Abstract
Duplication of the genome during the S phase of the cell cycle does not occur simultaneously; rather, different sequences are replicated at different times. The replication timing of specific sequences can change during development; however, the determinants of this dynamic process are poorly understood. To gain insights into the contribution of developmental state, genomic sequence, and transcriptional activity to replication timing, we investigated the timing of DNA replication at high resolution along an entire human chromosome (chromosome 22) in two different cell types. The pattern of replication timing was correlated with respect to annotated genes, gene expression, novel transcribed regions of unknown function, sequence composition, and cytological features. We observed that chromosome 22 contains regions of early- and late-replicating domains of 100 kb to 2 Mb, many (but not all) of which are associated with previously described chromosomal bands. In both cell types, expressed sequences are replicated earlier than nontranscribed regions. However, several highly transcribed regions replicate late. Overall, the DNA replication-timing profiles of the two different cell types are remarkably similar, with only nine regions of difference observed. In one case, this difference reflects the differential expression of an annotated gene that resides in this region. Novel transcribed regions with low coding potential exhibit a strong propensity for early DNA replication. Although the cellular function of such transcripts is poorly understood, our results suggest that their activity is linked to the replication-timing program.
Eukaryotic chromosomes initiate DNA replication from origins that fire at different times during the S phase of the cell cycle. Autoradiography experiments have revealed that mammalian DNA replication origins are spaced from 50 to 330 kb apart (1-3). Moreover, cytological and molecular studies have established that different chromosomal regions replicate at different times throughout the S phase (reviewed in ref. 4).
Several studies have correlated the timing of DNA replication with chromosomal features. Microscopic analysis of metaphase chromosomes suggested that gene-rich R bands replicate early and gene-poor G bands replicate late (5, 6). Although the exact nature of G and R band staining is unclear, R bands tend to be GC rich, and G bands tend to be GC poor. Thus, the differential replication timing of G and R bands is consistent with a recent report that replication timing correlates with GC content (7). However, it is not known whether R and G bands always correlate with DNA replication timing.
Analysis of a few mammalian loci has revealed that alterations in transcriptional activity can coincide with changes in replication timing (4). Thus, it has been speculated that replication timing and gene expression are functionally linked. A more comprehensive correlation of the timing of DNA replication with gene expression by using DNA microarrays has been performed in Drosophila and human cells with cDNAs and bacterial artificial chromosome arrays, respectively (7, 8). These studies have demonstrated that expressed regions tend to replicate early in metazoan genomes. In both studies, however, only one cell type was analyzed, and expression analysis was limited to annotated genes. Because transcriptional profiles are cell-type-specific, we have investigated the relationship between differential gene expression and replication timing in two cell types of different developmental origin.
Recent studies have revealed that many regions of the genome that lack known genes are nonetheless transcribed into polyadenylated RNA (9, 10), and it has been estimated that 30-50% of chromosomal transcription in a tissue or cell type is derived from such regions (11). Many of these transcriptionally active regions (TARs) have low coding potential and thus do not represent classical protein-encoding genes. The function of these RNAs is under intense investigation, and their abundance challenges much of our current thinking about transcriptional and post-transcriptional regulation. Importantly, it is not known how TAR activity relates to DNA replication.
The detection of TAR transcripts was made possible through the development of genomic microarrays, which cover large genomic regions with small tiled probes, regardless of previously assigned function. Using such microarrays, we have conducted an unbiased high-resolution analysis of DNA replication timing across human chromosome 22. DNA replication timing was compared with sequence characteristics including chromosome cytology, gene expression, and TAR activity. Moreover, we analyzed replication timing across a large genomic DNA region in cells of different developmental origin. Our studies indicate that (i) although, in general, transcribed regions are replicated early in the S phase, many exceptions are noted; (ii) most of chromosome 22 shows no cell-type-specific replication timing, whereas a small number of regions show cell-type replication-timing differences; and (iii) novel transcribed regions are positively correlated with early replication and tend to replicate even earlier than annotated genes.
Materials and Methods
Cell Culture. Human primary lung fibroblast HFL-1 cells [ATCC no. CCL-153 (12)] were obtained from the American Type Culture Collection and cultured in DMEM containing 10% FCS at 37°C and 5% CO2. Human B lymphoblastoid NC-NC cells [DSMZ-ACC 120 (13)] were obtained from DSMZ (Braunschweig, Germany) and cultured in RPMI medium 1640/10% FCS at 37°C/5% CO2.
Spectral Karyotyping (SKY). Cells were fixed in 3:1 methanol:acetic acid and dropped onto glass slides, which were hybridized with SKY reagent (Applied Spectral Imaging, Vista, CA) according to the manufacturer's specifications. Metaphase spreads were analyzed with skyview software (Applied Spectral Imaging).
BrdUrd Labeling. BrdUrd (Sigma) was added to the media of logarithmically growing cultures to a final concentration of 50 μM. Sixty minutes after addition of BrdUrd, cells were washed twice in cold PBS, resuspended in 2.5 ml of PBS, fixed by slowly adding 7.5 ml of cold ethanol, and stored at -20°C.
FACS and DNA Immunoprecipitation. Cell preparation and FACS were performed essentially as described in ref. 14, with minor modifications. Cells were sorted into S-phase fractions based on DNA content. Two gates were sorted, each representing roughly the first and last third of S phase. Cells (15,000 total) were collected from each gate directly into lysis buffer in the absence of salmon sperm DNA. DNA was purified as described in ref. 14 and was sonicated, denatured, and immunoprecipitated with a monoclonal antibody specific for BrdUrd (Becton Dickinson).
PCR Amplification and Fluorescent Labeling of BrdUrd-Enriched DNA. Amplification of the denatured and immunoprecipitated DNA was performed according to ref. 15, with minor modification (8). Size distribution and fluorescence of the product was confirmed by agarose gel electrophoresis followed by a fluorescence scan (Typhoon, Molecular Dynamics). Eight PCRs were performed for each of the fractions (early and late) and processed as described in ref. 16.
Control PCR. Primers to control for abundance in the early and late fraction were designed to amplify products of ≈400 bp. Sequences are available on request. Reactions were performed with 2-5 ng of DNA and 34 cycles under standard PCR conditions. PCR products were separated by agarose gel electrophoresis and visualized by ethidium bromide gel staining.
RNA Extraction. Total RNA was TRIzol-extracted (Invitrogen) from asynchronously growing cells of each type according to the manufacturer's protocol.
Expression Profiling on Affymetrix GeneChip Arrays. Total RNA (10 μg) was reverse-transcribed and labeled by using the Affymetrix cDNA and IVT kit per the manufacturer's instructions and hybridized to HG U133A GeneChips with washing and staining in an Affymetrix Fluidics Station 450. Samples were scanned in an Affymetrix GeneChip 3000 scanner. Each cell type was processed in triplicate.
Expression Profiling on Chromosomal Arrays. Total RNA (20 μg) was reverse-transcribed with Moloney murine leukemia virus reverse transcriptase by using oligonucleotide (dT) primers in the presence of amino-allyl dUTP. The resulting cDNA was labeled with Cy5 with an amino-allyl cDNA labeling kit (Ambion, Austin, TX) and hybridized to the microarray slides.
DNA Microarray Hybridization and Analysis. Chromosomal microarrays were prehybridized with 80 μl of prehybridization buffer [10× Denhardt's solution (0.02% polyvinylpyrrolidone/0.02% Ficoll/0.02% BSA)/5× SSC/0.1% SDS/1% BSA/25% form-amide (vol/vol)] for 2-3 h at 42°C. Coverslips were removed and microarrays were washed with H2O and dried by centrifugation at 600 × g for 5 min. Cy3- and Cy5-labeled probes were resuspended in 80 μl of hybridization buffer for 10 min at 37°C and placed at 100°C for 1 min. Labeled DNA (10 μg) was hybridized in duplicate to each of the three slides of the chromosome 22 microarray and hybridized for 16 h at 42°C. Coverslips were removed in wash buffer solution 1 (0.57× SSC/0.3% SDS) and washed by submerging 20 times into solution 1 and 20 times in solution 2 (0.057× SSC). Microarrays were then dried by centrifugation at 600 × g for 5 min and scanned immediately with a 4000A scanner (Axon Instruments, Union City, CA); images were analyzed with genepix pro 3.0 (Axon Instruments). All microarray data are available at http://array.mbb.yale.edu/chr22. A modified version of the express-yourself (17) array-analysis package was used for background correction and intensity normalization. Intraslide normalization was performed as described in ref. 18. Interslide scaling was done separately for each channel. To compare both cell lines, the intensities on the different slides were adjusted to have identical mean and SDs (ref. 17 and http://array.mbb.yale.edu/analysis). The log2 ratios of early vs. late intensities were calculated for each feature, and mean and SD of replicates were calculated. Fragments with an SD of >0.7 were removed. Only array features with calculable early/late replications ratios obtained from at least half of the microarrays were plotted. Replication profiles of chromosome 22 were generated as described in ref. 8. Outliers were handled by a standard loess procedure (18).
RNA Analysis from Affymetrix Microarrays. Expression values were estimated by using Robust Multichip Analysis [RMA algorithm (19)]. Subsequent analysis was performed with genespring 6.2 (Silicon Genetics, Redwood City, CA). Genes were required to have a minimum expression level of 50 in at least one cell line, differ in expression by at least 2-fold, and pass a t test (P < 0.05) with a Benjamini and Hochberg multiple-testing correction. The resulting datasets are available at the ArrayExpress archive of the European Bioinformatics Institute (www.ebi.ac.uk/arrayexpress); the accession no. is E-MEXP-184.
RNA Analysis from Chromosomal Microarrays. Only features that were measurable (non-flagged) in at least three of the four repeat experiments were analyzed for expression. A fragment was considered expressed if the average foreground-over-background ratio was >2.5 and the signal was >200 pixels. RNA expression vs. DNA replication-timing plots were generated as described in ref. 8. The chromosome-wide expression profile was created as described in ref. 9.
Results
Construction of DNA Replication-Timing Profiles for Two Cell Types. To construct a high-resolution DNA replication profile of a mammalian chromosome and compare its replication timing in different cell lines, we used a human chromosome 22 genomic DNA microarray (9). This DNA microarray contains continuously tiled fragments that represent >90% of nonrepetitive chromosome 22 DNA; the average length of the fragments is ≈800 bp. The array is ordered from the centromere to the end of the long arm of chromosome 22. To examine potential changes in DNA replication timing and gene expression in distinct epigenetic backgrounds, cells derived from two discrete tissues were analyzed. We used primary fibroblast cells [HFL-1 (12)] and the lymphoblastoid cell line NC-NC (13). To exclude the possibility that differences in replication timing between the two cell lines might be a consequence of chromosomal abnormalities, we performed detailed spectral karyotyping (19). In both cases, normal autosomal karyotypes without rearrangements or translocations were observed (Fig. 1).
Fig. 1.
Karyotypic analysis of primary fibroblast cells (HFL-1) and the lymphoblastoid NC-NC cell line. Spectral karyotyping was used to identify chromosome number and integrity. No abnormality was detected on the autosomes, whereas a subfraction of the female NC-NC cells lacks the inactive X chromosome as described in ref. 13.
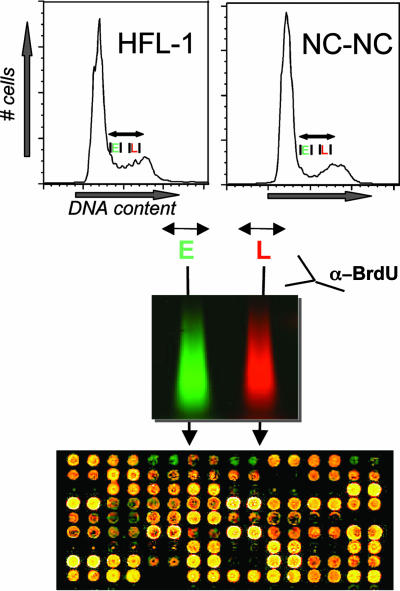
The protocol for constructing a DNA replication-timing profile is described in Fig. 2. Briefly, asynchronously growing cells were pulse-labeled with BrdUrd for 1 h, ethanol-fixed, and sorted by FACS analysis according to DNA content (20). A representative FACS plot for each cell type is shown. Cells from the first and last third of S phase were collected. After DNA isolation, sonication, and denaturation, BrdUrd-containing DNA was enriched by immunoprecipitation. BrdUrd-labeled DNA from the first third of S phase was amplified by PCR with Cy5 end-labeled primers, and immunoprecipitated DNA from the last third was labeled with Cy3 (8). Equal amounts of the two labeled probes were cohybridized to the chromosome 22 genomic DNA microarray. In addition, “dye-swap” experiments, in which early-replicating DNA was labeled with Cy3 and late-replicating DNA was labeled with Cy5, were performed to account for any influence of the individual fluorophores on experimental results. In total, three and four independent experiments were performed for HFL-1 and NC-NC, respectively. Each independent experiment consisted of four separate hybridizations, two of which were dye swaps.
Fig. 2.
Experimental strategy. Shown are histogram plots of DNA content for each cell type. Asynchronously replicating cells were labeled with BrdUrd for 1 h and sorted by FACS for DNA content. DNA from cells in the first and last thirds of S phase was extracted, and newly replicated DNA was immunoprecipitated with α-BrdUrd antibody. Immunoprecipitated DNA was labeled with Cy3 or Cy5 and hybridized to the chromosome 22 DNA microarray.
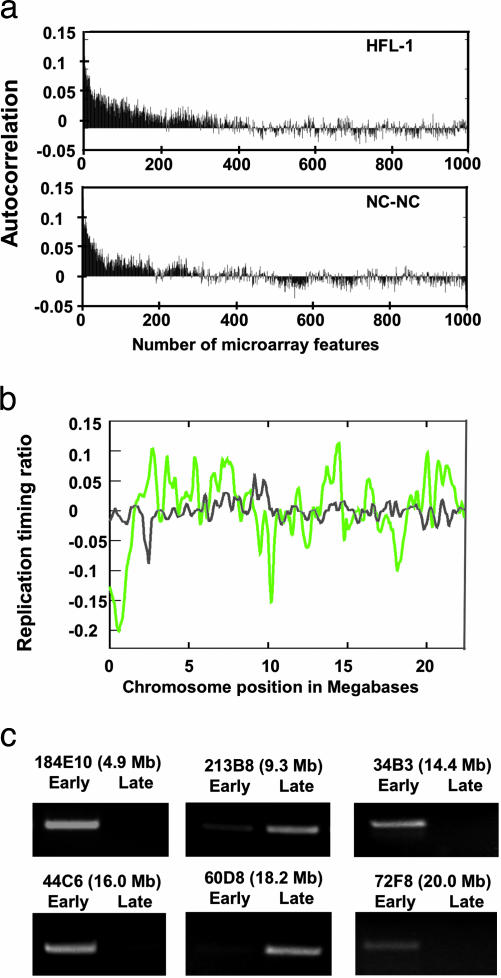
The resulting array data were analyzed by using express-yourself, and features were normalized by total fluorescence intensity normalization (17, 18). To construct a chromosomal map of replication timing, the log ratio of early/late replication was calculated and plotted according to genomic position. Early-replicating regions are indicated as a positive log ratio; late-replicating regions are indicated by a negative log ratio. In this analysis, neighboring genomic probes tended to have comparable ratios, indicating that they are replicated at similar times in S phase. This observation was validated by autocorrelation analysis, which revealed significant autocorrelation (>0.05) for up to 58 and 35 fragments for HFL-1 and NC-NC lymphoid cells, respectively (Fig. 3A). The average spacing of 2 kb from the start of one fragment to the next fragment corresponds to an autocorrelation of >116 and >70 kb for HFL-1 and NC-NC cells, respectively. As a control, we pooled early- and late-replicating DNA and subsequently labeled and then hybridized this DNA to the array. As shown in Fig. 3B, no considerable enrichment of early- or late-replicating DNA was observed with this sample. Thus, we conclude that the observed enrichments in early- and late-replication timing are significant. As a separate control, we performed single-gene PCR on a subset of sequences. This analysis revealed that early-replicating regions detected by microarray are correspondingly abundant in the early S-phase fraction when assayed by PCR, whereas late-replicating regions are more abundant in the late S-phase fraction (Fig. 3C). To generate the complete profile for both lines as shown in Fig. 4, loess smoothing was performed to fit the replication-timing dataset along a best fit (21).
Fig. 3.
Control experiments. (a) Autocorrelation analysis to measure the chromosomal extent of similar replication timing. The autocorrelation function is calculated for an increasing number of neighboring sequences. Positive autocorrelation exists if neighboring spots tend to be alike, which reflects replication at a similar time in S phase. The autocorrelation plot of replication-timing ratios before smoothing (see Fig. 4) is shown to identify the lengths of significantly similar (>0.05) DNA replication timing. (b) DNA replication-timing profile of HFL-1 compared with control hybridization. The DNA replication-timing profile of HFL-1 (green) is plotted with the mixed early and late S-phase DNA control (gray). Shown are the first 20 Mb of chromosome 22 and the chromosomal positions of amplicons used in single-gene controls. (c) Single-gene control PCR of early- and late-replicating regions for the HFL-1 cell type. Amplicon names refer to microarray features from which ≈400-bp fragments were amplified. In each case, the control reaction confirms early- or late-replication timing as revealed by the microarray analysis.
Fig. 4.
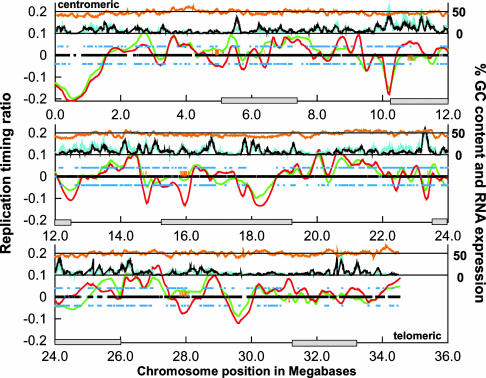
DNA replication profiles for HFL-1 and NC-NC. DNA replication-timing ratios (log2-transformed) were plotted according to chromosomal position. Shown is the nonrepetitive part of chromosome 22 extending from the centromere (Upper Left) to the telomere of the q arm (Lower Right). Data were smoothed by loess smoothing, and the best-fit plots are shown for HFL-1 (green line) and NC-NC (red line). The black mark along the baseline indicates regions of DNA replication timing that were present on the microarray and compared between both cell types. Associated orange hash marks indicate regions of significant difference (P < 0.05) in DNA replication timing. The percentage of chromosomal GC content is plotted as an orange line near the top of the plot as a sliding 100-kb window. The percentage of transcribed sequences for both cell types is plotted just below GC content as a sliding 100-kb window along the chromosome for HFL-1 (cyan line) and NC-NC (black line). Annotated genes that are expressed in HFL-1 and NC-NC are indicated as blue squares above (HFL-1) and below (NC-NC) the baseline. Locations of cytological G bands are indicated by gray boxes along the chromosome-position axis (22). (A larger version of this figure is available at http://array.mbb.yale.edu/chr22.)
DNA Replication-Timing Profile of Primary Human Lung Fibroblasts. Hybridizations of labeled BrdUrd-immunoprecipitated DNA to the chromosome 22 microarray produced a high-resolution replication-timing profile (Fig. 4). Analysis of the replication-timing profile of HFL-1 fibroblast cells revealed that both early- and late-replicating regions are present throughout chromosome 22. A total of 24 early-replicating and 24 late-replicating regions were identified (defined as segments above or below the baseline); the sizes of these regions vary from ≈100 kb to 2 Mb. Of these regions, 14 early- and 9 late-replicating regions are statistically significant, when a one SD cutoff (both above and below the median replication-timing ratio) is applied (Fig. 4).
DNA Replication-Timing Patterns Often, but Not Always, Correlate with Chromosome Cytology. Correlation of the different replicating regions in human lung fibroblasts with chromosome cytology reveals that several late-replicating regions overlap with expected G bands at the 850-band resolution (22). In particular, regions 10.0-13.0 Mb and 15.5-19.0 Mb exhibit strong overlap. These two regions also show a corresponding decrease in percentage of chromosomal GC content (Fig. 4). Two other G bands located at 5.2-7.2 Mb and 31-33.1 Mb occur in regions of the chromosome where the replication profile decreases and nears the baseline, suggesting that not all G bands replicate late. Interestingly, these regions do not have a lowered GC content. Indeed, it has been observed that the degree of GC content directly correlates with G-band replication timing (3, 23). In addition, the G band located at 23.4-25.9 Mb was not clearly identified as late-replicating. However, our DNA microarray contains few features in the region between 25 and 26 Mb, and our replication-timing profile identified a late-replicating, GC-poor region around 24 Mb at the centromeric end of this G band. As expected, pericentromeric DNA, composed of large regions of heterochromatin, replicates late in S phase. Finally, consistent with previous data (7, 24), the subtelomeric region replicates early in S phase.
Comparison of DNA Replication-Timing Profiles for the Lymphoblast and Fibroblast Cell Lines. Analysis of the DNA replication timing of lymphoblastoid NC-NC cells revealed extensive similarity to that of the fibroblasts. Similar to the fibroblast line, 26 regions of both early- and late-replication timing were observed with 15 and 13 peaks above and below 1 SD from the baseline, respectively. A t test of the replication profiles indicated that >99% of the chromosome fragments replicated at a similar time in lymphoblast and fibroblast cells. Interestingly, nine regions that differed in replication timing (P < 0.05) were observed. Six of these regions replicated earlier in the HFL-1 cells than in the NC-NC cells, whereas the remaining three replicated later. Thus, the timing of replication in several chromosomal regions is specific for a distinct cell type.
Correlation of DNA Replication Timing with Transcription. Total RNA was prepared from both cell lines, reverse-transcribed with oligonucleotide (dT) primers, labeled with Cy5, and hybridized to the chromosome 22 microarray. The results were analyzed both globally and as a function of chromosome position.
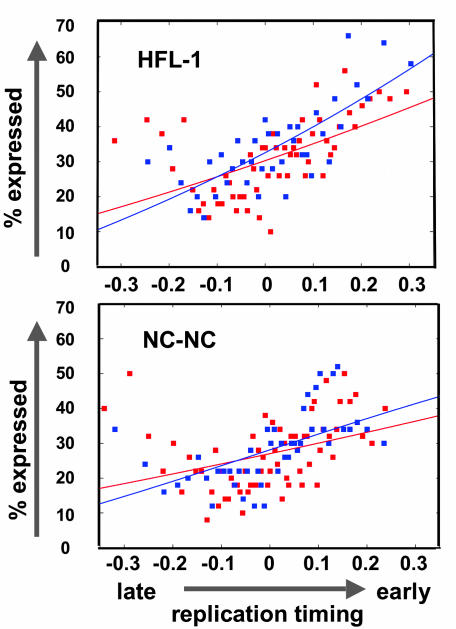
For the global analysis, microarray hybridization results from probes of annotated genes (Sanger data release 2.3 and Ensembl version 18.34.1) were binned into groups of 50 similar replication-timing ratios, and the percentage of features that showed at least a 2.5-fold increase in fluorescence from RNA over background (with a cutoff of 200 pixels) in each bin was calculated and plotted as a likelihood of expression relative to DNA replication timing (Fig. 5). A logistic regression curve was fitted to the data points. The strongly positive slope of the fibroblast and lymphoblast regression curves indicates that early-replicated DNA is more likely to be expressed than later-replicated DNA. Interestingly, in both cell types, very late-replicating DNA is a clear exception to this trend, because a higher percentage of this very late-replicating fraction is transcribed (note the increase at -0.3 in Fig. 5).
Fig. 5.
Relation of replication timing to transcription. mRNA expression and novel TARs correlate with early DNA replication timing. Microarray features were binned in groups of 50 features with similar replication-timing ratios. The percentage of expressed features in each bin was calculated and plotted according to DNA replication-timing ratio. A logistic regression curve was fitted to the data points. Red data points represent those fragments overlapping with annotated genes and blue data points represent those fragments that do not overlap with annotated genes
Novel Transcribed Sequences Replicate Early. In addition to studying known genes, we were interested in investigating the DNA replication timing of previously unidentified and unannotated novel transcribed sequences. These TARs constitute more than one-third of chromosomal transcription. Most of the transcripts have low coding potential, and neither their function nor whether they behave similar to annotated genes with respect to DNA replication timing is known. To address this question, we plotted the total percentage of RNA expression against DNA replication timing separately for annotated and unannotated sequences (Fig. 5). This global analysis revealed that TARs tend to replicate earlier than nontranscribed sequences, suggesting that their activity is linked to replication timing in a manner similar to that of annotated genes. Interestingly, in both cell types, the correlation of TARs and early replication is more pronounced than in annotated genes.
Differential Replication Timing of an Ig Locus Correlates with Expression. In addition to the global comparison, we examined in detail those regions that showed differential replication timing or expression in the two cell types. In the majority of differentially replicated regions, we did not detect significant differences in gene expression. However, the Ig λ-like polypeptide 3 locus (IGLL3) is an exception. This gene was more highly expressed in the lymphoblastoid cells than in the fibroblasts, and the region in which it lies (9.0-9.5 Mb) is replicated significantly earlier in the lymphoblastoid cells. Thus, in this instance, replication timing correlates with differential gene expression. To investigate this observation further, we performed the “reverse” experiment and compared the replication timing of 55 genes that were differentially expressed as measured in both our chromosomal array and Affymetrix cDNA arrays (see Materials and Methods). These genes showed no differences in replication timing at any resolution, suggesting that in the genomic regions analyzed, changes in expression are associated with changes in replication at only a small subset of loci.
Highly Transcribed Late-Replicating Regions. We also examined DNA replication timing and expression as a function of chromosome position. DNA replication timing and total chromosomal RNA expression for both cell types was plotted as a sliding window of the percentage of transcribed microarray fragments in 100 kb, as described in ref. 9 (Fig. 4). Significant unannotated transcription covering an extensive region was found in the late-replicating region at 18.0 Mb and between 11.0 and 11.5 Mb (9). Although, in general, gene expression correlates well with early-replication timing along the chromosome, about six regions of late-replicating DNA demonstrated a high percentage of transcribed sequences. The regions, between 10 and 10.5, 18.0 and 18.2, and 23.0 and 23.5 Mb, replicated very late yet were rich in annotated, expressed genes. In addition, the late-replicating regions at ≈14.9, 17.8, and 24.1 Mb were also heavily transcribed. Thus, although our experiments revealed a global correlation between expression and early replication, there are specific regions that show an opposite trend, indicating that early replication is not a simple consequence of transcriptional activity, and vice versa.
Discussion
The regulation of replication timing, as well as its function in transcriptional regulation, is not well understood, and current models are largely based on the examination of a few genes. To examine this relationship more comprehensively, we have generated a high-resolution DNA replication-timing map for a mammalian chromosome and, in addition, compared replication timing in two cell types of different developmental origin. Our analysis reveals that human chromosome 22 contains 24-26 early- and late-replication regions ranging in size from 100 kb to 2 Mb. Many of these regions agree well with early- and late-replicating R and G bands, and, in general, we found the G bands of lowest GC to be the latest replicating, consistent with the work of others (3, 23).
Previous studies in both Drosophila and human cells have demonstrated that early timing of DNA replication correlates with gene expression on a global scale. Our study supports this finding but also extends this observation to previously unidentified TARs, which reside outside of annotated genes and constitute up to one-third of nuclear transcription (11). Interestingly, we find that TARs replicate even earlier than annotated genes. Transcribed regions have an open chromatin conformation (25), and it is tempting to speculate that TAR activity might facilitate early replication, perhaps by mediating a chromatin structure necessary to initiate early origin firing (26). Alternatively, early replication itself could influence the transcriptional activity of these sequences.
Although our global analysis of transcribed sequences revealed a correlation between transcribed sequences and replication timing, examination of all chromosomal sequences reveals that some highly transcribed regions are replicated late. Only a few examples of this class of genes have been reported previously for mammalian cells (27, 28). Currently, the basis of this exception is not known, and we can only hypothesize that late replication might be involved or required for the proper regulation of this subset of sequences.
It has been observed that gene expression correlates with DNA replication timing. As gene expression can differ significantly among cell types in higher eukaryotes, chromosomal regions might be expected to show significant differences in replication timing among different cell types. However, our comparison of DNA replication timing in human fibroblast and lymphoblast cells revealed that replication timing of chromosome 22 is largely indistinguishable between these cell types; only nine areas of significant differences in timing were observed. One of these areas contains IGLL3, an Ig gene, which is expressed at higher levels and replicates earlier in the lymphoblastoid cells. We also observed 55 regions in which gene expression differs significantly between the two cells types that did not exhibit differences in replication timing, regardless of the window size examined. Thus, differences in replication timing do not necessarily correlate with differences in gene expression (29).
Can the results we obtained on chromosome 22 be extrapolated to the remainder of the human genome? Chromosome 22 replicates earlier than other chromosomes (7), has a high gene density, and contains many housekeeping genes. The expression of these annotated genes, in combination with the high TAR activity, may promote early replication by mediating a high percentage of active transcription throughout the chromosome, which might mask the potential effects of differentially expressed genes on replication. Consequently, replication-timing differences might be more pronounced in chromosomes or chromosomal regions that have a lower gene density. This possibility could be addressed by examining the replication timing of other human chromosomes by using a similar approach to the one presented here for chromosome 22.
Acknowledgments
We thank M. Smith for critical reading of the manuscript and Christiane Wirbelauer for technical assistance. This work was supported by Ruth L. Kirschstein National Institutes of Health-National Human Genome Research Institute Postdoctoral Fellowship HG002715 (to E.J.W.), a Knut and Alice Wallenberg Foundation Postdoctoral Fellowship (to O.E.), National Institutes of Health Grants DK44746 and HL65440 (to M.G.), the Novartis Research Foundation (E.J.O. and D.S.), and National Institutes of Health-National Human Genome Research Institute Center for Excellence in Genome Sciences Grant HG02357 (to the laboratories of M.S., M.G., and S.W.). S.K. is a fellow of the Jane Coffin Childs Memorial Fund for Biomedical Research.
Abbreviation: TAR, transcriptionally active region.
References
- 1.Huberman, J. A. & Riggs, A. D. (1968) J. Mol. Biol. 32, 327-341. [DOI] [PubMed] [Google Scholar]
- 2.Hand, R. (1978) Cell 15, 317-325. [DOI] [PubMed] [Google Scholar]
- 3.Dutrillaux, B., Couturier, J., Richer, C. L. & Viegas-Pequinot, E. (1976) Chromosoma 58, 51-61. [DOI] [PubMed] [Google Scholar]
- 4.Cimbora, D. M. & Groudine, M. (2001) Cell 104, 643-646. [PubMed] [Google Scholar]
- 5.Drouin, R., Holmquist, G. P. & Richer, C. L. (1994) Adv. Hum. Genet. 22, 47-115. [DOI] [PubMed] [Google Scholar]
- 6.Federico, C., Saccone, S. & Bernardi, G. (1998) Cytogenet. Cell Genet. 80, 83-88. [DOI] [PubMed] [Google Scholar]
- 7.Woodfine, K., Fiegler, H., Beare, D. M., Collins, J. E., McCann, O. T., Young, B. D., Debernadi, S., Mott, R., Dunham, I. & Carter, N. P. (2004) Hum. Mol. Genet. 13, 191-202. [DOI] [PubMed] [Google Scholar]
- 8.Schubeler, D., Scalzo, D., Kooperberg, C., van Steensel, B., Delrow, J. & Groudine, M. (2002) Nat. Genet. 32, 438-442. [DOI] [PubMed] [Google Scholar]
- 9.Rinn, J. L., Euskirchen, G., Bertone, P., Martone, R., Luscombe, N. M., Hartman, S., Harrison, P. M., Nelson, F. K., Miller, P., Gerstein, M., et al. (2003) Genes Dev. 17, 529-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kapranov, P., Cawley, S. E., Drenkow, J., Bekiranov, S., Strausberg, R. L., Fodor, S. P. & Gingeras, T. R. (2002) Science 296, 916-919. [DOI] [PubMed] [Google Scholar]
- 11.Kampa, D., Cheng, J., Kapranov, P., Yamanaka, M., Brubaker, S., Cawley, S., Drenkow, J., Piccolboni, A., Bekiranov, S., Helt, G., et al. (2004) Genome Res. 14, 331-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bruel, S. D., Bradley, K. H., Hance, A. J., Schafer, M. P., Berg, R. A. & Crystal, R. G. (1980) J. Biol. Chem. 255, 5250-5260. [PubMed] [Google Scholar]
- 13.MacLeod, R. A. & Bryant, P. E. (1992) Mutagenesis 7, 285-290. [DOI] [PubMed] [Google Scholar]
- 14.Cimbora, D. M., Schubeler, D., Reik, A., Hamilton, J., Francastel, C., Epner, E. M. & Groudine, M. (2000) Mol. Cell. Biol. 20, 5581-5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lieb, J. D., Liu, X., Botstein, D. & Brown, P. O. (2001) Nat. Genet. 28, 327-334. [DOI] [PubMed] [Google Scholar]
- 16.van Steensel, B., Delrow, J. & Henikoff, S. (2001) Nat. Genet. 27, 304-308. [DOI] [PubMed] [Google Scholar]
- 17.Luscombe, N. M., Royce, T. E., Bertone, P., Echols, N., Horak, C. E., Chang, J. T., Snyder, M. & Gerstein, M. (2003) Nucleic Acids Res. 31, 3477-3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quackenbush, J. (2002) Nat. Genet. 32, 496-501. [DOI] [PubMed] [Google Scholar]
- 19.Schrock, E., DuManoir, S., Veldman, T., Schoell, B., Wienberg, J., Ferguson-Smith, M. A., Ning, Y., Ledbetter, D. H., Bar-Am, I., Soenksen, D., et al. (1996) Science 273, 494-497. [DOI] [PubMed] [Google Scholar]
- 20.Hansen, R. S., Canfield, T. K., Lamb, M. M., Gartler, S. M. & Laird, C. D. (1993) Cell 73, 1403-1409. [DOI] [PubMed] [Google Scholar]
- 21.Cleveland, W. S. & Devlin, S. J. (1988) J. Am. Stat. Assoc. 83, 596-610. [Google Scholar]
- 22.Francke, U. (1994) Cytogenet. Cell Genet. 6, 206-219. [DOI] [PubMed] [Google Scholar]
- 23.Saccone, S. & Bernardi, G. (2001) Methods Cell Sci. 23, 7-15. [PubMed] [Google Scholar]
- 24.Smith, Z. E. & Higgs, D. R. (1999) Hum. Mol. Genet. 8, 1373-1386. [DOI] [PubMed] [Google Scholar]
- 25.Schubeler, D., MacAlpine, D. M., Scalzo, D., Wirbelauer, C., Kooperberg, C., van Leeuwen, F., Gottschling, D. E., O'Neill, L. P., Turner, B. M., Delrow, J., et al. (2004) Genes Dev. 18, 1263-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gilbert, D. (2002) Curr. Opin. Cell Biol. 14, 377-383. [DOI] [PubMed] [Google Scholar]
- 27.Hatton, K. S., Dhar, V., Brown, E. H., Iqbal, M. A., Stuart, S., Didamo, V. T. & Schildkraut, C. L. (1988) Mol. Cell. Biol. 8, 2149-2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gartler, S. M., Goldstein, L., Tyler-Freer, S. E. & Hansen, R. S. (1999) Hum. Mol. Genet. 8, 1085-1089. [DOI] [PubMed] [Google Scholar]
- 29.Azuara, V., Brown, K. E., Williams, R. R., Webb, N., Dillon, N., Festenstein, R., Buckle, V., Merkenschlager, M. & Fisher, A. G. (2003) Nat. Cell Biol. 5, 668-674. [DOI] [PubMed] [Google Scholar]