Summary
Chronic biofilm-associated infections caused by Staphylococcus aureus often lead to significant increases in morbidity and mortality, particularly when associated with indwelling medical devices. This has triggered a great deal of research attempting to understand the molecular mechanisms that control S. aureus biofilm formation and the basis for the recalcitrance of these multicellular structures to antibiotic therapy. The purpose of this review is to summarize our current understanding of S. aureus biofilm development, focusing on the description of a newly-defined, five-stage model of biofilm development and the mechanisms required for each stage. Importantly, this model includes an alternate view of the processes involved in microcolony formation in S. aureus and suggests that these structures originate as a result of stochastically regulated metabolic heterogeneity and proliferation within a maturing biofilm population, rather than a subtractive process involving the release of cell clusters from a thick, unstructured biofilm. Importantly, it is proposed that this new model of biofilm development involves the genetically programmed generation of metabolically distinct subpopulations of cells, resulting in an overall population that is better able to adapt to rapidly changing environmental conditions.
Graphical Abstract
Novel technological advances that combine microfluidic flow-cell systems with time-lapse microscopy have greatly enhanced the visualization of biofilm development. Using this technology, our laboratory has revealed a more detailed view of the morphological stages and differential gene expression that occurs during Staphylococcus aureus biofilm development. Here, we review the complex molecular mechanisms that are required for each developmental stage, and describe a new model for the formation of structure during biofilm maturation.
Introduction
In contrast to microbiology laboratory conditions where bacteria are often grown planktonically in nutrient-rich conditions, bacteria found in the environment almost exclusively grow in nutrient-deficient conditions where they form multicellular aggregations called biofilms (Costerton et al., 1987, Hall-Stoodley et al., 2004). In order to form biofilms, bacteria generate a self-produced extracellular matrix (ECM) composed of proteins, carbohydrates, and/or extracellular DNA (eDNA) (Flemming & Wingender, 2010), which encases the cells within a sticky matrix that facilitates survival in hostile or extreme environments. In recent years, bacterial biofilms produced by human pathogens have become particularly important to study due to their increased recalcitrance to not only the host immune system (Otto, 2006), but also to antibiotics (Costerton et al., 1999, Donlan & Costerton, 2002).
The biofilm-producing pathogen, Staphylococcus aureus, has become notorious for causing chronic infections due to its ability to resist therapeutic treatment by forming biofilms on indwelling medical devices, including implanted artificial heart valves, catheters and joint prosthetics (McConoughey et al., 2014, Ribeiro et al., 2012). Indeed, biofilm-related infections are associated with increased morbidity and mortality, with infected medical devices often requiring surgical removal and increased durations of hospitalization. As a result, the prevalence of these and other staphylococcal diseases has led to a significant increase in expenses associated with S. aureus infections over the past decade, with estimated annual costs near $450 million (Parvizi et al., 2010, Song et al., 2010). Consequently, a better understanding of the development of staphylococcal biofilms at the molecular level is imperative to generate new treatment strategies for biofilm-associated infections and to reduce the significant burdens caused by this pathogen. Here, we discuss recent advances in our understanding of the different stages of S. aureus biofilm development that have resulted from the use of state-of-the-art, time-lapse microscopic technology, and where possible, describe the molecular components that modulate each stage of this complex process.
Redefining the Stages of S. aureus Biofilm Development
While the molecular constituents involved in bacterial biofilm development vary amongst bacterial species, a basic model that is widely recognized consists of three sequential stages: 1) attachment, 2) accumulation/maturation, and 3) detachment/dispersal (Kostakioti et al., 2013, O’Toole et al., 2000, Hall-Stoodley et al., 2004). During the attachment stage, planktonic cells adhere to biotic or abiotic surfaces and proliferate into sticky aggregations called microcolonies (also known as towers or mushroom-like structures). As these microcolonies develop, bacterial cells produce an ECM that serves as a scaffold essential for establishing this three-dimensional architecture. Upon reaching a specific cell density, a mechanism is triggered to initiate ECM degradation that releases cells embedded within the biofilm to disperse and reinitiate biofilm development at distal sites.
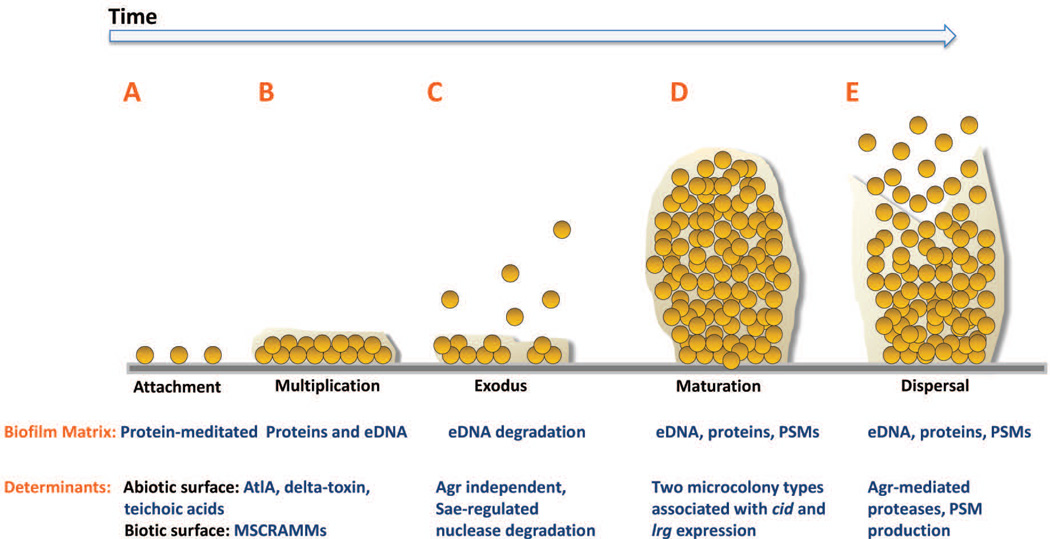
Like other bacterial species, S. aureus has been proposed to possess similar stages of biofilm development (Otto, 2013, Le et al., 2014). In fact, several biofilm studies suggest that S. aureus biofilms mature into thick layers of cells at which point detachment mechanisms are triggered and subpopulations of the biofilm are dispersed carving out microcolonies in the biomass that remains (Yarwood et al., 2004, Periasamy et al., 2012, Boles & Horswill, 2008). While these studies have provided immense insight into the molecular components that determine the biofilm architecture, new biofilm assays combining microfluidic flow-cell systems and time-lapse microscopy have revealed a more detailed view of the stages of S. aureus biofilm development. Indeed, the recent use of the BioFlux1000 system, a microfluidic flow-cell device integrated with a fluorescence microscope, has allowed S. aureus biofilm development to be evaluated in a nearly real-time manner (Benoit et al., 2010, Moormeier et al., 2013, Moormeier et al., 2014, Lehman et al., 2015, Vanhommerig et al., 2014, McCourt et al., 2014). Using this system, S. aureus biofilm development has been shown to proceed through a five-stage developmental process including: 1) attachment, 2) multiplication, 3) exodus, 4) maturation, and 5) dispersal (Figure 1) (Moormeier et al., 2014).
Figure 1. Model of Staphylococcus aureus biofilm development.
S. aureus biofilm development is described in five stages: A) attachment, B) multiplication, C) exodus, D) maturation, and E) dispersal. A. S. aureus cells attach to abiotic or biotic surfaces via hydrophobic interactions or MSCRAMMs, respectively. B. After cells attach, the biofilm develops into a confluent ‘mat’ of cells composed of an eDNA and proteinaceous matrix. C. Upon reaching confluency, a period of mass exodus of cells occurs in which a subpopulation of cells is released from the biofilm via Sae-regulated nuclease-mediated eDNA degradation to allow for the formation of three-dimensional microcolonies. D. Microcolonies form from distinct foci of cells that have remained attached during the exodus stage. This stage is characterized by rapid cell division that forms robust aggregations composed of proteins including PSMs and eDNA. E. Activated Agr-mediated quorum sensing initiates biofilm matrix modulation and dispersal of cells via protease activation and/or PSM production. AtlA, autolysin A; MSCRAMM, microbial surface components recognizing adhesive matrix molecules; eDNA, extracellular DNA; PSM, phenol soluble modulins; Agr, accessory gene regulator.
Attachment
To initiate biofilm formation on biotic materials, planktonic S. aureus cells first attach to a surface (Figure 1A) utilizing a variety of cell wall-anchored (CWA) proteins specific for different host matrix substrates. Part of this well-characterized group of surface attached proteins are the microbial surface components recognizing adhesive matrix molecules (MSCRAMMs), several of which share a common cell wall targeting motif (LPXTG; see Navarre & Schneewind, 1994) and Marraffini et al., 2006), but have different binding specificities for host matrix components such as fibronectin, fibrinogen, collagen, and cytokeratin (Speziale et al., 2009). Numerous MSCRAMMs such as fibronectin-binding proteins (FnBPA and FnBPB) (O’Neill et al., 2008, McCourt et al., 2014), serine-aspartate repeat family proteins (SdrC, SdrD, and SdrE) (Corrigan et al., 2009, Josefsson et al., 1998, O’Brien et al., 2002), clumping factors (ClfA and ClfB) (McDevitt et al., 1994, Ni Eidhin et al., 1998), collagen adhesin (Zong et al., 2005), Protein A (Nguyen et al., 2000), plasmin sensitive protein (Pls) (Huesca et al., 2002), SasG (Roche et al., 2003), iron-regulated surface determinants (IsdA, IsdB, IsdC, and IsdH) (Mazmanian et al., 2003, Miajlovic et al., 2010, Dryla et al., 2003), and bone sialoprotein (Bbp) (Vazquez et al., 2011) have been implicated in binding host matrix components to initiate cell adherence and/or biofilm development. Importantly, the attachment of most of these proteins to the bacterial cell wall is reliant on the membrane-associated protein, sortase A, which catalyzes the covalent attachment of these proteins to the penta-glycine cross-linker component of the peptidoglycan (Mazmanian et al., 1999). For a more comprehensive overview on the structures and functions of the different MSCRAMMs and other CWA proteins, the reader is referred to an outstanding recent review of this subject (Foster et al., 2014).
Although S. aureus is well equipped to bind multiple host matrix proteins that quickly coat implanted devices during biofilm-associated infections, recent findings suggest that these proteins play a minimal role when attaching directly to abiotic materials. In the absence of matrix molecules, such as under the conditions used in our Bioflux studies, S. aureus may attach to abiotic surfaces through electrostatic and hydrophobic interactions in static biofilm assays where differently charged polystyrene surfaces result in drastic alterations in attachment and overall biofilm development (Kennedy & O’Gara, 2004). Furthermore, the negatively charged teichoic acids have also been implicated in attachment to polystyrene and glass surfaces (Gross et al., 2001), in addition to the major autolysin, AtlA, which has been shown to aid in cell attachment to hydrophilic and hydrophobic polystyrene surfaces (Biswas et al., 2006, Houston et al., 2011).
Recently, the cell wall-associated protein-independent nature of binding to abiotic surfaces was further supported by testing mutants from the Nebraska Transposon Mutant Library (Fey et al., 2013). In these experiments, mutations affecting expression of CWA proteins previously described to have a function in biofilm development, including several of the MSCRAMMs listed above, agrA, atlA, and the cell wall anchoring enzymes, sortase A and B, were screened for their effect on the early stages of biofilm formation. Interestingly, only the agr and atlA mutants affected biofilm formation, demonstrating increased and decreased levels of attachment and multiplication, respectively (Moormeier et al., 2014). Indeed, the Agr quorum sensing circuit in S. aureus has been shown to play a role in biofilm adherence by regulating the phenol soluble modulin (PSM) peptides (Periasamy et al., 2012) (further discussed below). One of the PSMs, δ-toxin, was previously shown to inhibit attachment to polystyrene by preventing hydrophobic interactions between the cell and the polymer surface (Vuong et al., 2000). While AtlA binds to fibronectin (Houston et al., 2011), the demonstration that the autolytic activity of this enzyme is required for biofilm formation (Bose et al., 2012) suggests that the specific role of AtlA in early biofilm development may involve multiple functions.
Multiplication
After attaching to a surface, and in the presence of a sufficient nutrient source, the adherent S. aureus cells will begin to divide and accumulate. However, prior to the production of an ECM in which to embed, the newly formed daughter cells are vulnerable to detachment, especially in the presence of the shear forces associated with fluid flow. To maintain stability of this immature biofilm, S. aureus cells are known to produce a variety of factors that help to stabilize cell-to-cell interactions. It is this time of cell division and accumulation that we have termed the multiplication stage (Figure 1B).
Staphylococci produce several extracellular proteins that could facilitate biofilm accumulation by promoting intercellular binding shortly after initial attachment. Some of these proteins CWA proteins categorized as MSCRAMMs (see above), like the FnBPs, ClfB, and SdrC proteins, play dual roles in both attachment and accumulation (Speziale et al., 2014). Other CWA proteins such as the Staphylococcus epidermidis accumulation-associated protein (Aap) (Conlon et al., 2014, Schaeffer et al., 2015) and the S. aureus homolog, SasG (Geoghegan et al., 2010), have also been implicated in attachment and early accumulation. In addition, CWA proteins like Protein A (Merino et al., 2009), SasC (Schroeder et al., 2009), and Bap (Cucarella et al., 2001), have all shown a propensity to aid in biofilm accumulation. While these proteins appear to have a role in the multiplication stage of biofilm development, their function during this stage was not apparent in flow-cell experiments in the absence of matrix components (Moormeier et al., 2014). Similarly, polysaccharide intracellular adhesin (PIA) has been shown to function as an ECM component during early S. aureus biofilm formation (Cramton et al., 1999, Cramton et al., 2001, O’Gara, 2007), however, the production of this matrix molecule appears to be strain- or condition-specific (Fitzpatrick et al., 2005, Toledo-Arana et al., 2005, Brooks & Jefferson, 2014), O’Neill et al., 2007, Rohde et al., 2007, Boles et al., 2010). Indeed, icaA mutant (gene encoding a N-glycosyltransferase that is essential for PIA production) derivatives of UAMS-1 and USA300 JE2 strains demonstrated normal accumulation during the multiplication stage (Moormeier et al., 2014).
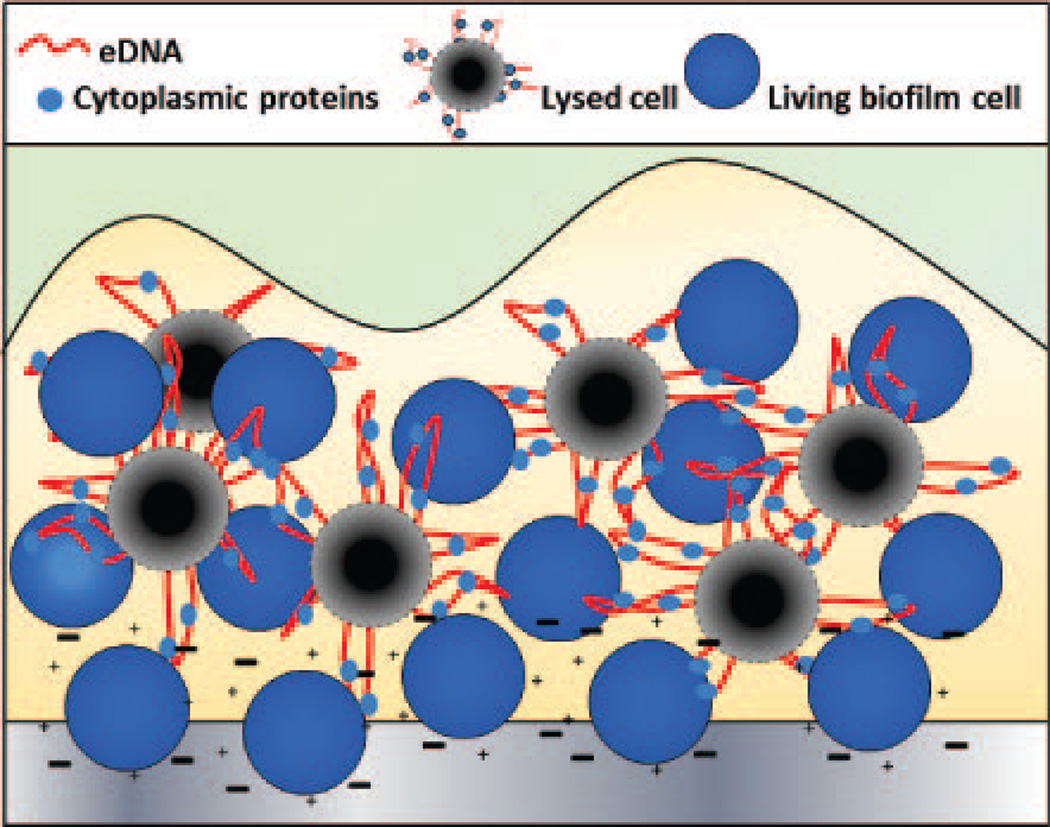
Although recent results indicate that cell wall-associated proteins are not involved during the multiplication stage, protease addition during this stage was shown to abrogate biofilm formation (Moormeier et al., 2014), indicating that the accumulation of cells involves a proteinaceous component. Interestingly, this is consistent with recent findings demonstrating that S. aureus biofilms utilize cytoplasmic proteins as matrix components (Foulston et al., 2014). In these studies, enolase and GAPDH, which are not typically recognized as biofilm-related proteins, were shown to “moonlight” as biofilm matrix components by attaching to the surface of cells in response to the decreasing pH of the biofilm environment (Foulston et al., 2014). Although the mechanisms used by cytoplasmic proteins devoid of a signal peptide can be transported to the extracellular milieu have not been described, the authors speculate that the release of these proteins is mediated by “regulated autolysis” similar to that described for the release of eDNA during biofilm development (Sadykov & Bayles, 2012, Bayles, 2014), which may establish an early ECM during the multiplication stage (Figure 2). Indeed, this may involve the binding of enolase and GAPDH to eDNA under low pH conditions as proposed by Dengler et al. (Dengler et al., 2015). Likewise, other extracellular proteins such as PSMs (Schwartz et al., 2016), beta-toxin (Hlb) (Huseby et al., 2010), and the immunodominant surface antigen B (IsaB) (Mackey-Lawrence et al., 2009) have been shown to bind eDNA and potentially function to stabilize the ECM. In addition, results also suggest that cytoplasmic nucleoid-associated proteins (NAPs), typically used for chromosomal structuring, may serve as an ECM component protein by binding eDNA (Goodman et al., 2011). Given these data and the results that demonstrate early-stage biofilms are protease-sensitive (Moormeier et al., 2014), cytoplasmic proteins that bind to eDNA may be important during the multiplication stage of biofilm formation before the matrix components have had a chance to accumulate.
Figure 2. Model of cellular interactions during the multiplication stage of biofilm development.
During the initial stages of S. aureus biofilm development, planktonic cells attach to a surface through electrostatic interactions (indicated by + and – symbols) involving teichoic acids, PSMs, and autolysin A. As biofilm development progresses into the multiplication stage, a subpopulation of cells dies and lyses (black circles) releasing extracellular DNA (red lines) and cytoplasmic proteins (light blue ovals) into the extracellular milieu, encasing the existing living cells (blue circles) in a mixture of cytoplasmic proteins and genomic DNA.
Exodus
One of the observations of biofilm development made using time-lapse microscopy was a distinct and coordinated release of the cells approximately six hours after the initiation of the multiplication stage. This, so-called, “exodus” stage of biofilm development is an early dispersal event that coincides with microcolony formation and results in the restructuring of the biofilm (Figure 1C). Importantly, exodus is mediated by nuclease-dependent degradation of eDNA and is independent of the Agr-dispersal mechanism that occurs after microcolony development (discussed below). Degradation of eDNA within the biofilm matrix by a self-produced, secreted nuclease has repeatedly been shown to reduce the total biomass of S. aureus biofilms (Mann et al., 2009, Kiedrowski et al., 2011, Moormeier et al., 2014, Kiedrowski et al., 2014, Beenken et al., 2012). However, it was not until recent studies using time-lapse microscopy that it was determined that the Nuc-mediated eDNA degradation occurs very early during biofilm development and mediates the exodus event (Figure 1C). This is a tightly regulated phase of biofilm development where only a subpopulation of cells within the biofilm expresses nuc resulting in the secretion of nuclease that mediates the detachment of the majority of the accumulated biofilm population (Moormeier et al., 2014). Additionally, the subpopulation of nuc-expressing cells was absent in a sae mutant (Moormeier et al., 2014), consistent with a previous study demonstrating the Sae-dependent control of nuc expression (Olson et al., 2013). Interestingly, another Sae-regulated gene, coa (whose product, coagulase, converts fibrinogen to fibrin), known to aid in biofilm formation when grown in the presence of host matrix proteins (Zapotoczna et al., 2015) suggests the potential for coordinated expression of biofilm effectors during biofilm development. Although several external stimuli have been shown to induce Sae-mediated signal transduction, including the presence of antimicrobial peptides (Flack et al., 2014), the signaling events at play during biofilm growth (which lack the presence of antimicrobial peptides) remains to be elucidated.
In addition to exposing a new stage in biofilm development, the studies conducted by Moormeier et al. (Moormeier et al., 2014) reveal a drastic shift in ECM as biofilm integrity progresses from a reliance on protein components only (attachment and multiplication stages, Figure 1A and 1B) to a dependence on both DNA and proteins (exodus stage, Figure 1C). Thus, the concept of exclusively PIA-based, protein-based, and eDNA-based biofilms should be replaced with a more dynamic model of biofilm development where the composition of the ECM changes both temporally and spatially as the biofilm develops. This model is similar to that described by Ma et al. (Ma et al., 2009) where significant changes in the matrix components were observed during P. aeruginosa biofilm development. Hence, studies aimed at further dissecting the molecular events mediating both Sae and Agr signaling are warranted.
What is the biological role of the exodus stage? Although the answer to this question is not known, it may be significant that the more complex developmental bacterium, Myxococcus xanthus, also exhibits a “restructuring” event as part of its developmental cycle. Indeed, prior to fruiting body formation, the M. xanthus population is reduced approximately 80% through a process that may involve the function of a toxin-antitoxin system (Wireman & Dworkin, 1977, Nariya & Inouye, 2008). Thus, it is possible that the reduction in the cellular population during early biofilm development (either through cell death or exodus) is a prerequisite to the formation of secondary structure (see below). Indeed, in the absence of exodus, such as in a S. aureus nuc mutant, microcolony formation is not observed (Moormeier et al., 2014). Clearly, additional studies are required to provide a more complete understanding of the role exodus plays in biofilm development, as well as the mechanisms controlling how S. aureus modulates its biofilm ECM as the biofilm transitions from one stage to the next.
Maturation
A key aspect of biofilm maturation for any bacterial species is the formation of microcolony structures that provide increased surface area for nutrient exchange and waste removal, as well as to promote the dissemination of the biofilm cells to distal sites (Hall-Stoodley et al., 2004, Stewart & Franklin, 2008). Like other bacterial species, there have been numerous studies reporting on the formation of microcolony-like structures during S. aureus biofilm development (Mann et al., 2009, Moormeier et al., 2013, Moormeier et al., 2014, Thomas et al., 2014, Thurlow et al., 2011, Yarwood et al., 2004, Periasamy et al., 2012), however, the mechanism that promotes their formation is still being elucidated.
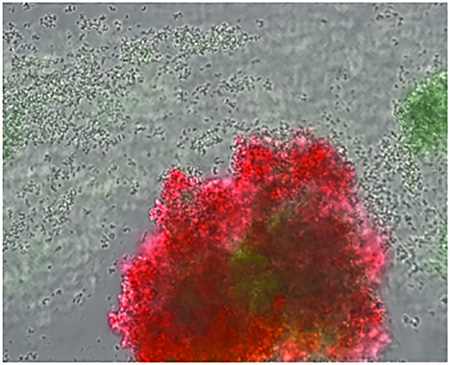
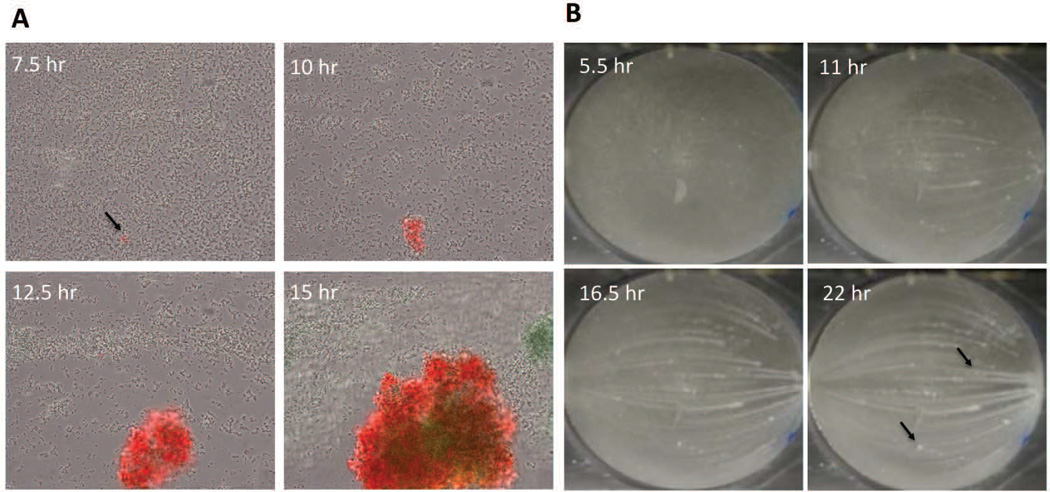
One model described previously (Periasamy et al., 2012) posits the formation of biofilm microcolony structures as a subtractive process, carving out channels from a thick mat of biofilm cells as a result of the PSM-mediated dispersal. However, observations of biofilm development using time-lapse microscopy clearly reveal the formation of microcolonies from distinct foci of cells that remain in the basal layer shortly after the commencement of exodus (Figure 3A). In a separate experiment, in which biofilm formation was monitored at a lower magnification, microcolony emergence from a basal layer of cells was also observed (Figure 3B). Thus, in contrast to the previous model, we envision an additive process where rapidly growing microcolonies emerge from a basal layer of slower growing cells (Moormeier et al., 2013). Interestingly, these studies also showed the emergence of different microcolony types that grow at different rates and with different gene expression patterns and physical properties. For example, using fluorescent reporters fused to the promoters of the cidABC and lrgAB cell death-associated operons, two different microcolony types were clearly delineated. The first was a rapid growing microcolony that exhibited constitutive lrgAB expression but delayed cidABC expression, presumably in response to the hypoxic nature of the microcolony as it increases in size (Figure 3A). The second microcolony type appeared to grow at a slower rate and expressed cidABC constitutively, with no observable lrgAB expression. In addition, the rapidly growing microcolonies were found to stain positive with propidium iodide (a DNA intercalating dye used to detect dead cells and/or eDNA), unlike the slower growing microcolonies (Moormeier et al., 2013). Interestingly, the two microcolony types also exhibited what appeared to be differences in dispersal rates (Moormeier et al., 2013), a difference that is readily observed in macroscopic imaging (Figure 3B), where dispersal can be seen as “streaking” of biofilm growth emerging from some, but not all, microcolonies formed.
Figure 3. Microcolony initiation.
A) S. aureus cells containing an lrgAB::gfp promoter fusion plasmid were inoculated into a Bioflux1000 microfluidics system and allowed to form a biofilm over a time-course experiment in which epifluorescence images were acquired at regular time points. Shown are images collected at regular intervals after the initiation of medium flow. Note the emergence of the microcolony originating from what appears to be a single (or relatively few) lrgAB-expressing (green) cells. (B) Macroscopic images of S. aureus biofilm grown in an FC flow-cell system. Shown are images collected at 5.5, 11, 16.5, and 22 hrs after the initiation of medium flow. Note the emergence of microcolonies from a basal layer of cell starting at 5.5 hrs, as well as the presence of streaking downstream of most (but not all; see arrows) of the microcolonies.
Given the differential expression of the cidABC and lrgAB operons within the two recently described microcolony types, it is conceivable that S. aureus biofilms undergo some level of metabolic diversification, where select cells within the developing biofilm are “programmed” to differentiate into different microcolony types that exhibit distinct metabolic activities. Consistent with this is the observation that microcolony formation during P. aeruginosa biofilm development is linked to pyruvate metabolism. For example, inactivation of genes involved in pyruvate utilization, as well as the depletion of pyruvate from the growth medium, were found to abrogate microcolony development (Petrova et al., 2012), suggesting that pyruvate metabolism is a distinct feature of microcolony physiology. Similarly, disruption of the S. aureus ackA and pta genes involved in the conversion of pyruvate to acetate both resulted in a dramatic shift in the types of biofilm microcolonies that were formed (manuscript in preparation). Likewise, growth of the biofilm under anaerobic conditions, which prevent respiratory activity (in the absence of a terminal electron acceptor), resulted in a similar shift in the types of microcolonies that were formed. Collectively, the results of these studies suggest the existence of a mechanism underlying the formation of structure during biofilm development that involves the metabolic differentiation of cells and the emergence of microcolonies from a basal layer that remain after the exodus stage. Importantly, this does not exclude the possibility that further modification of the biofilm structure is also mediated by the function of PSMs as envisioned by Periasamy et al. (Periasamy et al., 2012).
The observation that metabolically distinct microcolony types emerge during the maturation stage suggests another possible role for this stage – to provide diversity in preparation for the inevitable onslaught of unanticipated environmental stresses. Indeed, diversity is the name of the game in most healthy biological systems. For example, a diversified forest is better able to withstand the damaging effects of drought and disease if it is comprised of a diverse array of different tree species (Haas et al., 2011). Although the S. aureus biofilms under study in our laboratory (and likely associated with many implant-related infections) are genetically identical, they are still able to diversify through the coordinated expression of genes that control the metabolic status of the individual cells. The presence of metabolically diverse subpopulations may not only lessen (or eliminate) the time required to adapt to nutrient and oxygen stress, it may also provide an important metabolic context to resist antibacterial factors (e.g. via drug tolerance and/or promoting persister cell formation) present within the environment. Of course, if the stress becomes too great, the biofilm has a mechanism that promotes the dispersal of biofilm cells that enhances the chances that these cells encounter a more habitable environment.
Dispersal
Most S. aureus biofilm studies have focused on trying to understand the constituents that enable biofilm attachment and accumulation. However in recent years, there have been several studies examining the factors that contribute to the control of the biofilm dispersal. Dispersal of S. aureus biofilms has largely been shown to be under the control of Agr quorum sensing (Vuong et al., 2000, Yarwood et al., 2004, Periasamy et al., 2012) (Figure 1E), and like other quorum sensing systems, the Agr system is dependent on cell density and the accumulation of signal molecules called autoinducers. In S. aureus, an octapeptide pheromone called auto-inducing peptide (AIP) (Tong et al.) accumulates in the culture medium, and upon reaching a threshold concentration, binds to and activates the histidine kinase, AgrC. Once activated, AgrC phosphorylates the response regulator, AgrA, which then initiates transcription from the P3 promoter of the agr operon, producing a regulatory RNA molecule (RNAIII) that regulates expression of several virulence factors and biofilm-associated genes (Novick & Geisinger, 2008, Abdelnour et al., 1993, Dunman et al., 2001).
The first studies examining the contribution of the Agr system in S. aureus biofilm development demonstrated that agr-deficient strains formed more robust biofilms when compared to their wild-type counterparts (Vuong et al., 2000). Yet, it was not until flow-cell studies evaluating the function of Agr quorum sensing during S. aureus biofilm development that demonstrated P3 promoter expression in a subpopulation of cells located primarily within microcolonies that appeared to oscillate in waves over time coinciding with detached cells of the biofilm (Yarwood et al., 2004). Since then, this has been further corroborated by a study demonstrating increased P3 expression within microcolonies under low flow rates due to accumulation of AIP (Kim et al., 2016), as well as similar results using the BioFlux1000 system (Figure 4).
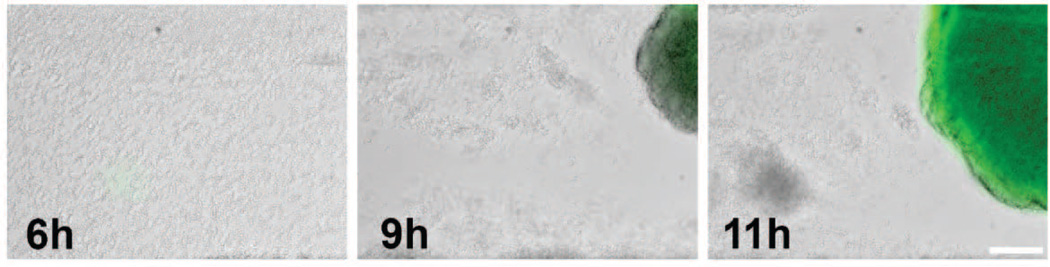
Figure 4. Agr expression in microcolonies.
S. aureus cells containing a agr-p3::gfp promoter fusion plasmid were inoculated into a Bioflux1000 microfluidics system and allowed to form a biofilm over a time-course experiment in which epifluorescence images were acquired at 0, 6, 9, and 11 hrs after the initiation of medium flow. Note the emergence of Agr expression presumably after the AIP octapeptide reaches a threshold density required for induction of P3 expression.
While the initial report provided support that Agr activity has a function in dispersal of biofilms, the Agr-regulated factors that mediate dispersal were not identified. However, two subsequent studies provided evidence of contrasting modes of Agr-mediated dispersal mechanisms. In one study, a direct correlation was demonstrated between P3 activation and dispersal of intact biofilms, which they propose was due to increased protease activity and subsequent degradation of the protein-based ECM (Boles & Horswill, 2008). While this provides a link to protease activity, Agr is not the only known regulator of the secreted proteases. Indeed, several other S. aureus transcriptional regulators, such as SarA, SigB, SaeRS, and Rot, and the newly defined msaABCR operon, have all been shown to mediate protease activity and biofilm maturation (Tsang et al., 2008, Lauderdale et al., 2009, Mootz et al., 2013, Mrak et al., 2012, Mootz et al., 2015, Sahukhal et al., 2015).
In a different study, Agr-dependent dispersal was proposed to involve the production of the phenol soluble modulin (PSM) peptides. These short amphipathic, α-helical peptides have been shown to be under the regulatory control of the Agr system through direct binding of AgrA to the psm operon promoters, and have been implicated in dispersing staphylococcal biofilms (Wang et al., 2007, Periasamy et al., 2012). Indeed, S. aureus isogenic mutants defective in the production of PSMα, PSMβ, or δ-toxin resulted in thicker biofilms (Periasamy et al., 2012). Like the Yarwood et al. study (Yarwood et al., 2004), it was also shown that the induction of Agr and the psm operon promoters correlated with waves of dispersal during late stages of biofilm development (Periasamy et al., 2012).
To mediate dispersal, PSMs are thought to function as surfactants disrupting molecular interactions within the biofilm matrix (Otto, 2013, Peschel & Otto, 2013). While the surfactant-like properties of the PSMs may indeed play a major role in biofilm dispersal, there is also contrasting evidence suggesting that the aggregation of the PSMs into nonsoluble amyloid-like fibers might abrogate biofilm dispersal and contribute to the maintenance of biofilm structure (Schwartz et al., 2012). Hence, the production of PSMs may not initiate dispersal, but rather the state in which the PSMs are assembled may contribute directly to biofilm integrity. Furthermore, it has become apparent that the presence of eDNA promotes the formation of these amyloid-like structures (Schwartz et al., 2016) suggesting a necessity for the production and interplay between ECM components to allow proper biofilm development. Clearly, more studies are needed to understand the interplay between the amyloid- and surfactant-like properties of the PSMs in biofilm stabilization and dispersal.
Conclusions
The use of new microfluidics technology to visualize S. aureus biofilm development has provided an enhanced perspective on the specific events that occur during this ill-defined process. These events appear to be more complex than previously appreciated and involve metabolic heterogeneity and differential gene expression that may be a hallmark of biofilms produced by all bacterial species. Based on this new perspective, we defined five stages of S. aureus biofilm development: 1) attachment, 2) multiplication, 3) exodus, 4) maturation, and 5) dispersal (Figure 1), and discussed the molecular mechanisms that contribute to each stage. In addition, we argue that the categorization of biofilm based on matrix types (PIA, protein, and eDNA) does not provide an accurate representation of S. aureus biofilm; rather, we envision biofilm development as a dynamic process involving the contributions of multiple matrix components. The emergence of distinct microcolony types exhibiting differential gene expression and different ECM components suggests the existence of temporal changes in the ECM composition during S. aureus biofilm development, and also the presence of spatial variations in ECM composition within the same biofilm. Given the heterogeneity of the different stages of biofilm development, it is important to incorporate real-time and time-lapse assays in conjunction with endpoint and static biofilm assays to best evaluate biofilm development. Ultimately, studies of this nature will lead to a more comprehensive understanding of the complexity of biofilm development and will enhance our ability to generate new therapeutic strategies to combat infections caused by these sophisticated multicellular communities.
Acknowledgments
We thank Ms. Jennifer Endres for assistance and support during the preparation of this manuscript. This work was supported by grants from the National Institutes of Health (PO1-AI83211 and R01-AI038901), as well as by a University of Nebraska Medical Center Graduate Student Fellowship (D.E.M.).
References
- Abdelnour A, Arvidson S, Bremell T, Ryden C, Tarkowski A. The accessory gene regulator (agr) controls Staphylococcus aureus virulence in a murine arthritis model. Infection and immunity. 1993;61:3879–3885. doi: 10.1128/iai.61.9.3879-3885.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayles KW. Bacterial programmed cell death: making sense of a paradox. Nat Rev Microbiol. 2014;12:63–69. doi: 10.1038/nrmicro3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beenken KE, Spencer H, Griffin LM, Smeltzer MS. Impact of extracellular nuclease production on the biofilm phenotype of Staphylococcus aureus under in vitro and in vivo conditions. Infection and immunity. 2012;80:1634–1638. doi: 10.1128/IAI.06134-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit MR, Conant CG, Ionescu-Zanetti C, Schwartz M, Matin A. New device for high-throughput viability screening of flow biofilms. Appl Environ Microbiol. 2010;76:4136–4142. doi: 10.1128/AEM.03065-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas R, Voggu L, Simon UK, Hentschel P, Thumm G, Gotz F. Activity of the major staphylococcal autolysin Atl. FEMS Microbiol Lett. 2006;259:260–268. doi: 10.1111/j.1574-6968.2006.00281.x. [DOI] [PubMed] [Google Scholar]
- Boles BR, Horswill AR. agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 2008;4:e1000052. doi: 10.1371/journal.ppat.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boles BR, Thoendel M, Roth AJ, Horswill AR. Identification of genes involved in polysaccharide-independent Staphylococcus aureus biofilm formation. PloS one. 2010;5:e10146. doi: 10.1371/journal.pone.0010146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose JL, Lehman MK, Fey PD, Bayles KW. Contribution of the Staphylococcus aureus Atl AM and GL murein hydrolase activities in cell division, autolysis, and biofilm formation. PloS one. 2012;7:e42244. doi: 10.1371/journal.pone.0042244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks JL, Jefferson KK. Phase variation of poly-N-acetylglucosamine expression in Staphylococcus aureus . PLoS Pathog. 2014;10:e1004292. doi: 10.1371/journal.ppat.1004292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon BP, Geoghegan JA, Waters EM, McCarthy H, Rowe SE, Davies JR, Schaeffer CR, Foster TJ, Fey PD, O’Gara JP. Role for the A domain of unprocessed accumulation-associated protein (Aap) in the attachment phase of the Staphylococcus epidermidis biofilm phenotype. J Bacteriol. 2014;196:4268–4275. doi: 10.1128/JB.01946-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan RM, Miajlovic H, Foster TJ. Surface proteins that promote adherence of Staphylococcus aureus to human desquamated nasal epithelial cells. BMC microbiology. 2009;9:22. doi: 10.1186/1471-2180-9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton JW, Cheng KJ, Geesey GG, Ladd TI, Nickel JC, Dasgupta M, Marrie TJ. Bacterial biofilms in nature and disease. Annual review of microbiology. 1987;41:435–464. doi: 10.1146/annurev.mi.41.100187.002251. [DOI] [PubMed] [Google Scholar]
- Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- Cramton SE, Gerke C, Schnell NF, Nichols WW, Gotz F. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infection and immunity. 1999;67:5427–5433. doi: 10.1128/iai.67.10.5427-5433.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramton SE, Ulrich M, Gotz F, Doring G. Anaerobic conditions induce expression of polysaccharide intercellular adhesin in Staphylococcus aureus and Staphylococcus epidermidis . Infection and immunity. 2001;69:4079–4085. doi: 10.1128/IAI.69.6.4079-4085.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucarella C, Solano C, Valle J, Amorena B, Lasa I, Penades JR. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J Bacteriol. 2001;183:2888–2896. doi: 10.1128/JB.183.9.2888-2896.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dengler V, Foulston L, DeFrancesco AS, Losick R. An Electrostatic Net Model for the Role of Extracellular DNA in Biofilm Formation by Staphylococcus aureus . Journal of bacteriology. 2015;197:3779–3787. doi: 10.1128/JB.00726-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev. 2002;15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryla A, Gelbmann D, von Gabain A, Nagy E. Identification of a novel iron regulated staphylococcal surface protein with haptoglobin-haemoglobin binding activity. Mol Microbiol. 2003;49:37–53. doi: 10.1046/j.1365-2958.2003.03542.x. [DOI] [PubMed] [Google Scholar]
- Dunman PM, Murphy E, Haney S, Palacios D, Tucker-Kellogg G, Wu S, Brown EL, Zagursky RJ, Shlaes D, Projan SJ. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. Journal of bacteriology. 2001;183:7341–7353. doi: 10.1128/JB.183.24.7341-7353.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fey PD, Endres JL, Yajjala VK, Widhelm TJ, Boissy RJ, Bose JL, Bayles KW. A Genetic Resource for Rapid and Comprehensive Phenotype Screening of Nonessential Staphylococcus aureus Genes. MBio. 2013;4 doi: 10.1128/mBio.00537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick F, Humphreys H, O’Gara JP. Evidence for icaADBC-independent biofilm development mechanism in methicillin-resistant Staphylococcus aureus clinical isolates. J Clin Microbiol. 2005;43:1973–1976. doi: 10.1128/JCM.43.4.1973-1976.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flack CE, Zurek OW, Meishery DD, Pallister KB, Malone CL, Horswill AR, Voyich JM. Differential regulation of staphylococcal virulence by the sensor kinase SaeS in response to neutrophil-derived stimuli. Proc Natl Acad Sci U S A. 2014;111:E2037–E2045. doi: 10.1073/pnas.1322125111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming HC, Wingender J. The biofilm matrix. Nature reviews. Microbiology. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- Foster TJ, Geoghegan JA, Ganesh VK, Hook M. Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus . Nat Rev Microbiol. 2014;12:49–62. doi: 10.1038/nrmicro3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulston L, Elsholz AK, DeFrancesco AS, Losick R. The extracellular matrix of Staphylococcus aureus biofilms comprises cytoplasmic proteins that associate with the cell surface in response to decreasing pH. MBio. 2014;5:e01667–e01614. doi: 10.1128/mBio.01667-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoghegan JA, Corrigan RM, Gruszka DT, Speziale P, O’Gara JP, Potts JR, Foster TJ. Role of surface protein SasG in biofilm formation by Staphylococcus aureus . J Bacteriol. 2010;192:5663–5673. doi: 10.1128/JB.00628-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman SD, Obergfell KP, Jurcisek JA, Novotny LA, Downey JS, Ayala EA, Tjokro N, Li B, Justice SS, Bakaletz LO. Biofilms can be dispersed by focusing the immune system on a common family of bacterial nucleoid-associated proteins. Mucosal Immunol. 2011;4:625–637. doi: 10.1038/mi.2011.27. [DOI] [PubMed] [Google Scholar]
- Gross M, Cramton SE, Gotz F, Peschel A. Key role of teichoic acid net charge in Staphylococcus aureus colonization of artificial surfaces. Infect Immun. 2001;69:3423–3426. doi: 10.1128/IAI.69.5.3423-3426.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas SE, Hooten MB, Rizzo DM, Meentemeyer RK. Forest species diversity reduces disease risk in a generalist plant pathogen invasion. Ecol Lett. 2011;14:1108–1116. doi: 10.1111/j.1461-0248.2011.01679.x. [DOI] [PubMed] [Google Scholar]
- Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- Houston P, Rowe SE, Pozzi C, Waters EM, O’Gara JP. Essential role for the major autolysin in the fibronectin-binding protein-mediated Staphylococcus aureus biofilm phenotype. Infect Immun. 2011;79:1153–1165. doi: 10.1128/IAI.00364-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huesca M, Peralta R, Sauder DN, Simor AE, McGavin MJ. Adhesion and virulence properties of epidemic Canadian methicillin-resistant Staphylococcus aureus strain 1: identification of novel adhesion functions associated with plasmin-sensitive surface protein. J Infect Dis. 2002;185:1285–1296. doi: 10.1086/340123. [DOI] [PubMed] [Google Scholar]
- Huseby MJ, Kruse AC, Digre J, Kohler PL, Vocke JA, Mann EE, Bayles KW, Bohach GA, Schlievert PM, Ohlendorf DH, Earhart CA. Beta toxin catalyzes formation of nucleoprotein matrix in staphylococcal biofilms. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:14407–14412. doi: 10.1073/pnas.0911032107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefsson E, McCrea KW, Eidhin DNi, O’Connell D, Cox J, Hook M, Foster TJ. Three new members of the serine-aspartate repeat protein multigene family of Staphylococcus aureus . Microbiology. 1998;144(Pt 12):3387–3395. doi: 10.1099/00221287-144-12-3387. [DOI] [PubMed] [Google Scholar]
- Kennedy CA, O’Gara JP. Contribution of culture media and chemical properties of polystyrene tissue culture plates to biofilm development by Staphylococcus aureus . J Med Microbiol. 2004;53:1171–1173. doi: 10.1099/jmm.0.45764-0. [DOI] [PubMed] [Google Scholar]
- Kiedrowski MR, Crosby HA, Hernandez FJ, Malone CL, McNamara JO, 2nd, Horswill AR. Staphylococcus aureus Nuc2 is a functional, surface-attached extracellular nuclease. PloS one. 2014;9:e95574. doi: 10.1371/journal.pone.0095574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiedrowski MR, Kavanaugh JS, Malone CL, Mootz JM, Voyich JM, Smeltzer MS, Bayles KW, Horswill AR. Nuclease modulates biofilm formation in community-associated methicillin-resistant Staphylococcus aureus . PloS one. 2011;6:e26714. doi: 10.1371/journal.pone.0026714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MK, Ingremeau F, Zhao A, Bassler BL, Stone HA. Local and global consequences of flow on bacterial quorum sensing. Nature Microbiology. 2016;1:15005. doi: 10.1038/nmicrobiol.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostakioti M, Hadjifrangiskou M, Hultgren SJ. Bacterial biofilms: development, dispersal, and therapeutic strategies in the dawn of the postantibiotic era. Cold Spring Harb Perspect Med. 2013;3:a010306. doi: 10.1101/cshperspect.a010306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauderdale KJ, Boles BR, Cheung AL, Horswill AR. Interconnections between Sigma B, agr, and proteolytic activity in Staphylococcus aureus biofilm maturation. Infection and immunity. 2009;77:1623–1635. doi: 10.1128/IAI.01036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le KY, Dastgheyb S, Ho TV, Otto M. Molecular determinants of staphylococcal biofilm dispersal and structuring. Front Cell Infect Microbiol. 2014;4:167. doi: 10.3389/fcimb.2014.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman MK, Bose JL, Sharma-Kuinkel BK, Moormeier DE, Endres JL, Sadykov MR, Biswas I, Bayles KW. Identification of the amino acids essential for LytSR-mediated signal transduction in Staphylococcus aureus and their roles in biofilm-specific gene expression. Molecular microbiology. 2015;95:723–737. doi: 10.1111/mmi.12902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Conover M, Lu H, Parsek MR, Bayles K, Wozniak DJ. Assembly and development of the Pseudomonas aeruginosa biofilm matrix. PLoS Pathog. 2009;5:e1000354. doi: 10.1371/journal.ppat.1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey-Lawrence NM, Potter DE, Cerca N, Jefferson KK. Staphylococcus aureus immunodominant surface antigen B is a cell-surface associated nucleic acid binding protein. BMC microbiology. 2009;9:61. doi: 10.1186/1471-2180-9-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann EE, Rice KC, Boles BR, Endres JL, Ranjit D, Chandramohan L, Tsang LH, Smeltzer MS, Horswill AR, Bayles KW. Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS One. 2009;4:e5822. doi: 10.1371/journal.pone.0005822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, DeDent AC, Schneewind O. Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. Microbiol Mol Biol R. 2006;70 doi: 10.1128/MMBR.70.1.192-221.2006. 192-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian SK, Liu G, Ton-That H, Schneewind O. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science. 1999;285:760–763. doi: 10.1126/science.285.5428.760. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Skaar EP, Gaspar AH, Humayun M, Gornicki P, Jelenska J, Joachmiak A, Missiakas DM, Schneewind O. Passage of heme-iron across the envelope of Staphylococcus aureus . Science. 2003;299:906–909. doi: 10.1126/science.1081147. [DOI] [PubMed] [Google Scholar]
- McConoughey SJ, Howlin R, Granger JF, Manring MM, Calhoun JH, Shirtliff M, Kathju S, Stoodley P. Biofilms in periprosthetic orthopedic infections. Future Microbiol. 2014;9:987–1007. doi: 10.2217/fmb.14.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCourt J, O’Halloran DP, McCarthy H, O’Gara JP, Geoghegan JA. Fibronectin-binding proteins are required for biofilm formation by community-associated methicillin-resistant Staphylococcus aureus strain LAC. FEMS Microbiol Lett. 2014;353:157–164. doi: 10.1111/1574-6968.12424. [DOI] [PubMed] [Google Scholar]
- McDevitt D, Francois P, Vaudaux P, Foster TJ. Molecular characterization of the clumping factor (fibrinogen receptor) of Staphylococcus aureus . Molecular microbiology. 1994;11:237–248. doi: 10.1111/j.1365-2958.1994.tb00304.x. [DOI] [PubMed] [Google Scholar]
- Merino N, Toledo-Arana A, Vergara-Irigaray M, Valle J, Solano C, Calvo E, Lopez JA, Foster TJ, Penades JR, Lasa I. Protein A-mediated multicellular behavior in Staphylococcus aureus . J Bacteriol. 2009;191:832–843. doi: 10.1128/JB.01222-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miajlovic H, Zapotoczna M, Geoghegan JA, Kerrigan SW, Speziale P, Foster TJ. Direct interaction of iron-regulated surface determinant IsdB of Staphylococcus aureus with the GPIIb/IIIa receptor on platelets. Microbiology. 2010;156:920–928. doi: 10.1099/mic.0.036673-0. [DOI] [PubMed] [Google Scholar]
- Moormeier DE, Bose JL, Horswill AR, Bayles KW. Temporal and stochastic control of Staphylococcus aureus biofilm development. MBio. 2014;5:e01341–e01314. doi: 10.1128/mBio.01341-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moormeier DE, Endres JL, Mann EE, Sadykov MR, Horswill AR, Rice KC, Fey PD, Bayles KW. Use of microfluidic technology to analyze gene expression during Staphylococcus aureus biofilm formation reveals distinct physiological niches. Appl Environ Microbiol. 2013;79:3413–3424. doi: 10.1128/AEM.00395-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootz JM, Benson MA, Heim CE, Crosby HA, Kavanaugh JS, Dunman PM, Kielian T, Torres VJ, Horswill AR. Rot is a key regulator of Staphylococcus aureus biofilm formation. Molecular microbiology. 2015;96:388–404. doi: 10.1111/mmi.12943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootz JM, Malone CL, Shaw LN, Horswill AR. Staphopains modulate Staphylococcus aureus biofilm integrity. Infection and immunity. 2013;81:3227–3238. doi: 10.1128/IAI.00377-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrak LN, Zielinska AK, Beenken KE, Mrak IN, Atwood DN, Griffin LM, Lee CY, Smeltzer MS. saeRS and sarA act synergistically to repress protease production and promote biofilm formation in Staphylococcus aureus . PloS one. 2012;7:e38453. doi: 10.1371/journal.pone.0038453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nariya H, Inouye M. MazF, an mRNA interferase, mediates programmed cell death during multicellular Myxococcus development. Cell. 2008;132:55–66. doi: 10.1016/j.cell.2007.11.044. [DOI] [PubMed] [Google Scholar]
- Navarre WW, Schneewind O. Proteolytic cleavage and cell wall anchoring at the LPXTG motif of surface proteins in gram-positive bacteria. Mol Microbiol. 1994;14:115–121. doi: 10.1111/j.1365-2958.1994.tb01271.x. [DOI] [PubMed] [Google Scholar]
- Nguyen T, Ghebrehiwet B, Peerschke EI. Staphylococcus aureus protein A recognizes platelet gC1qR/p33: a novel mechanism for staphylococcal interactions with platelets. Infect Immun. 2000;68:2061–2068. doi: 10.1128/iai.68.4.2061-2068.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Eidhin D, Perkins S, Francois P, Vaudaux P, Hook M, Foster TJ. Clumping factor B (ClfB), a new surface-located fibrinogen-binding adhesin of Staphylococcus aureus . Molecular microbiology. 1998;30:245–257. doi: 10.1046/j.1365-2958.1998.01050.x. [DOI] [PubMed] [Google Scholar]
- Novick RP, Geisinger E. Quorum sensing in staphylococci. Annu Rev Genet. 2008;42:541–564. doi: 10.1146/annurev.genet.42.110807.091640. [DOI] [PubMed] [Google Scholar]
- O’Brien L, Kerrigan SW, Kaw G, Hogan M, Penades J, Litt D, Fitzgerald DJ, Foster TJ, Cox D. Multiple mechanisms for the activation of human platelet aggregation by Staphylococcus aureus: roles for the clumping factors ClfA and ClfB, the serine-aspartate repeat protein SdrE and protein A. Mol Microbiol. 2002;44:1033–1044. doi: 10.1046/j.1365-2958.2002.02935.x. [DOI] [PubMed] [Google Scholar]
- O’Gara JP. ica and beyond: biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus. FEMS Microbiol Lett. 2007;270:179–188. doi: 10.1111/j.1574-6968.2007.00688.x. [DOI] [PubMed] [Google Scholar]
- O’Neill E, Pozzi C, Houston P, Humphreys H, Robinson DA, Loughman A, Foster TJ, O’Gara JP. A novel Staphylococcus aureus biofilm phenotype mediated by the fibronectin-binding proteins, FnBPA and FnBPB. Journal of bacteriology. 2008;190:3835–3850. doi: 10.1128/JB.00167-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill E, Pozzi C, Houston P, Smyth D, Humphreys H, Robinson DA, O’Gara JP. Association between methicillin susceptibility and biofilm regulation in Staphylococcus aureus isolates from device-related infections. J Clin Microbiol. 2007;45:1379–1388. doi: 10.1128/JCM.02280-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole G, Kaplan HB, Kolter R. Biofilm formation as microbial development. Annu Rev Microbiol. 2000;54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- Olson ME, Nygaard TK, Ackermann L, Watkins RL, Zurek OW, Pallister KB, Griffith S, Kiedrowski MR, Flack CE, Kavanaugh JS, Kreiswirth BN, Horswill AR, Voyich JM. Staphylococcus aureus nuclease is an SaeRS-dependent virulence factor. Infection and immunity. 2013;81:1316–1324. doi: 10.1128/IAI.01242-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto M. Bacterial evasion of antimicrobial peptides by biofilm formation. Curr Top Microbiol Immunol. 2006;306:251–258. doi: 10.1007/3-540-29916-5_10. [DOI] [PubMed] [Google Scholar]
- Otto M. Staphylococcal infections: mechanisms of biofilm maturation and detachment as critical determinants of pathogenicity. Annu Rev Med. 2013;64:175–188. doi: 10.1146/annurev-med-042711-140023. [DOI] [PubMed] [Google Scholar]
- Parvizi J, Pawasarat IM, Azzam KA, Joshi A, Hansen EN, Bozic KJ. Periprosthetic joint infection: the economic impact of methicillin-resistant infections. J Arthroplasty. 2010;25:103–107. doi: 10.1016/j.arth.2010.04.011. [DOI] [PubMed] [Google Scholar]
- Periasamy S, Joo HS, Duong AC, Bach TH, Tan VY, Chatterjee SS, Cheung GY, Otto M. How Staphylococcus aureus biofilms develop their characteristic structure. Proc Natl Acad Sci U S A. 2012;109:1281–1286. doi: 10.1073/pnas.1115006109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschel A, Otto M. Phenol-soluble modulins and staphylococcal infection. Nature reviews. Microbiology. 2013;11:667–673. doi: 10.1038/nrmicro3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova OE, Schurr JR, Schurr MJ, Sauer K. Microcolony formation by the opportunistic pathogen Pseudomonas aeruginosa requires pyruvate and pyruvate fermentation. Mol Microbiol. 2012;86:819–835. doi: 10.1111/mmi.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro M, Monteiro FJ, Ferraz MP. Infection of orthopedic implants with emphasis on bacterial adhesion process and techniques used in studying bacterial-material interactions. Biomatter. 2012;2:176–194. doi: 10.4161/biom.22905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche FM, Meehan M, Foster TJ. The Staphylococcus aureus surface protein SasG and its homologues promote bacterial adherence to human desquamated nasal epithelial cells. Microbiology. 2003;149:2759–2767. doi: 10.1099/mic.0.26412-0. [DOI] [PubMed] [Google Scholar]
- Rohde H, Burandt EC, Siemssen N, Frommelt L, Burdelski C, Wurster S, Scherpe S, Davies AP, Harris LG, Horstkotte MA, Knobloch JK, Ragunath C, Kaplan JB, Mack D. Polysaccharide intercellular adhesin or protein factors in biofilm accumulation of Staphylococcus epidermidis and Staphylococcus aureus isolated from prosthetic hip and knee joint infections. Biomaterials. 2007;28:1711–1720. doi: 10.1016/j.biomaterials.2006.11.046. [DOI] [PubMed] [Google Scholar]
- Sadykov MR, Bayles KW. The control of death and lysis in staphylococcal biofilms: a coordination of physiological signals. Curr Opin Microbiol. 2012;15:211–215. doi: 10.1016/j.mib.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahukhal GS, Batte JL, Elasri MO. msaABCR operon positively regulates biofilm development by repressing proteases and autolysis in Staphylococcus aureus . FEMS Microbiol Lett. 2015;362 doi: 10.1093/femsle/fnv006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer CR, Woods KM, Longo GM, Kiedrowski MR, Paharik AE, Buttner H, Christner M, Boissy RJ, Horswill AR, Rohde H, Fey PD. Accumulation-associated protein enhances Staphylococcus epidermidis biofilm formation under dynamic conditions and is required for infection in a rat catheter model. Infect Immun. 2015;83:214–226. doi: 10.1128/IAI.02177-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder K, Jularic M, Horsburgh SM, Hirschhausen N, Neumann C, Bertling A, Schulte A, Foster S, Kehrel BE, Peters G, Heilmann C. Molecular characterization of a novel Staphylococcus aureus surface protein (SasC) involved in cell aggregation and biofilm accumulation. PLoS One. 2009;4:e7567. doi: 10.1371/journal.pone.0007567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz K, Ganesan M, Payne DE, Solomon MJ, Boles BR. Extracellular DNA facilitates the formation of functional amyloids in Staphylococcus aureus biofilms. Molecular Microbiology. 2016;99:123–134. doi: 10.1111/mmi.13219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz K, Syed AK, Stephenson RE, Rickard AH, Boles BR. Functional amyloids composed of phenol soluble modulins stabilize Staphylococcus aureus biofilms. PLoS Pathog. 2012;8:e1002744. doi: 10.1371/journal.ppat.1002744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Perencevich E, Campos J, Short BL, Singh N. Clinical and economic impact of methicillin-resistant Staphylococcus aureus colonization or infection on neonates in intensive care units. Infect Control Hosp Epidemiol. 2010;31:177–182. doi: 10.1086/649797. [DOI] [PubMed] [Google Scholar]
- Speziale P, Pietrocola G, Foster TJ, Geoghegan JA. Protein-based biofilm matrices in Staphylococci. Front Cell Infect Microbiol. 2014;4:171. doi: 10.3389/fcimb.2014.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speziale P, Pietrocola G, Rindi S, Provenzano M, Provenza G, Poto ADi, Visai L, Arciola CR. Structural and functional role of Staphylococcus aureus surface components recognizing adhesive matrix molecules of the host. Future Microbiol. 2009;4:1337–1352. doi: 10.2217/fmb.09.102. [DOI] [PubMed] [Google Scholar]
- Stewart PS, Franklin MJ. Physiological heterogeneity in biofilms. Nat Rev Microbiol. 2008;6:199–210. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- Thomas VC, Sadykov MR, Chaudhari SS, Jones J, Endres JL, Widhelm TJ, Ahn JS, Jawa RS, Zimmerman MC, Bayles KW. A central role for carbon-overflow pathways in the modulation of bacterial cell death. PLoS Pathog. 2014;10:e1004205. doi: 10.1371/journal.ppat.1004205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurlow LR, Hanke ML, Fritz T, Angle A, Aldrich A, Williams SH, Engebretsen IL, Bayles KW, Horswill AR, Kielian T. Staphylococcus aureus biofilms prevent macrophage phagocytosis and attenuate inflammation in vivo. J Immunol. 2011;186:6585–6596. doi: 10.4049/jimmunol.1002794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Arana A, Merino N, Vergara-Irigaray M, Debarbouille M, Penades JR, Lasa I. Staphylococcus aureus develops an alternative, ica-independent biofilm in the absence of the arlRS two-component system. Journal of bacteriology. 2005;187:5318–5329. doi: 10.1128/JB.187.15.5318-5329.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong SY, Holden MT, Nickerson EK, Cooper BS, Koser CU, Cori A, Jombart T, Cauchemez S, Fraser C, Wuthiekanun V, Thaipadungpanit J, Hongsuwan M, Day NP, Limmathurotsakul D, Parkhill J, Peacock SJ. Genome sequencing defines phylogeny and spread of methicillin-resistant Staphylococcus aureus in a high transmission setting. Genome research. 2015;25:111–118. doi: 10.1101/gr.174730.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang LH, Cassat JE, Shaw LN, Beenken KE, Smeltzer MS. Factors contributing to the biofilm-deficient phenotype of Staphylococcus aureus sarA mutants. PloS one. 2008;3:e3361. doi: 10.1371/journal.pone.0003361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhommerig E, Moons P, Pirici D, Lammens C, Hernalsteens JP, Greve HDe, Kumar-Singh S, Goossens H, Malhotra-Kumar S. Comparison of biofilm formation between major clonal lineages of methicillin resistant Staphylococcus aureus . PloS one. 2014;9:e104561. doi: 10.1371/journal.pone.0104561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez V, Liang X, Horndahl JK, Ganesh VK, Smeds E, Foster TJ, Hook M. Fibrinogen is a ligand for the Staphylococcus aureus microbial surface components recognizing adhesive matrix molecules (MSCRAMM) bone sialoprotein-binding protein (Bbp) The Journal of biological chemistry. 2011;286:29797–29805. doi: 10.1074/jbc.M110.214981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong C, Saenz HL, Gotz F, Otto M. Impact of the agr quorum-sensing system on adherence to polystyrene in Staphylococcus aureus . J Infect Dis. 2000;182:1688–1693. doi: 10.1086/317606. [DOI] [PubMed] [Google Scholar]
- Wang R, Braughton KR, Kretschmer D, Bach TH, Queck SY, Li M, Kennedy AD, Dorward DW, Klebanoff SJ, Peschel A, DeLeo FR, Otto M. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat Med. 2007;13:1510–1514. doi: 10.1038/nm1656. [DOI] [PubMed] [Google Scholar]
- Wireman JW, Dworkin M. Developmentally induced autolysis during fruiting body formation by Myxococcus xanthus . J Bacteriol. 1977;129:798–802. doi: 10.1128/jb.129.2.798-802.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarwood JM, Bartels DJ, Volper EM, Greenberg EP. Quorum sensing in Staphylococcus aureus biofilms. J Bacteriol. 2004;186:1838–1850. doi: 10.1128/JB.186.6.1838-1850.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong Y, Xu Y, Liang X, Keene DR, Hook A, Gurusiddappa S, Hook M, Narayana SV. A ‘Collagen Hug’ model for Staphylococcus aureus CNA binding to collagen. EMBO J. 2005;24:4224–4236. doi: 10.1038/sj.emboj.7600888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapotoczna M, McCarthy H, Rudkin JK, O’Gara JP, O’Neill E. An essential role for coagulase in Staphylococcus aureus biofilm development reveals new therapeutic possibilities for device-related infections. J Infect Dis. 2015;212:1883–1893. doi: 10.1093/infdis/jiv319. [DOI] [PubMed] [Google Scholar]