target sequence |
|---|---|
| CCAATGCCGTGAAGTTGGAG | Oct4_F |
| TCCCAATTCCCTTCACTGCT | Oct4_R |
| GGGTCTGCTACTGAGATGCT | Nanog_F |
| TTACTGGGTTCTTCGGGGAC | Nanog_R |
| TGGCACCTTGGATTGAGAGT | STAT3_F |
| CCTGACTCAATGCTAAACCCC | STAT3_R |
| AGCCTATCTTGCCGTCCTTT | Klf2_F |
| CTGCACCCTGTAGCCTGGTA | Klf2_R |
| GAAGGGAGAAGACACTGCGT | Klf4_F |
| CCACCCCATCTGCAGAAATC | Klf4_R |
| CCGGACAAACTTCCAGAGTC | Tcf7_F |
| CGGTTCTAGCGCCTTCTTC | Tcf7_R |
| TAACATATGCGGGCGGGC | Sall4_F |
| CACACAGACGTCACACACC | Sall4_R |
| TGGTCAACGAATCGGAGAAT | Tcf7l1_F |
| CAGAGGCTGACCCTAGGAAT | Tcf7l1_R |
| CGGGGTACAGAGATGGTCAT | Tbx3_F |
| AGAGTCAGACAAAAGAGATGTGA | Tbx3_R |
| TCCAAGTGTTGTCCCCAAAT | Zfp42_F |
| GGAATAAAGGGACTGGCAGA | Zfp42_R |
| GCTCTCCTGCTAGCTACTCC | Gbx2_F |
| AACACACCAAGGACCCTCAA | Gbx2_R |
| CGCACCTGAGAACTCACACA | Klf13_F |
| GGTCCCAAGCAGTCCCTAGT | Klf13_R |
| TCTCGGCTGTACACGGTTTT | Rif1_F |
| TGAGGCAGTGCGAAGTCATT | Rif1_R |
| CCGCATCTCCAGGGTAAG | NFKB1_F |
| CTTGGGGCTCTCCCCTAGT | NFKB1_R |
| CTCATCCGGACCATCATTG | Eefsec_F |
| CAGGCCTAGGCTGTCAGG | Eefsec_R |
| ACTCAGAGCGGCTGTGTTTT | FGFRop_F |
| TACGGCATGAACAAGGTCTG | FGFRop_R |
| GCTGTCAGGAAGAAGCACAA | Septin15_F |
| GGTCTAACTGATTAAGCAGCCTGT | Septin15_R |
| AACTGACGAGTTGCGGGTA | Cct7_F |
| ACTGTAGACACTCCGCATGG | Cct7_R |
| TTGTCGCAAAACCTATGTGACC | Rps9_F |
| GCCGCCTTACGGATCTTGG | Rps9_R |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed
Acknowledgments
We thank Dr. Ka Yi Hui for providing us with the 7xSTAT3 lentiviral reporter and Dr. Jonathan Sobel for helping with bioinformatics analysis of the STAT3 binding sites.
Funding
This work was supported by the Carigest Foundation and the Swiss National Science Foundation (grant no. PP00P3_144828).
References
- [1].Egli D, Rosains J, Birkhoff G, Eggan K. Developmental reprogramming after chromosome transfer into mitotic mouse zygotes. Nature 2007; 447:679-85; PMID:17554301; http://dx.doi.org/ 10.1038/nature05879 [DOI] [PubMed] [Google Scholar]
- [2].Halley-Stott RP, Jullien J, Pasque V, Gurdon J. Mitosis gives a brief window of opportunity for a change in gene transcription. PLoS Biol 2014; 12:e1001914; PMID:25072650; http://dx.doi.org/ 10.1371/journal.pbio.1001914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Pauklin S, Madrigal P, Bertero A, Vallier L. Initiation of stem cell differentiation involves cell cycle-dependent regulation of developmental genes by Cyclin D. Genes Dev 2016; 30:421-33; PMID:26883361; http://dx.doi.org/ 10.1101/gad.271452.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Pauklin S, Vallier L. The cell-cycle state of stem cells determines cell fate propensity. Cell 2013; 155:135-47; PMID:24074866; http://dx.doi.org/ 10.1016/j.cell.2013.08.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Zaidi SK, Young DW, Montecino M, van Wijnen AJ, Stein JL, Lian JB, Stein GS. Bookmarking the genome: maintenance of epigenetic information. J Biol Chem 2011; 286:18355-61; PMID:21454629; http://dx.doi.org/ 10.1074/jbc.R110.197061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Blobel GA, Kadauke S, Wang E, Lau AW, Zuber J, Chou MM, Vakoc CR. A reconfigured pattern of MLL occupancy within mitotic chromatin promotes rapid transcriptional reactivation following mitotic exit. Mol Cell 2009; 36:970-83; PMID:20064463; http://dx.doi.org/ 10.1016/j.molcel.2009.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Zhao R, Nakamura T, Fu Y, Lazar Z, Spector DL. Gene bookmarking accelerates the kinetics of post-mitotic transcriptional re-activation. Nat Cell Biol 2011; 13:1295-304; PMID:21983563; http://dx.doi.org/ 10.1038/ncb2341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Chen D, Hinkley CS, Henry RW, Huang S. TBP dynamics in living human cells: constitutive association of TBP with mitotic chromosomes. Mol Biol Cell 2002; 13:276-84; PMID:11809839; http://dx.doi.org/ 10.1091/mbc.01-10-0523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kadauke S, Udugama MI, Pawlicki JM, Achtman JC, Jain DP, Cheng Y, Hardison RC, Blobel GA. Tissue-specific mitotic bookmarking by hematopoietic transcription factor GATA1. Cell 2012; 150:725-37; PMID:22901805; http://dx.doi.org/ 10.1016/j.cell.2012.06.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Caravaca JM, Donahue G, Becker JS, He X, Vinson C, Zaret KS. Bookmarking by specific and nonspecific binding of FoxA1 pioneer factor to mitotic chromosomes. Genes Dev 2013; 27:251-60; PMID:23355396; http://dx.doi.org/ 10.1101/gad.206458.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Young DW, Hassan MQ, Yang X-Q, Galindo M, Javed A, Zaidi SK, Furcinitti P, Lapointe D, Montecino M, Lian JB, et al.. Mitotic retention of gene expression patterns by the cell fate-determining transcription factor Runx2. Proc Natl Acad Sci U S A 2007; 104:3189-94; PMID:17360627; http://dx.doi.org/ 10.1073/pnas.0611419104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Festuccia N, Dubois A, Vandormael-Pournin S, Gallego Tejeda E, Mouren A, Bessonnard S, Mueller F, Proux C, Cohen-Tannoudji M, Navarro P. Mitotic binding of Esrrb marks key regulatory regions of the pluripotency network. Nat Cell Biol 2016; 18:1139-48; PMID:27723719; http://dx.doi.org/ 10.1038/ncb3418 [DOI] [PubMed] [Google Scholar]
- [13].Verdeguer F, Le Corre S, Fischer E, Callens C, Garbay S, Doyen A, Igarashi P, Terzi F, Pontoglio M. A mitotic transcriptional switch in polycystic kidney disease. Nat Med 2010; 16:106-10; PMID:19966811; http://dx.doi.org/ 10.1038/nm.2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lake RJ, Tsai PF, Choi I, Won KJ, Fan HY. RBPJ, the major transcriptional effector of notch signaling, remains associated with chromatin throughout mitosis, suggesting a role in mitotic bookmarking. PLoS Genet 2014; 10:e1004204; PMID:24603501; http://dx.doi.org/ 10.1371/journal.pgen.1004204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Deluz C, Friman ET, Strebinger D, Benke A, Raccaud M, Callegari A, Leleu M, Manley S, Suter DM. A role for mitotic bookmarking of SOX2 in pluripotency and differentiation. Genes Dev 2016; 30:2538-50; PMID:27920086; http://dx.doi.org/ 10.1101/gad.289256.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hsiung CC-S, Bartman CR, Huang P, Ginart P, Stonestrom AJ, Keller CA, Face C, Jahn KS, Evans P, Sankaranarayanan L, et al.. A hyperactive transcriptional state marks genome reactivation at the mitosis-G1 transition. Genes Dev 2016; 30:1423-39; PMID:27340175; http://dx.doi.org/ 10.1101/gad.280859.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Reményi A, Lins K, Nissen LJ, Reinbold R, Schöler HR, Wilmanns M. Crystal structure of a POU/HMG/DNA ternary complex suggests differential assembly of Oct4 and Sox2 on two enhancers. Genes Dev 2003; 17:2048-59; PMID:12923055; http://dx.doi.org/ 10.1101/gad.269303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Cole MF, Young RA. Mapping key features of transcriptional regulatory circuitry in embryonic stem cells. Cold Spring Harb Symp Quant Biol 2008; 73:183-93; PMID:19022761; http://dx.doi.org/ 10.1101/sqb.2008.73.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Niwa H, Burdon T, Chambers I, Smith A. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev 1998; 12:2048-60; PMID:9649508; http://dx.doi.org/ 10.1101/gad.12.13.2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ballabeni A, Park IH, Zhao R, Wang W, Lerou PH, Daley GQ, Kirschner MW. Cell cycle adaptations of embryonic stem cells. Proc Natl Acad Sci U A 2011; 108:19252-7; PMID:22084091; http://dx.doi.org/ 10.1073/pnas.1116794108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Gebhardt JC, Suter DM, Roy R, Zhao ZW, Chapman AR, Basu S, Maniatis T, Xie XS. Single-molecule imaging of transcription factor binding to DNA in live mammalian cells. Nat Methods 2013; 10:421-6; PMID:23524394; http://dx.doi.org/ 10.1038/nmeth.2411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Teves SS, An L, Hansen AS, Xie L, Darzacq X, Tjian R. A dynamic mode of mitotic bookmarking by transcription factors. eLife 2016; 5:e22280; PMID:27855781; http://dx.doi.org/ 10.7554/eLife.22280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Booth DG, Beckett AJ, Molina O, Samejima I, Masumoto H, Kouprina N, Larionov V, Prior IA, Earnshaw WC. 3D-CLEM reveals that a major portion of mitotic chromosomes is not chromatin. Mol Cell 2016; 64:790-802; PMID:27840028; http://dx.doi.org/ 10.1016/j.molcel.2016.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Suter DM, Molina N, Gatfield D, Schneider K, Schibler U, Naef F. Mammalian genes are transcribed with widely different bursting kinetics. Science 2011; 332:472-4; PMID:21415320; http://dx.doi.org/ 10.1126/science.1198817 [DOI] [PubMed] [Google Scholar]