Abstract
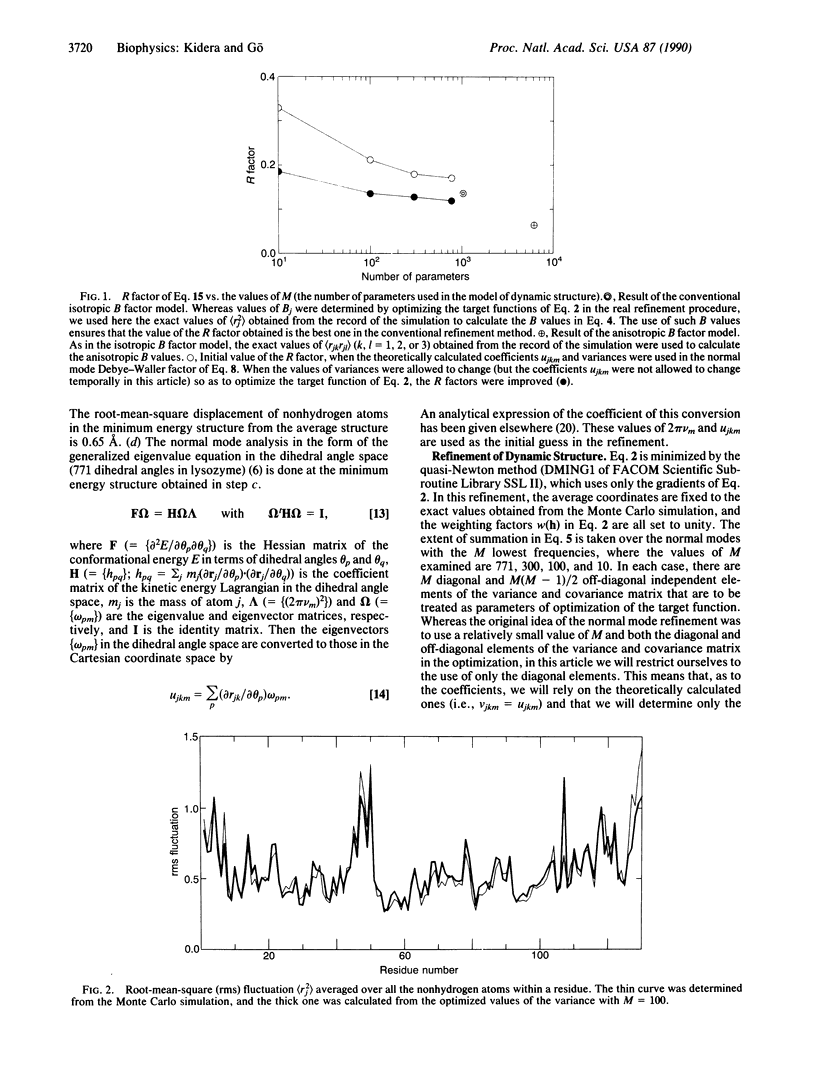
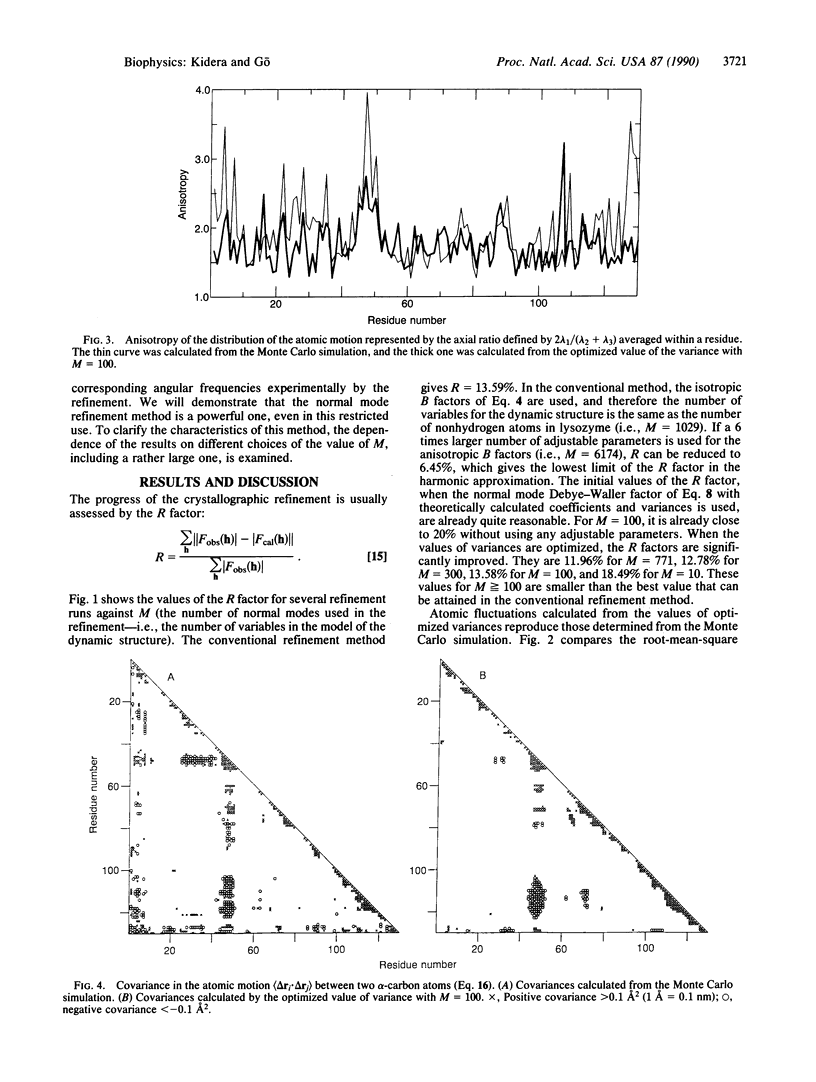
An x-ray crystallographic refinement method, referred to as the normal mode refinement, is proposed. The Debye-Waller factor is expanded in terms of the effective normal modes whose amplitudes and eigenvectors are experimentally determined by the crystallographic refinement. In contrast to the conventional method, the atomic motions are treated generally as anisotropic and concerted. This method is assessed by using the simulated x-ray data given by a Monte Carlo simulation of human lysozyme. In this article, we refine the dynamic structure by fixing the average static structure to exact coordinates. It is found that the normal mode refinement, using a smaller number of variables, gives a better R factor and more information on the dynamics (anisotropy and collectivity in the motion).
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blake C. C., Pulford W. C., Artymiuk P. J. X-ray studies of water in crystals of lysozyme. J Mol Biol. 1983 Jul 5;167(3):693–723. doi: 10.1016/s0022-2836(83)80105-3. [DOI] [PubMed] [Google Scholar]
- Brooks B., Karplus M. Harmonic dynamics of proteins: normal modes and fluctuations in bovine pancreatic trypsin inhibitor. Proc Natl Acad Sci U S A. 1983 Nov;80(21):6571–6575. doi: 10.1073/pnas.80.21.6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go N., Noguti T., Nishikawa T. Dynamics of a small globular protein in terms of low-frequency vibrational modes. Proc Natl Acad Sci U S A. 1983 Jun;80(12):3696–3700. doi: 10.1073/pnas.80.12.3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson W. A. Stereochemically restrained refinement of macromolecular structures. Methods Enzymol. 1985;115:252–270. doi: 10.1016/0076-6879(85)15021-4. [DOI] [PubMed] [Google Scholar]
- Kuriyan J., Petsko G. A., Levy R. M., Karplus M. Effect of anisotropy and anharmonicity on protein crystallographic refinement. An evaluation by molecular dynamics. J Mol Biol. 1986 Jul 20;190(2):227–254. doi: 10.1016/0022-2836(86)90295-0. [DOI] [PubMed] [Google Scholar]
- Levy R. M., Srinivasan A. R., Olson W. K., McCammon J. A. Quasi-harmonic method for studying very low frequency modes in proteins. Biopolymers. 1984 Jun;23(6):1099–1112. doi: 10.1002/bip.360230610. [DOI] [PubMed] [Google Scholar]
- Noguti T., Go N. Efficient Monte Carlo method for simulation of fluctuating conformations of native proteins. Biopolymers. 1985 Mar;24(3):527–546. doi: 10.1002/bip.360240308. [DOI] [PubMed] [Google Scholar]
- Noguti T., Go N. Structural basis of hierarchical multiple substates of a protein. II: Monte Carlo simulation of native thermal fluctuations and energy minimization. Proteins. 1989;5(2):104–112. doi: 10.1002/prot.340050204. [DOI] [PubMed] [Google Scholar]
- Noguti T., Go N. Structural basis of hierarchical multiple substates of a protein. III: Side chain and main chain local conformations. Proteins. 1989;5(2):113–124. doi: 10.1002/prot.340050205. [DOI] [PubMed] [Google Scholar]
- Noguti T., Go N. Structural basis of hierarchical multiple substates of a protein. IV: Rearrangements in atom packing and local deformations. Proteins. 1989;5(2):125–131. doi: 10.1002/prot.340050206. [DOI] [PubMed] [Google Scholar]
- Noguti T., Go N. Structural basis of hierarchical multiple substates of a protein. V: Nonlocal deformations. Proteins. 1989;5(2):132–138. doi: 10.1002/prot.340050207. [DOI] [PubMed] [Google Scholar]
- Petsko G. A., Ringe D. Fluctuations in protein structure from X-ray diffraction. Annu Rev Biophys Bioeng. 1984;13:331–371. doi: 10.1146/annurev.bb.13.060184.001555. [DOI] [PubMed] [Google Scholar]




