Abstract
Problem
Preservation of biospecimen quality is critical to accurately and reliably assessing genes and proteins. We evaluated the effect of preparation method and storage duration on RNA quality in placenta and decidua.
Method of Study
Aliquots of nine placentas and decidua were placed in RNAlater® (RL) or flash frozen (FF) within 30 minutes of delivery. RNA was extracted immediately (baseline), and from matched samples stored at −80°C for 1 and 8–10 months. RNA Integrity Number (RIN) and housekeeping gene expression were quantified.
Results
At both time points, RL placenta had RIN and housekeeping gene Ct values similar to baseline. However, FF placenta had significantly lower RIN and higher Ct values at 1 and 8–10 months. In RL and FF decidua, RIN was unchanged from baseline.
Conclusion
We found RNAlater more effectively and consistently preserved placenta, compared to flash freezing. However, for decidua, which is less dense than placenta, both modes yielded comparable RNA integrity over time.
Keywords: Decidua, housekeeping gene, RNA Integrity Number
Introduction
Adverse pregnancy outcomes, such as preeclampsia, miscarriage, and preterm birth, may result from abnormal placental development or function. Studies of such outcomes often include placenta tissue, collected over time from large cohorts, to evaluate changes and patterns in gene expression, metabolomics, and protein signaling. These investigations rely on high quality placenta samples, with appropriate preparation and storage, as sample degradation may impair analysis or lead to erroneous conclusions.
For studies involving genomic analysis, including microarrays and real-time quantitative RT-PCR (qRT-PCR), intact RNA is critical for accurately quantifying gene expression. The process of RNA degradation is not fully understood, but it is known to occur spontaneously in aqueous solution, and also results from ribonuclease (RNase) activity. The RIN Integrity Number (RIN) provides a standardized tool for assessing RNA, and accounts for several features of the total RNA electropherogram.(1) These features include the total RNA ratio (area of 18S and 28S peaks versus total area under the curve (AUC)), height of the 28S peak, and fast area ratio (area between 5S and 18S peaks versus total AUC). RIN is reported on a scale from 1 to 10, where 10 represents the most intact RNA. An RIN greater than 6 is considered acceptable for qRT-PCR, while an RIN over 8 is best for microarray analysis.
Traditionally, tissue samples have been preserved by flash freezing, or snap freezing, in liquid nitrogen, followed by storage at −80°C. However, stabilizing agents such as RNAlater® (Ambion, Inc., Austin, TX) provide an alternative method. These reagents are based in aqueous sulfate salt solutions at a controlled pH.(2) When tissue is placed in RNAlater, the reagent permeates cells, causing precipitation of RNases and other cellular proteins, and thereby decreasing RNA degradation. RNAlater allows for handling and short-term storage of samples at room temperature, and requires no additional equipment. However, RNAlater reagent costs can be high.
Several studies demonstrated that RNAlater yields higher RNA integrity in long-term storage of cancer tissue and brain tissue specimens, compared to flash freezing.(3–5) However, other tissues such as liver and those of the gastrointestinal tract were shown to have similar RNA quality with RNAlater or flash freezing.(6, 7) While recommendations exist for the use of RNAlater with placenta(8), there have been no systematic comparisons of RNAlater and flash freezing for long-term storage of placental tissue. In a study of 4 placentas, Wolfe et al found that RNAlater-treated samples had higher RIN values than flash frozen tissues, but did not assess the effect of storage duration on RNA integrity.(1, 9)
In this study, we systematically evaluated the efficacy of RNAlater and flash freezing in ensuring RNA quality over time, in matched placenta and decidua biospecimens, with RNA extracted from fresh tissue as our baseline.
Material and Methods
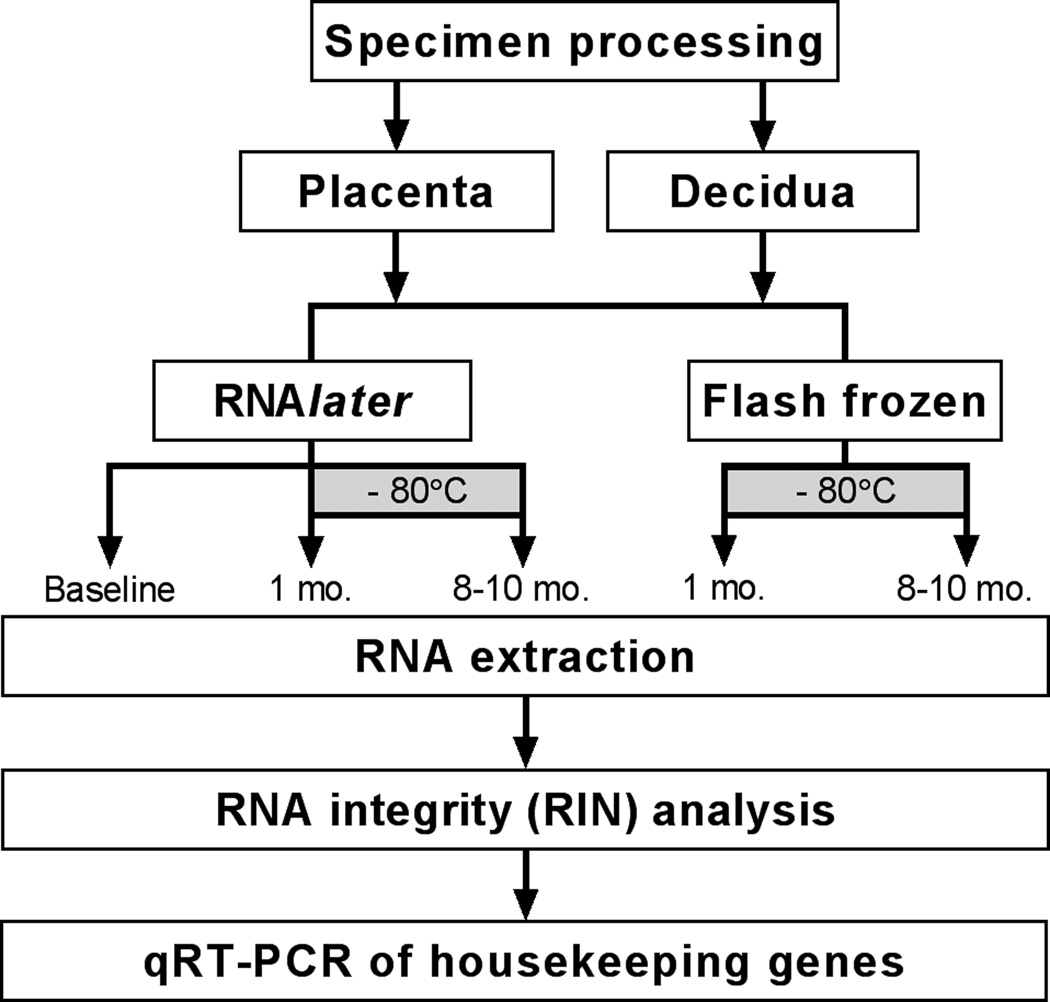
The study design is summarized in Figure 1, showing the handling and storage of placenta and decidua specimens.
Figure 1.
Overview of sample processing, storage, and analysis. Placenta and decidua specimens were obtained from 9 women by 8 mm punch biopsy. For each specimen, 3 aliquots were placed in RNAlater, and 2 aliquots were flash frozen by immersion in liquid nitrogen. RNA extraction from matched sample aliquots was performed immediately (baseline), after 1 month of storage, and after 8–10 months of storage. All stored samples were kept at −80°C. RNA was subsequently used for analysis of RIN values and housekeeping gene expression.
Biospecimen Collection
Placenta and decidua tissues were collected at the time of elective Caesarean section from nine women attending Yale-New Haven Hospital (YNHH). All patients provided written informed consent. The study was approved by the Yale University Human Investigations Committee. The time at which the whole placenta was removed from the patient was recorded. The placentas were immediately transported at room temperature to the laboratory, sectioned, and processed within 30 minutes of delivery. Placenta specimens were obtained by 8 mm punch biopsy of the maternal surface. Each placenta was inspected prior to sampling, to identify and exclude areas containing calcification or infarction. The location of placenta tissue sampling was otherwise randomized. The decidua was separated from the remainder of the fetal membranes, and an 8 mm punch biopsy was obtained from each decidua. All placenta and decidua samples were similarly rinsed with PBS before flash freezing or placement into RNAlater RNA Stabilization Reagent (Qiagen, Hilden, Germany) in order to remove maternal blood from the surface of the tissue, and minimize the impact of maternal leukocytes on analysis of placental gene expression.(8) For each placenta and decidua, a minimum of five aliquots were taken to enable tissue evaluation at three time points -- baseline, 1 month, and 8–10 months. Individual aliquots were preserved using one of two methods: 1) specimens were rinsed in PBS and placed in 1 mL of RNAlater, or 2) specimens were flash frozen in liquid nitrogen for a minimum of 15 minutes. All stabilized samples were stored at −20°C (for RNAlater samples) or in liquid nitrogen (for the flash frozen samples), until transfer to a −80°C freezer.
RNA Isolation and Quality Assessment
Placenta and decidua total tissue were homogenized in TRIzol® (Life Technologies, Carlsbad, CA). RNA was isolated with chloroform, precipitated with isopropanol, washed twice with 75% ethanol, and dissolved in RNase-free water. Samples were treated with RNase-free DNase, and RNA cleanup was performed with RNeasy® MinElute® Cleanup Kit (Qiagen). RNA concentration and ratios of absorbance at 260 nm and 280 nm were determined via NanoDrop™ 2000 Spectrophotometer (Thermo Scientific, Wilmington, DE). RNA quality was assessed by the W.M. Keck Foundation Biotechnology Resource Laboratory (Yale University, New Haven, CT), using Agilent Technologies 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). RNA Integrity Number (RIN) was assigned based on the electropherogram using the manufacturer software (Agilent Technologies, Santa Clara, CA).
Quantitative Real-Time Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
qRT-PCR was performed using 12.5 ng of cDNA template, reverse transcribed with iScript™ cDNA Synthesis Kit (Bio-Rad, Hercules, CA). The PCR assay was performed in triplicate using assay-specific primer concentrations and iQ™ SYBR® Green Supermix (Bio-Rad), and amplified using CFX Connect™ Real-Time PCR Detection System (Bio-Rad) under the following cycling conditions: 95°C for 3 minutes; 40 three-step cycles of 95°C for 15 seconds, 60°C for 30 seconds, and 72°C for 25 seconds; 95°C for 1 minute; 55°C for one minute; and an increase to 95°C by 0.5°C increments. Three genes – β-actin, tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta (YWHAZ), and DNA topoisomerase 1 (TOP1) – were selected after a literature review on housekeeping genes previously validated in placenta tissue.(10, 11) Primer sequences were designed using NCBI Primer-BLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast, NCBI, Bethesda, MD), and synthesized at the W.M. Keck Foundation Oligo Synthesis Resource (Yale University). Primer sequences are β-actin F-AGC CAT GTA CGT TGC TAT CCA, R-ACC GGA GTC CAT CAC GAT G; YWHAZ F-GGA ACC CTG GGG ACT ACG A, R-ATC CAT GAC TGG ATG TTC TTT T; and TOP1 F-AAG TTT GCC TGG GCC ATT GA, R-ACG CTG ACA AAT TCC CAT CCA. Optimal primer concentrations were determined by efficiency assays using various combinations of forward and reverse primer concentrations and two-fold serial dilutions of cDNA synthesized from human universal RNA (Qiagen Sciences, Germantown, MD). Primer efficiencies were 0.980 for β-actin at concentrations of F-0.5 µM, R-0.25 µM; 1.012 for YWHAZ at F-0.5 µM, R-0.25 µM; and 1.058 for TOP1 at F-0.5 µM, R-0.5 µM.
Statistical Analysis
RIN values are represented graphically as individual data points, with median and interquartile range (IQR). Ct values for housekeeping genes are represented graphically with median, first and third quartiles, and range. Data was matched within each tissue. Statistical significance was determined by Friedman’s one-way analysis of variance (ANOVA) test, using GraphPad Prism (GraphPad Software, Inc., La Jolla, CA). Significance was defined as p < 0.05, after a multiple-comparisons correction with Dunn’s test.
Results
RNA purity
Purity was assessed by 260/280 ratio for each sample. For the placenta samples, 98% had 260/280 ratios of 2.0 or greater, and 100% had ratios over 1.8. For decidua samples, 89% had 260/280 ratios of 2.0 or greater, and 89% had ratios over 1.8, and the lowest value was 1.45. This indicates our extraction and purification techniques yielded very pure RNA samples.
RNA integrity over time
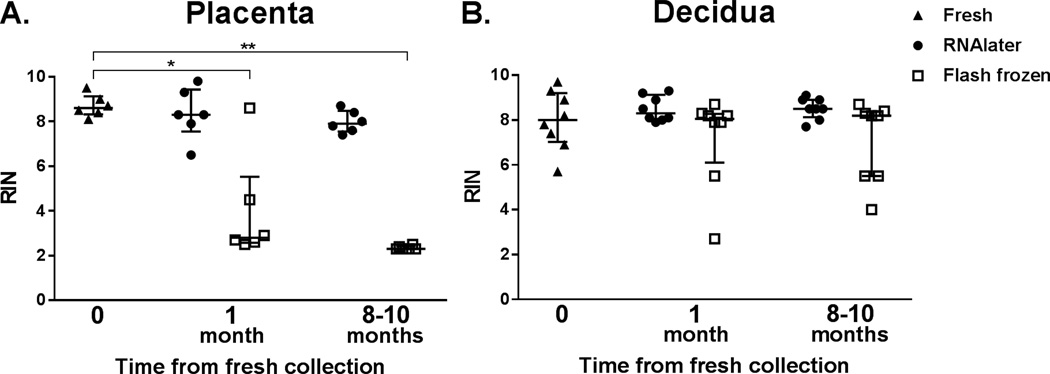
To assess RNA integrity in stored placenta and decidua aliquots, we compared RIN values for baseline samples to those flash frozen or stored in RNAlater for 1 and 8–10 months (Figure 2). RNA from 9 sets of matched samples was assessed with a bioanalyzer. However, RIN values were unable to be assigned by the software algorithm for 4 placenta time point samples and 1 decidua time point sample. Therefore, analysis of RIN values includes data from 6 placenta and 8 decidua specimens.
Figure 2.
RNA integrity over time. RNA Integrity Number (RIN) was determined for RNA extracted from (A) placenta (n = 6) and (B) decidua (n = 8) samples. For both placenta and decidua, RIN values from matched samples either stored in RNAlater or flash frozen for 1 month and 8–10 months were compared to baseline. Significance is denoted by * (p < 0.05) and ** (p < 0.001).
For baseline RNA, which was extracted immediately after delivery, RIN values were within ideal range for both placenta and decidua.(1) RIN values for baseline placenta RNA ranged from 8.1 to 9.5 (median 8.6). RIN values for placenta samples stored in RNAlater for 1 month (range 6.5 to 9.8; median 8.3) and 8–10 months (range 7.4 to 8.7; median 7.9) were similar to baseline (p=NS). However, after 1 month of storage, RNA from flash frozen placenta samples had RIN values ranging from 2.5 to 8.6, with a lower median (2.8) than baseline (p < 0.05). After 8–10 months of storage, the RIN values for flash frozen placenta decreased further (range 2.3 to 2.5, median 2.3), a significant change compared to baseline (p < 0.001).
RIN values for baseline decidua RNA ranged from 5.7 to 9.7 (median 8.0). RNA from decidua samples stored in RNAlater for 1 month (range 7.9 to 9.3, median 8.3) and 8–10 months (range 7.7 to 9.1, median 8.5) were similar to baseline (p=NS). In contrast to flash frozen placenta samples, the majority of RNA from flash frozen decidua samples had RIN values similar to baseline at both 1 month (range 2.7 to 8.7, median 8.1; p=NS) and 8–10 months (range 4.0 to 8.7, median 8.2; p=NS).
Housekeeping gene expression
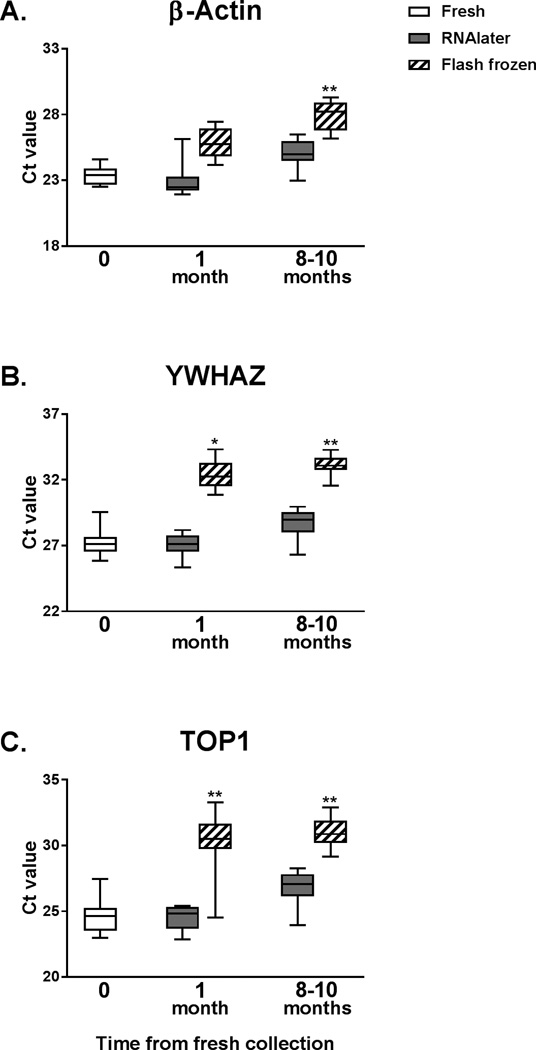
Because we found significant differences in RIN values for matched placenta samples based on storage method, we investigated whether these disparities in RNA integrity were reflected in gene expression. Three housekeeping genes, β-actin, YWHAZ, and TOP1, were quantified by qRT-PCR (Figure 3). For each of the 9 sets of placenta samples, the Ct values of RNAlater and flash frozen samples stored for 1 and 8–10 months were compared to baseline. For all three housekeeping genes, the Ct values for RNAlater samples showed no significant change from baseline at 1 month (p=NS) and 8–10 months (p=NS). For flash frozen samples, Ct values were significantly higher at 1 month for YWHAZ (p = 0.002) and TOP1 (p < 0.001), and at 8–10 months for all three genes (p < 0.001) relative to baseline.
Figure 3.
Housekeeping gene expression by qRT-PCR. Gene expression in placenta (n = 9) was quantified for (A) β-actin, (B) YWHAZ, and (C) TOP1. Ct values for matched samples stored in RNAlater or flash frozen for 1 month and 8–10 months were compared to baseline. Significance is denoted by * (p < 0.01) and ** (p < 0.001).
The qRT-PCR assay is sufficiently robust such that normalization of a target gene with a housekeeping gene many times preserves the ratio of gene expression, as previously found by other investigators.(12) The assumption is that the RNA for every gene will be similarly degraded in a sample with partially degraded RNA, as determined by the RIN value. In our dataset, when we normalized YWHAZ to β-actin, we found that the ΔCt and 2−ΔCt values were different between matched placenta samples processed by flash freezing or RNAlater. Lower RIN values were associated with higher ΔCt values (r=0.62) and lower 2−ΔCt values (r=0.53). In addition, the 2−ΔCt values for flash frozen placenta were significantly lower at both 1 or 8–10 month time points, relative to baseline (p<0.01, p < 0.05, respectively) (Supplemental Figure 1).
Based on these findings, we then examined β-actin, YWHAZ, and TOP1 expression in flash frozen decidua to examine whether the flash frozen decidua samples stored for 8–10 months, which have similar RIN values to baseline, also have similar Ct values to baseline. The average Ct values for β-actin, YWHAZ, and TOP1 in flash frozen samples were similar to baseline decidua (p=NS, respectively) (Supplemental Figure 2).
Discussion
In this systematic and controlled study, we found that placenta tissue stored in RNAlater yields higher RNA integrity over time, compared to flash frozen samples. Differences in placenta RNA integrity also translated to differences in gene expression, demonstrating the importance of quality preservation. For decidua, both RNAlater and flash freezing provide similar long-term preservation of RNA integrity. Decidua is a thin tissue with lower density than placenta, and thus, density may account for differences in preservation quality by flash-freezing. Surface area may also cause the differences in preservation quality. Since the decidua is flat and thin, all of its surface area is exposed and the RNAlater can more easily permeate the aliquot. Sample size may also alter preservation quality by flash freezing, although we did not examine size as a variable. The 8 mm punch biopsy tool used in this study produced aliquot volumes in the range of 0.25 cm3 to 0.6 cm3, which were well preserved in RNAlater. Wolfe et al also found that placenta sample volumes between ~0.1 and 1 cm3 were similar in RIN values when treated with RNAlater.(9)
RNAlater is convenient because it is effective at room temperature, and does not require special equipment. Of note, the manufacturer recommends incubating tissue samples at 2–8°C for 24 hours, followed by either storage in the reagent at −20°C or reagent removal before archival storage at −80°C(13). Our samples remained in RNAlater for the full duration of storage at −80°C, thus eliminating an additional step in processing. Our findings show appropriate quality is maintained. In our hands, we found the cost of RNAlater to be 25 times greater than flash freezing. RNAlater provides a convenient and highly effective, though costly, alternative to flash freezing placenta in liquid nitrogen or dry ice.
Supplementary Material
Acknowledgments
We would like to acknowledge the Yale University Reproductive Sciences (YURS) Biobank for their help in collecting tissue. This work was supported by YURS Biobank and a National Institutes of Health grant from the Eunice Kennedy Shriver National Institute of Child Health and Human Development K08HD071010 (to C.A.F.).
References
- 1.Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, et al. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Molecular Biology. 2006;7 doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mutter GL, Zahrieh D, Liu C, Neuberg D, Finkelstein D, Baker HE, et al. Comparison of frozen and RNALater solid tissue storage methods for use in RNA expression microarrays. BMC Genomics. 2004;5:88. doi: 10.1186/1471-2164-5-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hatzis C, Sun H, Yao H, Hubbard RE, Meric-Bernstam F, Babiera GV, et al. Effects of Tissue Handling on RNA Integrity and Microarray Measurements From Resected Breast Cancers. JNCI Journal of the National Cancer Institute. 2011;103(24) doi: 10.1093/jnci/djr438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sherwood KR, Head MW, Walker R, Smith C, Ironside JW, Fazakerley JK. RNA integrity in post mortem human variant Creutzfeldt–Jakob disease (vCJD) and control brain tissue. Neuropathology and Applied Neurobiology. 2011;37(6) doi: 10.1111/j.1365-2990.2011.01162.x. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y, Zheng H, Chen J, Zhong X, Wang Y, Wang Z, et al. The Impact of Different Preservation Conditions and Freezing-Thawing Cycles on Quality of RNA, DNA, and Proteins in Cancer Tissue. Biopreservation and biobanking. 2015;13(5) doi: 10.1089/bio.2015.0029. [DOI] [PubMed] [Google Scholar]
- 6.Lee SM, Schelcher C, Thasler R, Schiergens TS, Thasler WE. Pre-Analytical Determination of the Effect of Extended Warm or Cold Ischaemia on RNA Stability in the Human Ileum Mucosa. PLoS One. 2015;10(9):e0138214. doi: 10.1371/journal.pone.0138214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee SML, Schelcher C, Gashi S, Schreiber S, Thasler RMK, Jauch K-W, et al. RNA Stability in Human Liver: Comparison of Different Processing Times, Temperatures and Methods. Molecular Biotechnology. 2012;53(1):1–8. doi: 10.1007/s12033-011-9493-4. [DOI] [PubMed] [Google Scholar]
- 8.Burton GJ, Sebire NJ, Myatt L, Tannetta D, Wang YL, Sadovsky Y, et al. Optimising sample collection for placental research. Placenta. 2014;35(1) doi: 10.1016/j.placenta.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 9.Wolfe LM, Thiagarajan RD, Boscolo F, Taché V, Coleman RL, Kim J, et al. Banking placental tissue: An optimized collection procedure for genome-wide analysis of nucleic acids. Placenta. 2014;35(8) doi: 10.1016/j.placenta.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cleal JK, Day P, Hanson MA, Lewis RM. Measurement of Housekeeping Genes in Human Placenta. Placenta. 2009;30(11) doi: 10.1016/j.placenta.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 11.Lanoix D, Lacasse A-A, St-Pierre J, Taylor SC, Ethier-Chiasson M, Lafond J, et al. Quantitative PCR Pitfalls: The Case of the Human Placenta. Molecular Biotechnology. 2012;52(3) doi: 10.1007/s12033-012-9539-2. [DOI] [PubMed] [Google Scholar]
- 12.Fleige S, Pfaffl MW. RNA integrity and the effect on the real-time qRT-PCR performance. Molecular aspects of medicine. 2006;27(2–3):126–139. doi: 10.1016/j.mam.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 13.Qiagen RNAlater® Handbook [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.