Abstract
Hepatitis C infection is a common risk factor for the development of liver cancer. The molecular mechanisms underlying this effect are only partially understood. Here we show that the HCV protein NS5B directly binds to the tumor suppressor NORE1A (RASSF5) and promotes its proteosomal degradation. In addition, we show that NORE1A co-localizes to sites of HCV viral replication and suppresses the process. Thus, NORE1A has anti-viral activity which is specifically antagonized by NS5B. Moreover, the suppression of NORE1A protein levels correlated almost perfectly with elevation of Ras activity in primary human samples. Therefore, NORE1A inactivation by NS5B may be essential for maximal HCV replication and may make a major contribution to HCV induced liver cancer by shifting Ras signaling away from pro-senescent/apoptotic signaling pathways.
Conclusion
HCV uses NS5B to specifically suppress NORE1A facilitating viral replication and elevated Ras signaling.
Keywords: Hepatitis C virus (HCV), hepatoma cellular carcinoma (HCC), NORE1A, Ras, RNA dependent RNA polymerase (RdRp), Cancer
Despite recent advances in treatment, the blood-born pathogen hepatitis C virus (HCV) remains a serious public health concern with over 170 million cases worldwide (1, 2). Chronic HCV infection increases the risk of developing liver cirrhosis and hepatocellular carcinoma (HCC) (3), the third leading cause of cancer related deaths (4). In western countries, the increasing numbers of HCV infections is primarily responsible for the increased incidence of HCC (5). HCV is a member of the Flaviviridae family. Similar to other flaviviruses, replication of the plus-stranded RNA genome occurs exclusively in the cytoplasm of host cells. The 9.6 kb RNA is translated as a single polyprotein of approximately 3000 amino acids (6). The viral polyprotein is subsequently cleaved by a combination of host and viral proteases into four structural (core, E1, E2, and p7) and six nonstructural (NS2, NS3, NS4A, NS4B, NS5A, and NS5B) proteins (6). The mechanism whereby the cytoplasmically replicating HCV induces deregulation of the hepatocyte cell cycle leading to carcinogenesis is not well understood. While HCV infection induces inflammation, an event thought to contribute to the development of HCC, there are emerging lines of evidence that HCV proteins, either alone or in combination, contribute to carcinogenesis. For example, transgenic mice that express the HCV polyprotein develop HCC at higher incidence (7). Interestingly, these mice do not develop hepatic inflammation, which supports the involvement of HCV proteins in the development of HCC (7). Additionally, expression of the HCV core, NS3 and NS5A alter cellular proliferation and may contribute to hepatocarcinogenesis (8–13). Recently, the viral RNA-dependent RNA polymerase (RdRp) NS5B has also been implicated as an oncoprotein. Indeed, NS5B binds and induces the degradation of the tumor suppressor retinoblastoma (Rb) protein via recruitment of the E3 ubiquitin ligase, E6-associated protein (E6AP) (14).
The goal of this study was to identify and investigate the role of additional host cell proteins that modulate HCV replication and pathogenesis. Using a BacterioMatch two hybrid approach, we identified NORE1A, a Ras effector/tumor suppressor that mediates the pro-senescence and pro-apoptotic effects of the Ras oncoprotein as a potential binding partner of NS5B. Here we confirm the interaction is physiological and demonstrate that NORE1A can suppress HCV viral replication. In addition, we show that NS5B acts to promote the proteosomal degradation of NORE1A. These observations are likely to be physiologically relevant as we also show that loss of expression of NORE1A protein in primary HCC correlates well with HCV infection. Thus, we present the first evidence for anti-viral effects of RASSF family tumor suppressors and identify a novel mechanism by which NS5B can promote cancer during HCV infection by suppressing NORE1A.
MATERIALS AND METHODS
Plasmids
GFP-NORE1A and GFP-NORE1B plasmids have been described previously (15). GFP-BRaf was obtained from Addgene. HA-Ubiquitin plasmid was a kind gift from Dr. Chee Gun-Lee. pNTAP vector was purchased from Stratagene. The plasmids pNTAP-NS5B and pFLAG-CMV2-NS5B were generated by amplifying the NS5B coding sequence (1–591amino acids). The plasmid pNTAP-NS5BCΔ21 containing full-length NS5B truncated by 21 amino acid at the C terminus, was also generated by PCR.
Cell Culture
Huh-7 (human hepatocellular carcinoma) and Hek293 (human embryonic kidney) cells were cultured in Dulbecco Modified Eagles Medium (DMEM) (Sigma) supplemented with 10% Fetal Bovine Serum, L-Glutamine (500µM) (Invitrogen), Penicillin/Streptomycin (liquid, 1000x) (Invitrogen). NS5B stable 293 (termed as 2.3) cells were generated by transfection of the plasmid encoding Flag-NS5BCΔ21. HCV subgenomic replicon cells (MH-14) (kind gift of Dr. Kunitada Shimotohno) and full length HCV genomic cell line (C-5B) (obtained from Dr. Stanley Lemon) were cultured in above medium supplemented with 500µg/µl G418 (Invitrogen). Plasmid transfection was performed by LipoD293 reagent (Signa Gen Laboratories) in accordance with the manufacturer’s protocol.
Antibodies
Anti-NS5B (Rabbit Polyclonal) antibody was generated at Covance against NS5BCΔ21 protein as antigen. NORE1A (Rabbit Polyclonal) has been described previously (16). Antibodies against NORE1A (Mouse Monoclonal), HA (Mouse Monoclonal), GFP (Mouse Monoclonal), Actin (Rabbit Polyclonal) and GAPDH (Mouse Monoclonal) were purchased from Santa Cruz. Anti-BrU (Rabbit polyclonal) was purchased from Thermofisher. Anti-Ubiquitin. Rabbit polyclonal was a kind gift from Dr. Melissa Rogers. HRP-tagged anti-Rabbit, anti-Mouse antibodies and FITC-tagged anti-Rabbit, Cy3-tagged anti-Mouse antibodies were purchased from Jackson Immunoresearch.
Cell Transfection, Immunoprecipitation and Immunoblotting
For immunoprecipitation analysis, or protein pull down (GST/GST-NS5B) studies, cells (transfected/control) in 10 cm dish were washed twice with chilled PBS (pH 7.4) and lysed with 400µl of lysis buffer A (50mM Tris-Cl [pH 7.5], 120mM NaCl, 1% Triton X-100) supplemented with 1X Protease inhibitor cocktail (Roche Applied Science) followed by centrifugation at 13,000 rpm for 10 minute. Cleared lysates were incubated either with GST proteins or immunoprecipitated with antibodies overnight at 4°C, and precipitates were collected by incubating with glutathione beads (Sigma) or protein G/A beads (Sigma) respectively for 1 hour at 4°C. pNTAP-tagged NS5BCΔ21 proteins with SBP motif were affinity purified by binding with streptavidin beads for 2 hours at 4°C. Beads were washed three times with 0.1% lysis buffer A and precipitates were then boiled 5 minutes in laemmli sample buffer and stored at −20°C. Sample were resolved on Polyacrylamide gel and transferred on Nitrocellulose membranes (Millipore) for Western blotting. Ras activivty in primary samples was determined using a Ras G-LISA® kit from Cytoskeleton Inc. (Denver CO).
shRNA Knockdown and Colony Formation Assay
MH14 cells were seeded in 10 cm dishes at 30–40% confluence and 16 hours post seeding, transfected with 500ng NORE1 shRNA or shRNA pRS vector in triplicate using lipofectamine reagent. Two 10cm dish were mock transected. Plates were incubated for 24 hours at 37°C. 24 hours post transfection medium was replaced with complete medium containing G418 (300µg/ml) and puromycin (1µg/ml) in transfected plates. Untransfected plates were treated with either G418 alone and or with Puromycin +G418.
cDNA Preparation and Real Time PCR
Total RNA was prepared from cells using RNAeasy mini kit (Qiagen), and quantified by nanodrop. Quality of RNA was verified by running on agarose gel. Equal amounts of RNA from each sample was subjected to first strand cDNA synthesis by using a first strand cDNA synthesis kit (Applied Biosystem) and gene specific reverse primer for 5’NTR cDNA or random primers for NORE1A and NORE1B and Actin cDNA preparation. Real Time PCR mixture was prepared by combining 1/10 of cDNA prepared, 2X SYBR Green Mix (Applied Biosystems), forward and reverse primers as per manufacturer’s instructions. The fluorescent signals were analyzed by ABI 7500 software (Applied Biosystems). Primer pairs used were as follows: Forward 5’-CGG GAG AGC CAT AGT GG-3’ and Reverse; 5’- AGT ACC ACA AGG CCT TTC G-3’ for 5’NTR amplification and Forward; 5’-CGT GGG GCG CCC CAG GCA CCA-3’ and Reverse; 5’-TTG GCC TTC GGG TTC AGG GG- 3’ for Actin, Forward “CCG CGC TAT CTA CAG AGC” and Reverse “GGG GGC TCC AGG TTC” for NORE1A, Forward “GGC GGG CGC TGT GTC and Reverse “GGC GGG CGC TGT GTC TCC” for NORE1B amplification. For human specimens, NORE1A expression was assessed by using a primer pair specific for exon 2 (5’-ATCCAGTTGGACTGCAGTCA-3’) and exon 4 (5’-GGGATTGTCCACAACCATG-3’) to give an expected product size of 450 bp, as previously reported (Hesson et al., 2003). In all RT–PCR reactions the expression of β-Actin was used as a positive control. All experiments were performed in triplicate and analyzed using the ABI 7500 software (Applied Biosystems).
Dual Immunoflourescence Analysis
NS5B stable and replicon cells were seeded on a cover glass, fixed (4% formaldehyde), blocked and permeabilized incubating with a mixture of anti-NS5B (Mouse Monoclonal) and anti-NORE1A (Rabbit polyclonal) for one hour. Cells were washed with chilled 1X PBS and then subjected to antibody incubation with FITC-tagged Goat anti-Rabbit and Cy5-tagged Donkey anti-Mouse secondary antibodies respectively. A nucleus staining was performed with 1µg/ml DAPI at room temperature.
Proteinase K (PK) Digestion and 5’ BrU Labeling of Nascent HCV RNA
Localization of endogenous NORE1A and newly synthesized HCV RNA was performed as reported earlier for other host proteins in replicon cell based system (17).
Proteasome inhibitors & Cellular Ubiquitination Assays
Cells were seeded into 6 well plates, grown to 50% confluence and transfection was carried out using GFP-NORE1A plasmid. 24 hours post transfection cells were either treated with DMSO or 10µM MG132 or 1µM ALLN (Calbiochem, CA) and further incubated for 16 hours at 37°C. Cells were observed under fluorescent microscope and pictures were taken using SPOT advanced software. Samples were harvested, immunoblotted and analyzed using anti-GFP, anti-Ubiquitin, anti-GAPDH and anti-NS5B antibodies.
Co-immunoprecipitation of HCV NS5B with Ubiquitnated NORE1A
For detection of ubiquitinated NORE1A, Replicon cells (MH14/C-5B) and 2.3 Cells were transfected with GFP-NORE1A plasmid. 24 hours post transfection cells were treated with either DMSO or MG132 (10µM) and further incubated for 16 hours at 37°C. Cells were lysed in Buffer A (50mM Tris.Cl [pH 7.5], 120mM NaCl, 0.75% Triton X-100) and immunoprecipated using anti-GFP antibodies and immunoblotted using anti-GFP, anti-NS5B and anti-Ubiquitin antibodies. For detection of endogenous ubiquitinated NORE1A, cells were treated with DMSO or MG132 and immunoprecipitated using anti-NORE1A antibodies and samples were immunoblotted using anti-NORE1A, anti-NS5B and anti-Ubiquitin antibodies. For determination of association of NS5B with ubiquitinated NORE1A, Replicon cells, 2.3 Cells and control 293 cells were transfected with 2.0 µg of HA-Ubiquitin expression vector or control plasmid. At 36 hours post transfection cells were treated with MG132 and incubated further at 37°C for 16 hours. Cells were lysed in Buffer A (50 mM Tris.Cl [pH 7.5], 120 mM NaCl, 0.75% Triton X-100) and immunoprecipitated using anti-HA antibodies and immunoblotted using anti-NORE1A, anti-NS5B and anti-HA antibodies.
Pulse Chase Analysis
HCV replicon, 2.3 Cells, 293 and Huh7 cells were seeded in 6 cm dish at 50–60% confluence. After 24 hours post seeding cells were treated with 10µCi S35 in Methionine free medium for 30 minutes at 37°C. Medium was replaced with complete medium and cells were further incubated at 37°C. Cells were harvested in at 0 hour, 1 hour, 2 hour and 3 hours post treatment in Buffer A (50 mM Tris.Cl [pH 7.5], 120 mM NaCl, 0.75% Triton X-100). Lysates were immunoprecipitated using anti-NORE1A antibodies (Rabbit Polyclonal) and ran on SDS-PAGE. Gel was dried and subjected to Phosphor imaging for quantification. Intensity of bands were quantified using ImageQuant5.2 software.
Clinical Samples Details
Liver samples were kindly provided by Dr. Snorri Thorgeirsson (NCI, Bethesda, MD) and the Liver Tissue Procurement and Distribution System (LTPDS), which was funded by NIH contract no. N01-DK-9–2310. Institutional Review Board approval was obtained at participating hospitals in the LTPDS (University of Minnesota, Minneapolis, Minnesota, USA, and University of Pittsburgh, Pittsburgh, Pennsylvania, USA) and the NIH. Statistical analysis was via ANOVA.
RESULTS
HCV NS5B interacts with NORE1A
Using a BacterioMatch Two hybrid system we identified NORE1A as a NS5B binding partner protein. To validate this interaction in cells, we investigated NS5B-NORE1A complexes in C-5B cells containing the autonomously replicating HCV RNA. Endogenous NORE1A was detected in lysates of both Huh7.5 and C-5B cells, albeit to a relatively lower level in the latter (Figure 1A, input). When lysates were immunoprecipitated with anti-NS5B antibodies, a band corresponding to NORE1A was detected in C-5B lysates, but not in Huh7.5 lysates (Figure 1A, top). The specificity of this interaction was validated by observing the absence of NORE1A band when C-5B lysates were immunoprecipated with IgG (Figure 1A, bottom). We then performed the reverse co-immunoprecipitation in C-5B cells. NS5B was easily detected in anti-NORE1A precipitates but absent in reactions containing a nonspecific IgG (Figure 1A, middle).
Figure 1. Endogenous NORE1A interacts with and colocalizes with NS5B.
(A, B, C) Total lysates from Huh7.5, C-5B, MH14, 2.3, and 293 cells were immunoprecipitated (IP) with either anti-NS5B (top and bottom panels), anti-NORE1A antibodies (middle panel), or Isotype specific IgG antibodies (middle and bottom panels). Immunoprecipitated complexes were visualized by immunoblotting (IB) with NORE1A and NS5B specific antibodies. A portion of the lysate (input) was immunoblotted to verify expression of NORE1A and NS5B. (D) Indirect Immunofluorescence analysis of C-5B and 2.3 cells. Primary antibodies used were anti-NORE1A (Green) and anti-NS5B (Red). DAPI staining was used to visualize nuclei. Yellow denotes areas of NORE1A and NS5B colocalization.
We next analyzed this interaction in MH14 cells bearing the sub-genomic HCV replicons. NORE1A was detected in MH14 lysates immunoprecipitated with anti-NS5B antibody, but not in Huh7.5 lysates immunoprecipitated with anti-NS5B or when M14 lysates were immunoprecipitated with nonspecific IgG antibodies (Figure 1B top and bottom). Similarly, NS5B was detectable in MH14 lysates immunoprecipitated with anti-NORE1A antibodies (Figure 1B, middle). As the MH14 subgenomic replicon does not contain the HCV structural proteins, the results suggest that the NS5B-NORE1A interaction occurs in the absence of HCV structural proteins. To further rule out the possibility of other HCV proteins mediating the NS5B-NORE1A interaction, we repeated our experiments in 2.3 cells which stably expresses the NS5B protein (Figure 1C). Fluorescent microscopy was then used to confirm NORE1A co-localizes with NS5B in both C-5B and 2.3 cells (Figure 1D).
Expression of NS5B correlates with decreased levels of NORE1A
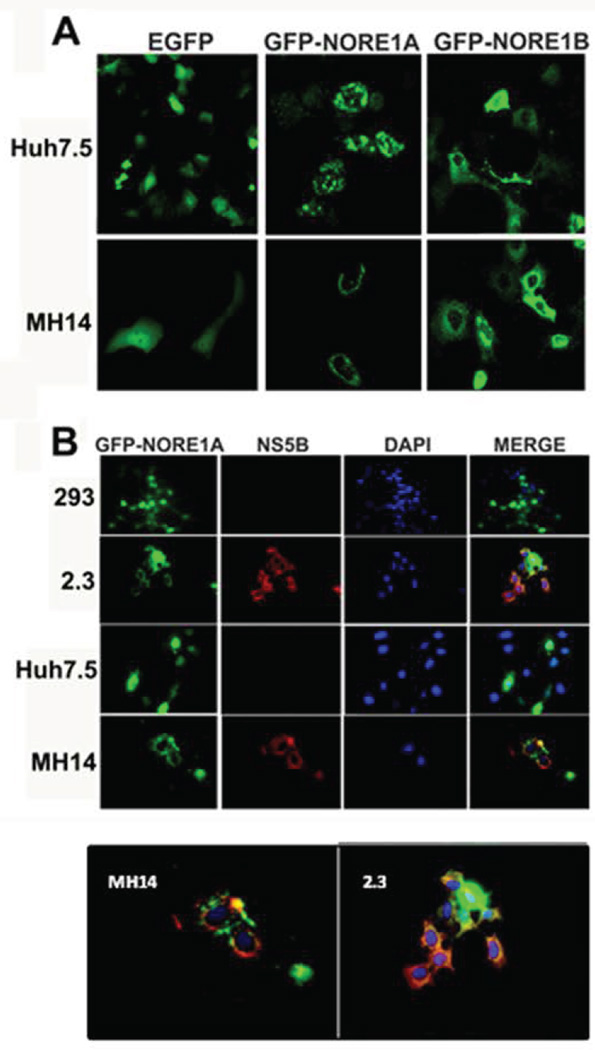
NS5B has been reported to associate with the tumor suppressor Rb, which leads to Rb degradation (14, 22). We therefore hypothesized that NS5B may exert a similar effect on the NORE1A tumor suppressor. Therefore, we ectopically expressed GFP-NORE1A or the smaller isoform GFP-NORE1B expression plasmids in Huh7.5 and MH14 cells and visualized GFP expression 36 h post-transfection (Figure 2A). Expression of GFP-NORE1A but not GFP-NORE1B was dramatically reduced in MH14 cells relative to Huh7.5 cells.
Figure 2. NS5B negatively regulates expression of NORE1A.
(A) Huh7.5 and MH14 cells were transfected with EGFP, GFP-NORE1A, or GFP-NORE1B. Following 36 hours incubation, cells were visualized for expression of GFP-NORE1A or GFP-NORE1B in each cell. ImageJ quantification of corrected total cell fluorescence (arbitrary units) is shown on each panel (B) Relative expression of NORE1A and NORE1B in Huh7.5 and MH14 cells was analyzed on a SDS-PAGE. Anti-GFP, -NS5B, and GAPDH were used to probe immunoblots (IB). GAPDH was employed as a loading control. (C) Lysates obtained from Huh7.5, C-5B, MH14, 293, or 2.3 cells were immunoblotted (IB) with anti-NORE1A, -NS5B, and –GAPDH to visualize relative levels of the endogneous proteins in each cellular lysate. GAPDH was employed as a loading control. (D) Huh7.5 cells were transfected with GFP-NORE1A expression plasmid along with pNTAP vector or pNTAP-NS5BCΔ21 plasmid. Following 36h hours incubation, cellular monolayers were lysed and immunoblot analysis was performed using anti-GFP, anti-NS5B, and anti-GAPDH antibodies.
To verify our results, we performed immunoblot analysis of lysates obtained from MH14 and Huh7.5 cells expressing GFP, GFP-NORE1A or GFP-NORE1B (Figure 2B). Consistent with our Figure 2A observation, the expression of GFP or NORE1B was unaffected in MH14 cells relative to Huh7.5. By contrast, the band corresponding to NORE1A was reduced by approximately 5.3-fold in lysates from MH14 cells when compared to Huh7.5 cells. We next examined the endogenous levels of NORE1A in C-5B, MH14 and 2.3 cells relative to their cognate control cells (Figure 2C). Similar to our data with ectopically expressed GFP-NORE1A, endogenous NORE1A levels were substantially reduced in both C-5B and MH14 cells when compared to Huh7.5 cells. Endogenous NORE1A levels were also sharply reduced in the NS5B stable cell line when compared to the parental 293 cells lacking NS5B (Figure 2C). To determine if HCV-NS5B protein alone was sufficient to alter NORE1A expression, we transfected 293 cells with a GFP-NORE1A expression plasmid along with NS5B expression plasmid or cognate vector, and analyzed their expression by immunoblot analysis. As seen in Figure 2D, the level of GFP-NORE1A was reduced in the presence of NS5B when compared to cells transfected with empty vector. Therefore, we conclude that NS5B down-regulates NORE1A expression in the absence of additional HCV proteins.
NS5B decreases NORE1A protein levels via degradation mediated by the host proteasome
Several studies have reported on the epigenetic down-regulation of NORE1 in cancer (23, 24). To determine if NS5B might decrease expression of the NORE1A gene, we compared NORE1A mRNA levels in cells expressing either NS5B or the HCV replicon to their parent cell lines. As shown in Figure 3A, NORE1A mRNA levels in 293 and 2.3 cells stably expressing NS5B were near equal. Similarly, Huh7.5 cells and C-5B cells exhibited equal expression of NORE1A mRNA. By contrast, NORE1A mRNA was approximately 3-fold higher in MH14 cells versus the Huh7.5 cells.
Figure 3. NS5B expression and HCV replication induces proteosomal degradation of NORE1A.
(A) Real time PCR analysis of NORE1A mRNA levels in 293, 2.3, Huh7.5, MH14, and C-5B cells. Results are shown relative to actin levels in each cell line. (B) Pulse chase analysis of NORE1A protein levels. Overnight cultured cells were subjected to 30 minutes pulse in 35S Methionine supplemented methionine free medium, cultured in complete medium for the indicated time, and immunoprecipitated with anti-NORE1A antibodies. The immunoprecipitated complexes were resolved on 10% SDS-PAGE, and subjected to autoradiography. The intensity of bands corresponding to NORE1A was quantified using ImageQuant 5.2 software. (C) MH14 or 2.3 cells were transfected with GFP-NORE1A expression plasmid. 24 hours post transfection, cells were treated with either DMSO or MG132 for 16 hours. Fluorescent microscopy was utilized to visualize the expression of GFP-NORE1A, quantification of fluorescence is shown top right of each panel in arbitrary units (ImageJ). (D) C-5B, MH14, 2.3, Huh7 and 293 cells were transfected with GFP-NORE1A expression plasmid and treated with either DMSO (−) or MG132 (+) as described above. Cellular lysates were resolved on 10% SDS-PAGE and anti-GFP, -NS5B, and –GAPDH specific antibodies were used for immunoblot analysis to visualize bands corresponding to NORE1A, NS5B, and GAPDH respectively.
Since, the HCV replicon or NS5B expression alone does not decrease NORE1A mRNA, we next performed a pulse-chase experiment to determine the stability of NORE1A protein in the presence of NS5B (2.3) or the MH14 HCV sub-genomic replicon. Compared to control cells, the levels of NORE1A protein decreased much faster in the presence of either NS5B or the MH14 replicon suggesting that NS5B may alter the stability of the NORE1A protein (Figure 3B). The half-life of NORE1A in 2.3 cells was approximately 30 minutes shorter than in 293 cells. The difference in the half-life was even more pronounced in MH14 vs. Huh7 cells (~2 hours vs. ~3 hours respectively). A previous study had reported that NORE1A is degraded by calpain-mediated proteolysis (25). However, treatment with the calpain inhibitor N-Acetyl-Leu-Leu-Nle-CHO (ALLN) failed to restore NORE1A levels in the presence of NS5B. NS5B-downregulation of Rb involves the host proteasome and treatment with proteasome inhibitors reverses NS5B-induced degradation of Rb (14). Therefore, we reasoned that NS5B might also induce NORE1A degradation in a process mediated by the host proteasome. To test our hypothesis, we visualized GFP-NORE1A protein levels in the presence or absence of the proteasome inhibitor MG132. Treatment with MG132 enhanced expression of GFP-NORE1A relative to DMSO treated controls, in both MH14 and NS5B stable cell lines (Figure 3C). This was clearly evident from immunoblot analysis of these cells subjected to MG132 treatment. Levels of GFP-tagged NORE1A increased dramatically upon treatment with MG132 in C-5B, MH14 and 2.3 cells relative to their cognate control cells (Figure 3D).
As ubiquitination is a key step in tagging proteins for degradation by the host proteasome, we tested NORE1A for ubiquitination in the presence and absence of NS5B. GFP-NORE1A was immunoprecipitated from lysates of MH14, C-5B, or the NS5B stable cell line that were treated with either DMSO or MG132. When the immunoprecipitated complexes were immunoblotted with anti-ubiquitin antibodies, a smear of higher molecular weight products were detected and these higher molecular weight bands were enhanced in cells treated with MG132 (Figure 4A). A similar result was obtained when probing anti-NORE1A-immunoprecipitated complexes with anti-ubiquitin antibodies (Figure 4B). To investigate if the high molecular weight species were ubiquitinated-NORE1A, we transfected Huh7.5, MH14, or C-5B cell lines with a HA-tagged ubiquitin expressing plasmid. When the cellular lysates were immunoprecipitated with anti-HA antibody to pull down the HA-tagged ubiquitin and subsequently immunoblotted with anti-NORE1A antibodies, a ladder of ubiquitinated NORE1A bands were visualized in C-5B and M14 cells, but not in Huh7.5 cells lacking the replicon suggesting that NORE1A ubiquitination occurs in the presence of NS5B (Figure 4C). As further proof NORE1A ubiquitination is mediated by NS5B and not additional HCV proteins, we performed a similar experiment in the NS5B stably expressing cell line. Similar to results obtained when using replicon cells, NORE1A ubiquitination was dependent on the presence of NS5B (Figure 4D). The effects of NS5B on NORE1A appeared to be specific as we found that Ns5B did not suppress the expression of a different Ras effector called B-Raf (Figure 4E).
Figure 4. NS5B expression induces the polyubiquitination of NORE1A.
(A and B) MH14, C-5B, and 2.3 cells were transfected with GFP-NORE1A expression vector, and 24 hours post-transfection cells were treated with either DMSO or 10 µM MG132 for an additional 16 hours. Lysates were immunoprecipitated (IP) with either anti-GFP (A) or anti-NORE1A antibodies. Immunoprecipitated complexes were resolved on SDS-PAGE and immunoblot (IB) analysis was performed using anti-ubiquitin, anti- NS5B, and anti–NORE1A antibodies. (C and D) Huh7.5, MH14, C-5B, 293, and 2.3 cells were transfected with HA-Ub expression vector. Following 36 hours incubation, cellular monolayers were lysed and immunoprecipitated (IP) with anti-HA antibodies. An aliquot of total lysate was resolved on SDS-PAGE (Input) and immunoblotted with anti-NS5B antibodies as loading control. To determine if the effect of NS5B was specific to the NORE1A Ras effector, we examined the ability of NS5B to suppress NORE1A protein expression in comparison to the Ras effector B-Raf. Transient transfections in HEK-293 cells showed that NS5B suppressed NORE1A expression almost completely but had little or no effect on B-Raf. NS5B expression was detected with a polyclonal antibody which gave rise to some non-specific bands (NS) in the control panel.
NORE1A negatively modulates HCV RNA replication
HCV infection can activate the Ras pathway (26). Further, NORE1A is involved in a variety of anti-proliferation and pro-apoptotic cellular responses to activated Ras (27). So it makes sense that HCV would need to suppress NORE1A function to avoid inducing death of its host cell. However, we wondered if there might be a more significant biological reason for the interaction of NORE1A with NS5B. Perhaps NORE1A might have anti-viral activity, and so HCV NS5B might induce NORE1A degradation as a means to enhance HCV replication. When we over-expressed GFP-tagged NORE1A in MH14 replicon containing cells NORE1A reduced the level of HCV RNA replication by approximately 78% (Figure 5A).
Figure 5. Expression levels of NORE1A inversely correlate with the level of HCV replication.
(A) MH14 cells were transfected with EGFP or NORE1A. Anti-GFP and anti-GAPDH antibodies were used for immunoblot analysis (top panel). (Bottom panel) Real Time PCR analysis of HCV RNA levels in Huh7.5 and MH14 cells transfected with NORE1A. Levels of HCV RNA replication were determined relative to levels of actin RNA. Levels of HCV RNA replication in vector transfected MH14 cells were set at 100%. (B) MH14 cells were either untransfected, lipofectamine treated, or transfected with 500 ng of either NORE1A shRNA or control Vector. 48 hours post transfection, cellular monolayers were lysed and immunoblotted with anti-NORE1A or –GAPDH antibodies to verify successful knockdown of NORE1A levels (top panel). (Bottom panel) Real time PCR analysis was performed as described above. The levels of HCV RNA under each condition are displayed relative to the level of HCV RNA in untransfected (U) MH14 cells (set at one).
To further confirm an antagonist role for NORE1A in modulating HCV replication, we used shRNA to knockdown NORE1A in MH14 replicon containing cells. Though the levels of NORE1A protein are lower in this cell line than in Huh7.5 cells, a further reduction in expression of NORE1A was achieved upon treatment of NORE1A specific shRNA (Figure 5B, top panel). Treatment of cells with the transfection reagent alone or scrambled shRNA had no detectable effect on HCV replication (Figure 5B, lower panel). However, treatment of replicon containing cells with NORE1A specific shRNA resulted in greater than a 2 fold enhancement of HCV RNA replication (Figure 5B, lower panel). This data suggests NORE1A antagonizes HCV replication and that HCV may induce the degradation of NORE1A in a process mediated by NS5B as a mechanism to promote viral replication.
NORE1A is re-distributed to the cytoplasm and perinuclear region in HCV replicons and NS5B stable cells
To investigate the cellular distribution of GFP-NORE1A in the presence of NS5B, we carried out live cell imaging of Huh7.5 and MH14 cells over-expressing GFP-NORE1A, GFP-NORE1B, or GFP alone (Figure 6A). NORE1A contains a nuclear localization signal and is reported to express predominantly in the nucleus, whereas the expression of NORE1B is cytoplasmic (28). Consistent with these reports, the expression of GFP-NORE1A and GFP-NORE1B was nuclear and cytoplasmic in Huh7.5 cells. By contrast, GFP-NORE1A re-distributed to the perinuclear region in MH14 replicons, while the localization of GFP-NORE1B was similar to that observed in Huh7.5 cells.
Figure 6. NORE1A redistributes around perinuclear region and colocalizes with NS5B.
(A) Huh7.5 and MH14 cells were transfected with either EGFP, GFP-NORE1A, or GFP-NORE1B plasmids. The localization of EGFP or GFP-NORE1A was visualized using confocal microscopy. (B) Expression of NS5B shifts NORE1A localization to the perinuclear region. 293, 2.3, Huh7.5, and MH14 cells were transfected with GFP-NORE1A expression plasmid. Thirty-six hours post transfection, cells fixed and subjected to immunofluorescence analysis using primary antibodies against NS5B (Red) and DAPI (Blue) to visualize nucleus. Images were merged and overlapping regions of red NS5B and green GFP-NORE1A are indicated in yellow.
We next carried out indirect immunofluorescence analysis to investigate whether GFP-NORE1A co-localized with NS5B in MH14 and 2.3 cells (Figure 6B). 293, 2.3, Huh7.5 and MH14 cells were transfected with GFP-NORE1A plasmid and its expression and co-localization with NS5B monitored 30 hours post-transfection. As expected, GFP-NORE1A exhibited a predominant nuclear localization in 293 and Huh7.5 cells. However, in both 2.3 and MH14 cells, GFP-NORE1A re-distributed from the nucleus to the perinuclear region and co-localized with NS5B. This suggests that the expression of NS5B alone is sufficient for the redistribution of NORE1A and that NS5B binding to NORE1A may prevent NORE1A from translocating to the nucleus.
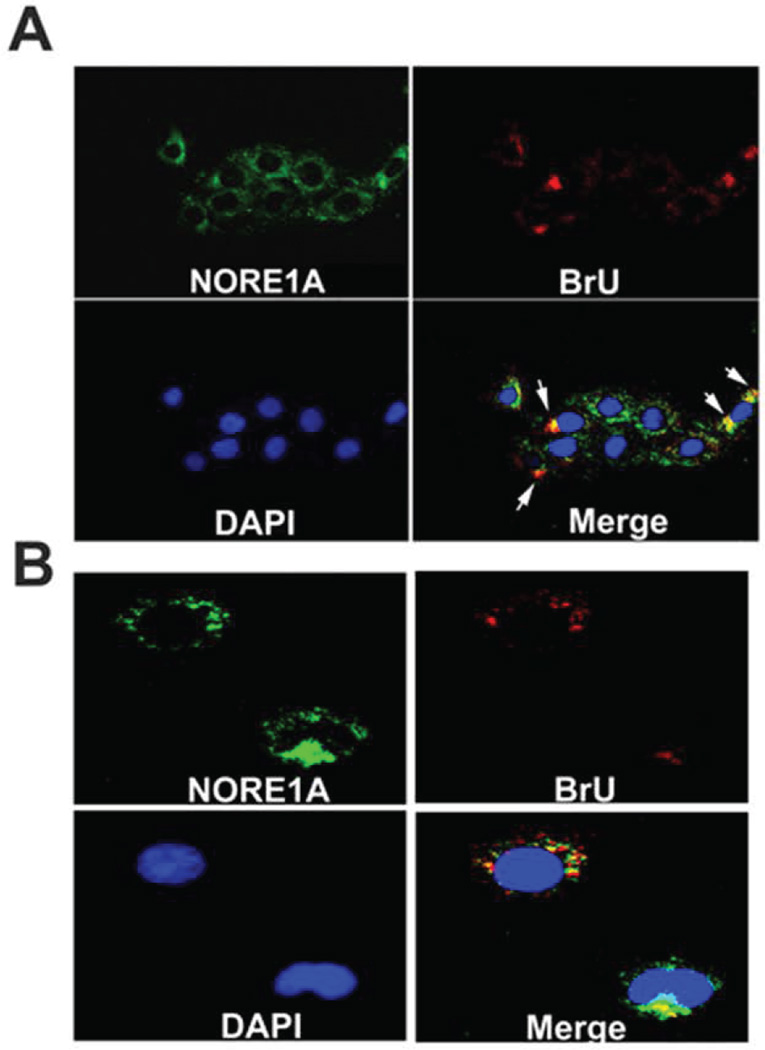
The observation that NORE1A localizes with NS5B to the perinuclear region in the presence of the HCV replicon supports a model of NORE1A and NS5B localizing to active HCV replication complexes (RC). To visualize nascent RNA synthesized in the RC in relationship to NS5B and NORE1A, we treated HCV C-5B/MH14 replicon cells with actinomycin D and BrU as described previously (17–20). We found NORE1A localizes with HCV RNA as indicated by a strong merged signal of NORE1A and BrU suggesting that NORE1A is present in the HCV replication complex (Figure 7A, white arrows). Similar experiments were performed on Huh7 cells and no perinuclear BrdU staining was detectable (data not shown). HCV RNA replication occurs in membranous web-like structures (21, 29–32), recently reported to be derived from the host secretory pathway (33). To determine if NORE1A may be present within these HCV generated intracellular membranes where RNA replication occurs, we treated cells with digitonin to disrupt the cellular membrane followed by treating cells with proteinase K (17, 21). This treatment results in the degradation of the majority of non-membrane bound proteins. Upon permeabilization with Triton X-100, fixing, and staining cells with anti-NORE1A and anti-BrU antibodies, we observed that a large portion of perinuclear NORE1A was resistant to proteinase K treatment suggesting that NORE1A is contained within intracellular membranes (Figure 7B). Upon merging NORE1A and anti-BrU, a strong co-localization, as indicated in yellow, was again observed in the perinuclear region of HCV replicon cells (Figure 7B). This observation indicates that a portion of NORE1A is present within HCV modified membranes.
Figure 7. NORE1A colocalizes with HCV RNA within intracellular membranes.
(A) NORE1A colocalizes with nascent HCV RNA. MH14 cells were treated with actinomycin D then labeled with BrU for 2 hours. Immunofluorescence analysis was performed with anti-NORE1A (Green) and anti-BrU (Red) antibodies. DAPI (Blue) was used to visualize nuclei. Images were merged and regions of green (NORE1A) and red (HCV RNA) overlap are displayed in yellow highlighted by white arrowheads. (B) NORE1A associated with replicating HCV RNA is impermeable to proteinase K. MH14 cells were treated with actinomycin D and BrU to label nascent HCV RNA. Prior to immunofluorescence analysis, replicon cells were treated with digitonin followed by proteinase K. Cells were subsequently permeabilized with Triton X-100, fixed, and stained using either anti-NORE1A or anti-BrU antibodies. Images were merged and regions of NORE1A and BrU signal overlap are indicated in yellow.
NORE1A protein levels are frequently downregulated in HCV infected human livers
If NS5B has a physiological role in suppressing NORE1A expression them we might expect that HCV infected humans would often exhibit suppressed NORE1A levels in the liver. We compared the expression levels of NORE1A in primary, non-tumorous (cirrhotic) liver samples associated with various etiologies to normal livers (Figure 8). An example of the experimental data is shown in the upper panel and a summary in the lower panels. None of the HBV-infected livers or cirrhotic livers associated with ethanol consumption showed reduced protein levels of NORE1A protein when compared with normal liver, In contrast, decreased protein levels of NORE1A were detected in 10 out of 18 (55%) of HCV-infected livers (Figure 8). Curiously, all the HCV infected samples that exhibited reduced NORE1A protein levels actually exhibited elevated NORE1A mRNA (left lower panel).
Figure 8. NORE1A protein levels are specifically suppressed in HCV infected human livers.
(A) Upper panel: Representative immunoblot analysis of NORE1A in human cirrhotic livers associated with HCV infection, HBV infection or ethanol consumption. β-Actin serves as a loading control.
(B) Lower Left panel: Summary of NORE1A mRNA levels in cirrhotic livers associated with HBV infection, ethanol, and HCV infection. Values were normalized to β-Actin. Lower right panel, quantification of NORE1A protein levels in the liver samples expressed in arbitrary units relative to β-Actin.
(C) Ras activity in the primary samples was assayed using a Ras G-LISA® kit (Cytoskeleton Inc, CO) using the manufacturer’s protocol.
Finally, as NORE1A is a pro-apoptotic, pro-senescent Ras oncoprotein effector, we examined the levels of active Ras in the clinical samples. We found an almost perfect correlation between reduction of NORE1A expression and elevation of Ras activity (Figure 8C).
Discussion
HCV infection induces numerous changes in cellular signaling networks and predisposes to the development of HCC (34). The mechanisms underlying the cancer promoting effects of HCV remain poorly characterized. HCV infection causes general inflammation, which likely contributes to the progression to HCC. However, recent evidence suggests that certain HCV proteins can bind and inactivate key regulators of cellular growth, such as Rb, to facilitate virus replication (14). The inactivation of such proteins has the additional effect of promoting cellular transformation. Here, we identify a new virus/host protein interaction system by showing that NS5B directly binds and promotes the degradation of the tumor suppressor NORE1A.
NORE1A is a member of the RASSF family of tumor suppressors that is frequently subjected to epigenetic inactivation in human tumors (35). NORE1A appears to serve as a major senescence effector of the Ras oncoprotein and couples Ras to the regulation of both p53 and Rb (27). We have previously found that aberrant NORE1A promoter methylation is common in malignant HCC and correlates with the loss of senescence markers (15, 36). Therefore, the NS5B-NORE1A interaction resulting in NORE1A protein degradation may be an additional factor that leads to the progression to HCC during chronic HCV infections.
To define the mechanism by which NS5B suppresses NORE1A, we examined mRNA expression, and protein stability. We found no evidence that HCV suppresses NORE1A epigenetically or transcriptionally. Indeed, NORE1A mRNA actually increases in infected primary liver samples while the protein levels are decreased. We observed NS5B induced ubiquitination of NORE1A and found that we could block the degradation of NORE1A by the use of proteasome inhibitors. Therefore, it appears that turnover of NORE1A is mediated by the host proteasome during HCV-infection. We speculate that the increase in NORE1A transcription observed in infected livers is an anti-viral response that is blocked by viral mediated degradation of the protein.
NS5B-induced degradation of Rb by NS5B (22) was found to involve recruitment of E6AP to NS5B-Rb complexes (14). Our preliminary data suggests that NS5B-NORE1A and E6AP complexes form in cells though further investigation is required before we are able to definitively implicate E6AP as involved in NS5B-induced degradation of NORE1A (data not shown).
One of the most obvious questions arising from this study is what role NORE1A plays during HCV infection. We observed over-expression of NORE1A antagonizes HCV RNA replication. Conversely, further knockdown of NORE1A levels in replicon expressing cells using shRNA resulted in greater than two-fold increase in HCV RNA replication. If NORE1A functions in an antiviral role, then it is not surprising that HCV has evolved a mechanism to counteract the effects of NORE1, specifically by binding NORE1A and inducing its degradation. How NORE1 might antagonize HCV replication will be the subject of future investigations. One plausible mechanism may lie in the role of NORE1 in Ras pathways. HCV-infection activates the Ras pathway, and it is believed that constitutive activation of Ras enhances HCV replication (26, 37). Further, HCV RNA replication was observed to be enhanced during S phase, and cap independent translation of the HCV polyprotein is more efficient in active replicating than in resting cells (38, 39). As NORE1 promotes apoptosis and/or senescence in the presence of activated Ras (16, 40–42), its presence may restrict the ability of host cells to support viral replication. In primary human liver samples we observed an almost perfect correlation with suppression of NORE1A protein by HCV and upregulation of Ras activity. Therefore, it is tempting to speculate that HCV NS5B binds and induces the degradation of NORE1A as a mechanism to shift Ras-signaling to proliferation and away from growth inhibition/cell death responses, thus ensuring survival and replication of the HCV genome while simultaneously promoting malignancy.
Alternatively, we cannot discount the possibility that NORE1A expression may directly antagonize the polymerase activity of NS5B or negatively impact an additional HCV replication process. NS5B interacts with a variety of host and HCV proteins to form the replicase complex. Further, oligomerization of NS5B is required for efficient polymerase activity (43–45). An interesting possibility might be that the NORE1A protein disrupts HCV replication complexes by inhibiting NS5B oligomerization and/or reducing the efficiency of the NS5B polymerase activity. In support, we observed NORE1A co-localizes with both NS5B and nascent HCV RNA in replicon cells. However, whether NORE1A is actively antagonizing the formation of replication complexes or if the presence of NORE1A at the replication complex is merely a consequence of NS5B recruitment remains to be tested.
In conclusion, HCV NS5B induces the degradation of the tumor suppressor NORE1A. Interestingly, we found that degradation of NORE1A occurred in some cirrhotic livers during chronic HCV infection but not in cirrhotic livers as a result of HBV infection or ethanol consumption. Down-regulation of NORE1A occurs during the development of many forms of cancer (35). Therefore, HCV-mediated down-regulation of NORE1A might be one of several preliminary steps in the progression to HCC during chronic HCV infection. Development of novel NS5B inhibitors that target the NS5B-NORE1 interaction may represent a novel strategy to inhibit HCV replication and also prevent chronic HCV infections from progressing to HCC.
Supplementary Material
Acknowledgments
Financial Support
This work was supported by the National Institute of Health Research Grant CA153147 and UMDNJ-NJMS research fund 105110 to N.K.-B and CA133171-01A2 to G.J.C.
Footnotes
Disclaimer: This work was prepared while Dr. Neerja Kaushik-Basu was employed at New Jersey Medical School, Rutgers University. The opinions expressed in this article are the author's own and do not reflect the view of the National Institutes of Health, the Department of Health and Human Services, or the United States government.
References
- 1.Wasley A, Alter MJ. Epidemiology of hepatitis C: geographic differences and temporal trends. Semin Liver Dis. 2000;20:1–16. doi: 10.1055/s-2000-9506. [DOI] [PubMed] [Google Scholar]
- 2.National Institutes of Health Consensus Development Conference Statement: Management of hepatitis C 2002 (June 10–12, 2002) Gastroenterology. 2002;123:2082–2099. doi: 10.1053/gast.2002.1232082. [DOI] [PubMed] [Google Scholar]
- 3.Saito I, Miyamura T, Ohbayashi A, Harada H, Katayama T, Kikuchi S, Watanabe Y, et al. Hepatitis C virus infection is associated with the development of hepatocellular carcinoma. Proc Natl Acad Sci U S A. 1990;87:6547–6549. doi: 10.1073/pnas.87.17.6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sangiovanni A, Del Ninno E, Fasani P, De Fazio C, Ronchi G, Romeo R, Morabito A, et al. Increased survival of cirrhotic patients with a hepatocellular carcinoma detected during surveillance. Gastroenterology. 2004;126:1005–1014. doi: 10.1053/j.gastro.2003.12.049. [DOI] [PubMed] [Google Scholar]
- 5.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 6.Brass V, Moradpour D, Blum HE. Molecular virology of hepatitis C virus (HCV): 2006 update. Int J Med Sci. 2006;3:29–34. doi: 10.7150/ijms.3.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lerat H, Honda M, Beard MR, Loesch K, Sun J, Yang Y, Okuda M, et al. Steatosis and liver cancer in transgenic mice expressing the structural and nonstructural proteins of hepatitis C virus. Gastroenterology. 2002;122:352–365. doi: 10.1053/gast.2002.31001. [DOI] [PubMed] [Google Scholar]
- 8.Moriya K, Fujie H, Shintani Y, Yotsuyanagi H, Tsutsumi T, Ishibashi K, Matsuura Y, et al. The core protein of hepatitis C virus induces hepatocellular carcinoma in transgenic mice. Nat Med. 1998;4:1065–1067. doi: 10.1038/2053. [DOI] [PubMed] [Google Scholar]
- 9.Honda M, Kaneko S, Shimazaki T, Matsushita E, Kobayashi K, Ping LH, Zhang HC, et al. Hepatitis C virus core protein induces apoptosis and impairs cell-cycle regulation in stably transformed Chinese hamster ovary cells. Hepatology. 2000;31:1351–1359. doi: 10.1053/jhep.2000.7985. [DOI] [PubMed] [Google Scholar]
- 10.Siavoshian S, Abraham JD, Kieny MP, Schuster C. HCV core, NS3, NS5A and NS5B proteins modulate cell proliferation independently from p53 expression in hepatocarcinoma cell lines. Arch Virol. 2004;149:323–336. doi: 10.1007/s00705-003-0205-7. [DOI] [PubMed] [Google Scholar]
- 11.Majumder M, Ghosh AK, Steele R, Ray R, Ray RB. Hepatitis C virus NS5A physically associates with p53 and regulates p21/waf1 gene expression in a p53-dependent manner. J Virol. 2001;75:1401–1407. doi: 10.1128/JVI.75.3.1401-1407.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwun HJ, Jung EY, Ahn JY, Lee MN, Jang KL. p53-dependent transcriptional repression of p21(waf1) by hepatitis C virus NS3. J Gen Virol. 2001;82:2235–2241. doi: 10.1099/0022-1317-82-9-2235. [DOI] [PubMed] [Google Scholar]
- 13.Arima N, Kao CY, Licht T, Padmanabhan R, Sasaguri Y. Modulation of cell growth by the hepatitis C virus nonstructural protein NS5A. J Biol Chem. 2001;276:12675–12684. doi: 10.1074/jbc.M008329200. [DOI] [PubMed] [Google Scholar]
- 14.Munakata T, Liang Y, Kim S, McGivern DR, Huibregtse J, Nomoto A, Lemon SM. Hepatitis C virus induces E6AP-dependent degradation of the retinoblastoma protein. PLoS Pathog. 2007;3:1335–1347. doi: 10.1371/journal.ppat.0030139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donninger H, Calvisi DF, Barnoud T, Clark J, Schmidt ML, Vos MD, Clark GJ. NORE1A is a Ras senescence effector that controls the apoptotic/senescent balance of p53 via HIPK2. J Cell Biol. 2015;208:777–789. doi: 10.1083/jcb.201408087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vos MD, Martinez A, Ellis CA, Vallecorsa T, Clark GJ. The pro-apoptotic Ras effector Nore1 may serve as a Ras-regulated tumor suppressor in the lung. J Biol Chem. 2003;278:21938–21943. doi: 10.1074/jbc.M211019200. [DOI] [PubMed] [Google Scholar]
- 17.Watashi K, Ishii N, Hijikata M, Inoue D, Murata T, Miyanari Y, Shimotohno K. Cyclophilin B is a functional regulator of hepatitis C virus RNA polymerase. Mol Cell. 2005;19:111–122. doi: 10.1016/j.molcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 18.El-Hage N, Luo G. Replication of hepatitis C virus RNA occurs in a membrane-bound replication complex containing nonstructural viral proteins and RNA. J Gen Virol. 2003;84:2761–2769. doi: 10.1099/vir.0.19305-0. [DOI] [PubMed] [Google Scholar]
- 19.Gosert R, Egger D, Lohmann V, Bartenschlager R, Blum HE, Bienz K, Moradpour D. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J Virol. 2003;77:5487–5492. doi: 10.1128/JVI.77.9.5487-5492.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Restrepo-Hartwig MA, Ahlquist P. Brome mosaic virus helicase- and polymerase-like proteins colocalize on the endoplasmic reticulum at sites of viral RNA synthesis. J Virol. 1996;70:8908–8916. doi: 10.1128/jvi.70.12.8908-8916.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyanari Y, Hijikata M, Yamaji M, Hosaka M, Takahashi H, Shimotohno K. Hepatitis C virus non-structural proteins in the probable membranous compartment function in viral genome replication. J Biol Chem. 2003;278:50301–50308. doi: 10.1074/jbc.M305684200. [DOI] [PubMed] [Google Scholar]
- 22.Munakata T, Nakamura M, Liang Y, Li K, Lemon SM. Down-regulation of the retinoblastoma tumor suppressor by the hepatitis C virus NS5B RNA-dependent RNA polymerase. Proc Natl Acad Sci U S A. 2005;102:18159–18164. doi: 10.1073/pnas.0505605102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van der Weyden L, Adams DJ. The Ras-association domain family (RASSF) members and their role in human tumourigenesis. Biochim Biophys Acta. 2007;1776:58–85. doi: 10.1016/j.bbcan.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Avruch J, Xavier R, Bardeesy N, Zhang XF, Praskova M, Zhou D, Xia F. Rassf family of tumor suppressor polypeptides. J Biol Chem. 2009;284:11001–11005. doi: 10.1074/jbc.R800073200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuznetsov S, Khokhlatchev AV. The growth and tumor suppressors NORE1A and RASSF1A are targets for calpain-mediated proteolysis. PLoS One. 2008;3:e3997. doi: 10.1371/journal.pone.0003997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mannova P, Beretta L. Activation of the N-Ras-PI3K–Akt-mTOR pathway by hepatitis C virus: control of cell survival and viral replication. J Virol. 2005;79:8742–8749. doi: 10.1128/JVI.79.14.8742-8749.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Donninger H, Barnoud T, Clark GJ. NORE1A is a double barreled Ras senescence effector that activates p53 and Rb. Cell Cycle. 2016:0. doi: 10.1080/15384101.2016.1152431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumari G, Singhal PK, Rao MR, Mahalingam S. Nuclear transport of Ras-associated tumor suppressor proteins: different transport receptor binding specificities for arginine-rich nuclear targeting signals. J Mol Biol. 2007;367:1294–1311. doi: 10.1016/j.jmb.2007.01.026. [DOI] [PubMed] [Google Scholar]
- 29.Aizaki H, Lee KJ, Sung VM, Ishiko H, Lai MM. Characterization of the hepatitis C virus RNA replication complex associated with lipid rafts. Virology. 2004;324:450–461. doi: 10.1016/j.virol.2004.03.034. [DOI] [PubMed] [Google Scholar]
- 30.Gao L, Aizaki H, He JW, Lai MM. Interactions between viral nonstructural proteins and host protein hVAP-33 mediate the formation of hepatitis C virus RNA replication complex on lipid raft. J Virol. 2004;78:3480–3488. doi: 10.1128/JVI.78.7.3480-3488.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hwang SB, Park KJ, Kim YS, Sung YC, Lai MM. Hepatitis C virus NS5B protein is a membrane-associated phosphoprotein with a predominantly perinuclear localization. Virology. 1997;227:439–446. doi: 10.1006/viro.1996.8357. [DOI] [PubMed] [Google Scholar]
- 32.Shi ST, Lee KJ, Aizaki H, Hwang SB, Lai MM. Hepatitis C virus RNA replication occurs on a detergent-resistant membrane that cofractionates with caveolin-2. J Virol. 2003;77:4160–4168. doi: 10.1128/JVI.77.7.4160-4168.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hsu NY, Ilnytska O, Belov G, Santiana M, Chen YH, Takvorian PM, Pau C, et al. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell. 2010;141:799–811. doi: 10.1016/j.cell.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sukowati CH, El-Khobar KE, Ie SI, Anfuso B, Muljono DH, Tiribelli C. Significance of hepatitis virus infection in the oncogenic initiation of hepatocellular carcinoma. World J Gastroenterol. 2016;22:1497–1512. doi: 10.3748/wjg.v22.i4.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Donninger H, Schmidt ML, Mezzanotte J, Barnoud T, Clark GJ. Ras signaling through RASSF proteins. Semin Cell Dev Biol. 2016 doi: 10.1016/j.semcdb.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Calvisi DF, Donninger H, Vos MD, Birrer MJ, Gordon L, Leaner V, Clark GJ. NORE1A tumor suppressor candidate modulates p21CIP1 via p53. Cancer Res. 2009;69:4629–4637. doi: 10.1158/0008-5472.CAN-08-3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Q, Gong R, Qu J, Zhou Y, Liu W, Chen M, Liu Y, et al. Activation of the Ras/Raf/MEK pathway facilitates HCV replication via attenuation of the IFN-JAK-STAT pathway. J Virol. 2011 doi: 10.1128/JVI.00688-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scholle F, Li K, Bodola F, Ikeda M, Luxon BA, Lemon SM. Virus-host cell interactions during hepatitis C virus RNA replication: impact of polyprotein expression on the cellular transcriptome and cell cycle association with viral RNA synthesis. J Virol. 2004;78:1513–1524. doi: 10.1128/JVI.78.3.1513-1524.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Honda M, Kaneko S, Matsushita E, Kobayashi K, Abell GA, Lemon SM. Cell cycle regulation of hepatitis C virus internal ribosomal entry site-directed translation. Gastroenterology. 2000;118:152–162. doi: 10.1016/s0016-5085(00)70424-0. [DOI] [PubMed] [Google Scholar]
- 40.Khokhlatchev A, Rabizadeh S, Xavier R, Nedwidek M, Chen T, Zhang XF, Seed B, et al. Identification of a novel Ras-regulated proapoptotic pathway. Curr Biol. 2002;12:253–265. doi: 10.1016/s0960-9822(02)00683-8. [DOI] [PubMed] [Google Scholar]
- 41.Hwang E, Ryu KS, Paakkonen K, Guntert P, Cheong HK, Lim DS, Lee JO, et al. Structural insight into dimeric interaction of the SARAH domains from Mst1 and RASSF family proteins in the apoptosis pathway. Proc Natl Acad Sci U S A. 2007;104:9236–9241. doi: 10.1073/pnas.0610716104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barnoud T, Donninger H, Clark GJ. Ras Regulates Rb via NORE1A. J Biol Chem. 2016;291:3114–3123. doi: 10.1074/jbc.M115.697557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang QM, Hockman MA, Staschke K, Johnson RB, Case KA, Lu J, Parsons S, et al. Oligomerization and cooperative RNA synthesis activity of hepatitis C virus RNA-dependent RNA polymerase. J Virol. 2002;76:3865–3872. doi: 10.1128/JVI.76.8.3865-3872.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qin W, Luo H, Nomura T, Hayashi N, Yamashita T, Murakami S. Oligomeric interaction of hepatitis C virus NS5B is critical for catalytic activity of RNA-dependent RNA polymerase. J Biol Chem. 2002;277:2132–2137. doi: 10.1074/jbc.M106880200. [DOI] [PubMed] [Google Scholar]
- 45.Gu B, Gutshall LL, Maley D, Pruss CM, Nguyen TT, Silverman CL, Lin-Goerke J, et al. Mapping cooperative activity of the hepatitis C virus RNA-dependent RNA polymerase using genotype 1a-1b chimeras. Biochem Biophys Res Commun. 2004;313:343–350. doi: 10.1016/j.bbrc.2003.11.118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.