Abstract
Using genome-wide information to understand holistically how cells function is a major challenge of the postgenomic era. Recent efforts to understand molecular pathway operation from a global perspective have lacked experimental data on phenotypic context, so insights concerning biologically relevant network characteristics of key genes or proteins have remained largely speculative. Here, we present a global network investigation of the genotype/phenotype data set we developed for the recovery of the yeast Saccharomyces cerevisiae from exposure to DNA-damaging agents, enabling explicit study of how protein–protein interaction network characteristics may be associated with phenotypic functional effects. We show that toxicity-modulating proteins have similar topological properties as essential proteins, suggesting that cells initiate highly coordinated responses to damage similar to those needed for vital cellular functions. We also identify toxicologically important protein complexes, pathways, and modules. These results have potential implications for understanding toxicity-modulating processes relevant to a number of human diseases, including cancer and aging.
Keywords: DNA damage, graph theory, DNA repair, signaling, systems and computational biology
Cells represent complex systems with thousands of proteins, carbohydrates, lipids, nucleic acids, and small molecules interacting to maintain growth and homeostasis. Such maintenance requires that cells appropriately respond to both endogenous and exogenous environmental cues. The recent completion of several genome projects provided us with parts lists of genes and proteins that contribute to maintaining growth and homeostasis. Our current challenge is to use this information, along with other extensive data sets, to understand how cells operate in an integrated manner to carry out phenotypic functions. Toward this goal, thousands of protein–protein interactions and genetic interactions have been mapped into complex networks for several organisms (1–10). Although some global network analysis has been performed on these interacting networks, such analyses have rarely been connected with systematic global genotype/phenotype information. Here we connect protein–protein interaction maps for the budding yeast Saccharomyces cerevisiae with a genomic-scale data set describing the phenotypic role of all nonessential yeast proteins in modulating toxicity after exposure to a number of DNA-damaging agents typical of those encountered in our endogenous and exogenous environment.
Graph theoretic approaches are now being used to study global properties of biological networks (11–23). However, network analyses are mostly carried out in the absence of functional information, and when functional information is present it is usually based on cataloged information assembled from an array of unrelated experiments. Tools permitting systematic network perturbations are crucial for establishing biologically meaningful network characteristics, and the S. cerevisiae single-gene deletion strain library provides a tool for such analyses. In principle, each gene deletion strain represents an engineered cell model in which one node (protein) and its corresponding edges (protein–protein interactions) have been removed from the yeast gene and protein network. High-throughput phenotypic studies that use gene deletion strains to identify associated phenotypic effects under specific experimental conditions [a procedure we have termed genomic phenotyping (24, 25)] provide biologically relevant data sets for network studies.
Methods
Genomic Phenotyping. Genomic phenotyping used 4,733 haploid S. cerevisiae single-gene deletion strains to identify deletions that affect growth (relative to wild type) upon exposure to the methylating agent methyl methanesulfonate (MMS), the bulky alkylating agent 4-nitroquinoline-N-oxide (4NQO), the oxidizing agent tert-butyl hydroperoxide (t-BuOOH), or 254-nm UV radiation. Only the 4,733 nonessential yeast genes could be examined because deletion of the essential genes in haploid strains is lethal, a priori. A sensitive, reproducible, multireplicate, and multidose screen was developed to monitor individual strain growth after exposure to the four DNA-damaging agents; results of this screen have been described (25). The term “toxicity-modulating protein” in this study corresponds to the product of the gene deleted in a strain displaying significantly more growth inhibition than wild type after exposure to one of the four DNA-damaging agents.
Phenotypic Annotation of the Yeast Interactome. We used 14,493 protein–protein interactions for 4,686 S. cerevisiae proteins found in the Database of Interacting Proteins (26) as of November 2002 to build the yeast protein interactome. A smaller network based on high-confidence interactions was also used (see Supporting Methods, which is published as supporting information on the PNAS web site). Essential, toxicity-modulating, and no-phenotype classifications were based on results obtained by the S. cerevisiae gene deletion consortium (Essential) and the high-throughput genomic phenotyping study (25). MMS-, t-BuOOH-, 4NQO-, and UV-modulating proteins correlate to gene deletion strains that exhibit impaired growth compared with wild type after agent treatment. The toxicity-modulating phenotype represents gene-deletion strain sensitivity to one of these DNA-damaging agents relative to wild type. The no-phenotype classification indicates that gene deletion strains had no relative growth defects after agent treatment. The resulting network structure represents a phenotypically annotated interactome of essential, toxicity-modulating, and no-phenotype proteins (Fig. 1A and Tables 3 and 4, which are published as supporting information on the PNAS web site). Networks specific for essential, toxicity-modulating, and no-phenotype proteins were also built in silico by identifying the proteins (nodes) in a given category and their associated protein–protein interactions (edges).
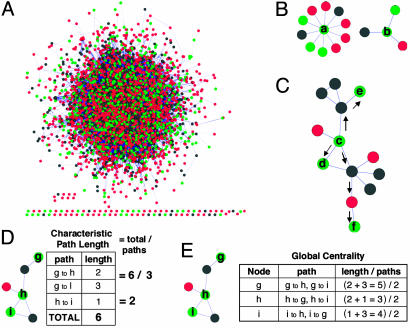
Fig. 1.
Yeast protein categories and global network measures. (A) Proteins (4,684) from yeast connected by 14,993 protein–protein interactions. Proteins that modulate toxicity are shown in green, essential proteins are shown in black, and proteins associated with the no-phenotype category are shown in red. (B) Degree of a node in a graph. As an example, the degree of protein a is 10, whereas the degree of protein b is 3. (C) Shortest path length. The shortest path between proteins c and d is 1; whereas, between nodes c and e, it is 2; and, for nodes c and f, it is 3. (D) Characteristic path length. (E) Global centrality. In the figure, protein h is most central relative to proteins i and g.
Randomizations, P Values, and Network Measures. Eight sets of randomizations based on the eight phenotypic categories were generated. In each set, 1,000 randomized networks were obtained based on 1,000 independent experiments consisting of randomly selected nodes from the full yeast network. To factor out bias introduced by essential genes, randomizations corresponding to the nonessential categories were generated by randomly selecting nodes from the nonessential yeast network. Based on these randomizations, P values were computed by using a two-sided hypothesis test with a Normal distribution assumption (27). Applying the randomizations to the overall underlying yeast protein network by random node selection contrasts with other randomization network studies (12, 28) in which networks are constructed through random connections between nodes. Biased randomizations were also performed. Details are described in Supporting Methods. Several network measures were computed and are discussed in detail in Results (Fig. 1; see Fig. 4A) and Supporting Methods.
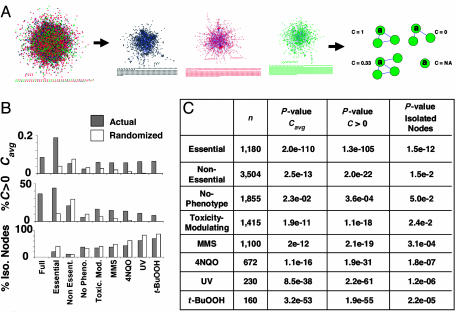
Fig. 4.
Newly defined networks and clustering coefficient analysis. (A) Derivations of new networks. Networks comprised of only essential proteins and connecting edges are shown in black, proteins that prevent agent-induced cell death and connecting edges are shown in green, and no-phenotype proteins and connecting edges are shown in red. The clustering coefficient, C, can be determined for each protein to identify the degree of connectivity between a given protein's neighbors. (B) Clustering coefficient analysis. The clustering coefficient average, Cavg, is computed for each network and compared with the average obtained from 1,000 randomized networks (randomized). The percent of nodes with a non-zero clustering coefficient (C > 0) as well as the percent of isolated nodes are also computed. (C) The number of proteins (n) in each specified network along with the P values.
Supporting Information. For further information, see Tables 3–13 and Figs. 6–9, which are published as supporting information on the PNAS web site.
Results
Global Properties of Selected Proteins Within the Full Yeast Interactome. We exploited two fundamental network metrics to extract phenotype-dependent global network characteristics for essential, toxicity-modulating, and no-phenotype proteins in the context of the full yeast network (Fig. 1A). We determined the degree of each node, which details the number of interacting partners for each node in the network (Fig. 1B), and the shortest-path distance between pairs of nodes, which details the shortest-edge distance between similarly categorized pairs of proteins, allowing for transitions through proteins in other categories (Fig. 1C). From the shortest-path distance we computed two additional measures: the characteristic path length (Fig. 1D) defined as the shortest-path distance averaged over all pairs of proteins and a global centrality measure (Fig. 1E), which, for a given protein, computes the average shortest-path distance to every other similarly categorized protein in the network. Each of these measures reveals insight into the architecture of toxicity-modulating pathways.
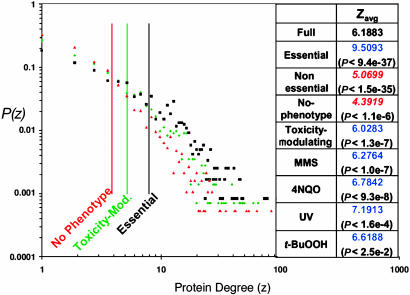
Degree distribution. The degree distributions and average degree for the nodes in the full yeast interactome and for the nodes in each phenotypic category are shown in Fig. 2. These distributions are characterized by a number of highly connected proteins, or hubs, as previously observed for a smaller yeast protein–protein interaction network (15, 17). When the full network is divided into essential and nonessential proteins, it is clear that essential proteins have a significantly higher average degree (P < 10-36). The results illustrated in Fig. 2 show that not only is the essential proteins distribution skewed toward higher degree, in agreement with previous results (15), but a similar trend is also true for toxicity-modulating proteins. Among the entire set of nonessential proteins, i.e., the set for which we have phenotypic data, toxicity-modulating proteins have a higher average degree than the entire set of nonessential proteins and the no-phenotype proteins. On average each category of toxicity-modulating proteins, including the collective toxicity-modulating category, contains significantly more direct interactions than randomly selected proteins, (P values range from <0.03 to <10-7) as well as nonessential and no-phenotype proteins. Moreover, the no-phenotype distribution shows the opposite characteristic, with significantly fewer direct interactions than the randomly selected proteins (P < 10-6), fewer direct interactions than the nonessential category, and fewer direct interactions than for the full network. A protein with high degree (>15 direct interactions) is two times more likely to be essential than a random protein in the yeast network. Furthermore, a nonessential protein with a high degree is one and a half times more likely to be important for toxicity modulation than a random nonessential yeast protein. In contrast, a high-degree protein is more than a third less likely to be involved in metabolism than a random protein in the yeast network (see Topological Organization of Other Functional Yeast Networks).
Fig. 2.
Degree distributions of selected proteins. The proportion of proteins normalized to 1, P(z), with a given number of interacting proteins, z, in the underlying full yeast network is plotted for the essential (black squares), toxicity-modulating (green diamonds), and no-phenotype (red triangles) proteins. The solid vertical lines represent the average degree. (Right) Average protein degree (zavg) for different categories of proteins along with P values. Blue font indicates an average greater than the corresponding randomized average, and red italic font indicates an average smaller than the corresponding randomized average.
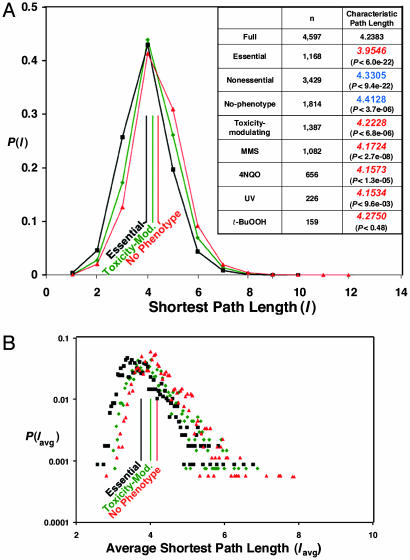
Shortest-path distribution, characteristic path length, and global centrality distribution. The existence of a few highly connected nodes (hubs) holding together a large number of lesser-connected nodes adds shortcuts into a network and creates a smaller average shortest-path length between any two nodes. We computed the shortest-path length distribution, the characteristic path length, and the global centrality distribution for the full yeast protein network and for each phenotypic category; in calculating the distance between two proteins in one particular category, the shortest path can pass through proteins belonging to other categories (see Fig. 1C). Results for three shortest-path-length and global-centrality distributions are shown in Fig. 3 A and B, respectively (the rest are in Fig. 6 and Table 6). Characteristic path length for all categories is given in the Fig. 3A Inset. These results follow similar significant trends as observed for the degree distributions and average degree. This shortest-path analysis may provide an idea of network navigability and of efficiency with which a perturbation can spread throughout the network. However, in analyses of this type, it is assumed that the connections between each node (i.e., the edges) are equivalent, which seems unlikely to be true in a biological system. Ultimately, metrics to describe the attributes of each edge in the protein–protein interaction network will be needed to be quantitative with respect to network navigability.
Fig. 3.
Shortest-path length, characteristic path length, and global centrality of selected proteins. (A) The proportion of pairs of proteins normalized to one with a given shortest-path length in the overall full yeast network, P(l), is plotted for the essential (black squares), toxicity-modulating (green diamonds), and no-phenotype (red triangles) proteins. The solid vertical lines give the characteristic path length. (Inset) The number of proteins (n) and the characteristic path length, along with P values. Blue font indicates an average greater than the corresponding randomized average, and red italic font indicates an average smaller than the corresponding randomized average. (B) The proportion of proteins with a given average shortest-path length (global centrality) is plotted for the essential (black squares), toxicity-modulating (green diamonds), and no-phenotype (red triangles) proteins.
Network properties of proteins with varying sensitivities to damage. The suggestion that the “importance” of a protein may be reflected by its connectivity degree was further investigated by quantitatively categorizing the toxicity-modulating proteins in high, medium, and low sensitivity categories and calculating the average degree and characteristic path length of each category (see Supporting Methods). Results are given in Table 7 and indicate that highly sensitive mutants are distinct in topological properties from their low-sensitivity counterparts. In most of the cases, high sensitivity corresponds to higher connectivity degree and shorter characteristic path length, further supporting the hypothesis that a protein with higher degree and greater centrality is on average more important for toxicity-modulation than a lowly connected and less central one.
Synthesis of Phenotypic Subnetworks. To gain further insight into the organization and local environment of proteins in each category, subnetworks were compiled composed solely of protein–protein interactions between proteins within a given phenotypic category; the newly defined subnetworks have nodes corresponding to proteins exhibiting a given phenotype and edges representing experimentally characterized protein–protein interactions. We thus generated seven new subnetwork structures. Fig. 4A illustrates the full network (as in Fig. 1A), the essential subnetwork, the no-phenotype subnetwork, and the collective toxicity-modulating subnetwork that includes MMS-, 4NQO-, UV-, and t-BuOOH-modulating proteins. The individual MMS, 4NQO, UV, and t-BuOOH subnetworks are shown in Fig. 7.
Network connectivity. The landscape of connected components in each newly defined network was explored to probe the connectivity of each structure. In each of the seven networks (except the t-BuOOH-modulating network) one large connected component emerged (Table 1 and Figs. 4A and 7). The size of these components was significantly larger for the essential and each of the toxicity-modulating subnetworks (except for t-BuOOH) than the size expected if the nodes were selected at random from the full or nonessential yeast network (P values from <0.009 to <10-8). These observations indicate that essential and toxicity-modulating proteins are relatively highly connected, suggesting that cohesive signaling pathways, protein complexes, and biochemical pathways are at least partially represented in these subnetworks. Local protein environments in the phenotypic subnetworks. We further investigated organization of the newly defined subnetworks by using clustering coefficient analysis, which measures whether direct, first-degree partners of a particular node interact with each other. Tendency to form protein clusters is significantly overemphasized in the essential and toxicity-modulating networks (Fig. 4B). Essential, MMS-modulating, and 4NQO-modulating networks are around five times more clustered than what would be expected from a random sampling of nodes from the full yeast network. Moreover, clustering in the UV-modulating network is more than one order of magnitude higher than expected by a random selection of nodes, whereas the t-BuOOH-modulating network is more than two orders of magnitude more clustered than the corresponding randomized network. P values for the enrichment of protein clusters in these phenotypically derived subnetworks range from <10-10 to <10-109. In contrast, the no-phenotype network is less clustered than the corresponding randomized network (P < 0.023).
Table 1. The large connected component (LC) size in newly defined subnetworks.
| Subnetworks | No. of proteins | No. of proteins in LC |
|---|---|---|
| Full | 4,684 | 4,597 |
| Essential | 1,180 | 914 (P < 1.4 × 10-9) |
| Nonessential | 3,504 | 2,967 (P < 1.8 × 10-2) |
| No-phenotype | 1,855 | 981 (P < 7.4 × 10-2) |
| Toxicity-modulating | 1,415 | 851 (P < 8.9 × 10-3) |
| MMS | 1,100 | 639 (P < 1.4 × 10-4) |
| 4NQO | 672 | 343 (P < 1.4 × 10-5) |
| UV | 230 | 31 (P < 3.8 × 10-5) |
| t-BuOOH | 160 | 8 (P < 0.3) |
The large connected component size in all subnetworks was larger than the average obtained from the corresponding randomized networks, except for the nonessential and no-phenotype subnetworks, which had smaller large connected component size than the average obtained from the randomized networks. P values are shown in parentheses below the values for the large connected component size.
The average clustering coefficient (Cavg) results suggest that phenotypic effects of the proteins in these subnetworks are governed by denser-than-normal, interconnected biochemical pathways, signaling pathways, and protein complexes. This notion is supported by a closer look at the distribution of the clustering coefficients (Fig. 4 B and C). The percentage of nodes having a non-zero clustering coefficient indicates that a certain degree of local clustering has transpired. This percentage is significantly higher for the essential and toxicity-modulating subnetworks and significantly smaller for the no-phenotype subnetwork, compared with randomized counterparts (P values range from <10-3 to <10-104). In corollary, the number of isolated nodes in the essential and toxicity-modulating subnetworks are significantly smaller than that expected from a random selection (P values range from <0.0024 to 10-11), whereas the no-phenotype subnetwork has a larger proportion of isolated nodes (P < 0.05).
We further investigated the extent to which the high degree of clustering could be a simple consequence of the high average node degree observed in the essential and toxicity-modulating networks by carrying out a new set of biased randomizations that preserve the proportion of high degree nodes in each network (see Supporting Methods). Results are shown in Table 9 and indicate that even when the randomized networks are constrained to have the same average degree as the tested network, the average clustering coefficient as well as the proportion of nodes with a non-zero clustering coefficient are significantly higher for the toxicity-modulating and essential networks than for the randomized networks. Thus, the degree of local clustering is not a consequence of the presence of higher degree nodes but a reflection of another underlying phenomenon related to protein complexes and dense signaling pathways.
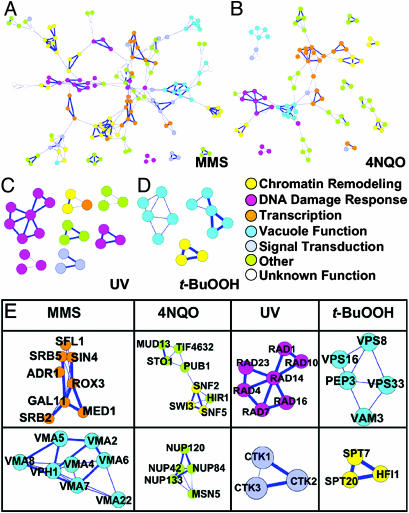
Identification of Toxicologically Important Protein Complexes and Signaling Pathways. Our clustering coefficient analysis identifies local neighbor interactions that we in turn have used to build higher order complexes in silico with the program cytoscape (29). Fig. 5 A–D contains every protein in each of the toxicity-modulating subnetworks that has a non-zero clustering coefficient; these proteins are displayed with their corresponding protein–protein interactions (edges) derived from each subnetwork; the edge is shown in bold where the protein–protein interaction has previously been characterized in a biological context [i.e., not just as part of a high throughput screen (www.yeastgenome.org)]. All of the proteins in Fig. 5 A–D are shown with gene names in Fig. 8 and Table 10. The bold edges in Fig. 5 indicate that previously recognized complexes, pathways, and signaling modules are represented, even though many of them were not previously recognized as being important for modulating toxicity after exposure to a DNA-damaging agent. [All of the protein nodes shown in Fig. 5 play a role in modulating toxicity, but they are colored to represent their cellular function (www.yeastgenome.org).] As expected, groups of proteins that participate in coordinated DNA damage responses (i.e., nucleotide excision repair, mismatch repair, or DNA damage checkpoints) were identified among the clustered proteins in the toxicity-modulating subnetworks (Fig. 5E). Groups of proteins involved in transcription regulation and chromatin remodeling are also well represented, and these include components of the Spt-Ada-Gcn5 acetyltransferase (SAGA) regulatory complex (MMSS, 4NQOS, and t-BuOOHS), RNA polymerase II complex (MMSS, 4NQOS, and UVS) and SWI/SNF complex (4NQOS). Signal transduction is represented for MMS and UV toxicity-modulation with the cyclin-dependent kinase that phosphorylates the C terminus of RNA polymerase II and contains CTK1, CTK2, and CTK3 as subunits (www.yeastgenome.org). Surprisingly, protein complexes and pathways involving the nuclear pore complex (NUP proteins), RNA metabolism (MUD, STO1, PUB, and TIF proteins), vacuolar function and targeting (VMA proteins) are also represented upon visualizing the proteins with non-zero clustering coefficients (i.e., C > 0) from the MMS-, 4NQO-, UV- and t-BuOOH-modulating subnetworks (Fig. 5E). Moreover, Fig. 5 suggests that other known and putative complexes await further investigation into their role in modulating toxicity after exposure to the DNA-damaging agents, roles that may involve protein complexes, biochemical pathways, or signaling modules.
Fig. 5.
MMS, 4NQO, UV, and t-BuOOH protein networks with C > 0. Subnetworks composed of MMS-modulating (A), 4NQO-modulating (B), UV-modulating (C), and t-BuOOH-modulating (D) proteins with a non-zero clustering coefficient. Thick blue lines represent previously reported protein complexes. Circles are color-coded to represent basic cellular processes carried out by each protein (for all protein names, see Fig. 8). (E) Selected complexes identified by using clustering coefficient analysis. (Upper) From left to right, RNA polymerase II holoenzyme, SWI/SNF complex, nucleotide excision repair pathway, and putative vacuolar sorting subnetwork. (Lower) From left to right, mediator complex and vacuolar H-ATP assembly complex, nuclear pore complex, C-terminal domain kinase I complex, and Spt-Ada-Gcn5 acetyltransferase transcriptional regulatory complex.
Topological Organization of Other Functional Yeast Networks. One might ask whether all biomolecular networks involved in cell functions should share these topological features described previously for essential proteins and now here for toxicity-modulating proteins. Therefore, we also applied our approach to the metabolic network reconstructed by Forster et al. (30), which is based on currently available genomic, biochemical, and physiological information. The network contains 708 structural ORFs, of which 508 are present in the yeast protein–protein interactome used in this study. Of the 508 proteins, 395 are nonessential proteins. Table 2 shows all network measures used in this study, computed for both the full metabolic network and for the metabolic network composed of only nonessential proteins; the phenotypic networks presented earlier are also shown for comparison. The results indicate that the metabolic network exhibits properties more similar to the randomized (and no-phenotype) networks than to the essential and toxicity-modulating networks. Thus, not all protein networks involved in important cell functions share the topological features of essential protein networks.
Table 2. Network measures for the metabolic network compared with the other networks.
| Networks | No. of proteins | Average degree | Characteristic path length | No. of proteins in LC | Cavg | C > 0, % | Isolated nodes, % |
|---|---|---|---|---|---|---|---|
| Full | 4,684 | 6.1883 | 4.2383 | 4,597 | 0.0846 | 36.3 | 0 |
| Essential | 1,180 | 9.5093 | 3.9546 | 914 | 0.1879 | 44.2 | 19.9 |
| (P < 9.4 × 10-37) | (P < 6.0 × 10-22) | (P < 1.4 × 10-9) | (P < 2.0 × 10-110) | (P < 1.3 × 10-105) | (P < 1.5 × 10-12) | ||
| Noness. | 3,504 | 5.0699 | 4.3305 | 2,967 | 0.0529 | 20.7 | 11.0 |
| (P < 1.5 × 10-35) | (P < 9.4 × 10-22) | (P < 1.8 × 10-2) | (P < 2.5 × 10-13) | (P < 2.0 × 10-22) | (P < 1.5 × 10-2) | ||
| Metabolic | 508 | 4.6024 | 4.2888 | 93 | 0.0276 | 3.5 | 64.8 |
| (P < 4 × 10-4) | (P < 0.3) | (P < 0.98) | (P < 0.15) | (P < 0.8) | (P < 0.68) | ||
| No-phenotype | 1,855 | 4.3919 | 4.4128 | 981 | 0.0227 | 5.9 | 36.4 |
| (P < 1.1 × 10-6) | (P < 3.7 × 10-6) | (P < 7.4 × 10-2) | (P < 2.3 × 10-2) | (P < 3.6 × 10-4) | (P < 5.0 × 10-2) | ||
| Toxicity-modulating | 1,415 | 6.0283 | 4.2228 | 851 | 0.0584 | 16.3 | 34.5 |
| (P < 1.3 × 10-7) | (P < 6.8 × 10-6) | (P < 8.9 × 10-3) | (P < 1.9 × 10-11) | (P < 1.1 × 10-18) | (P < 2.4 × 10-2) | ||
| Metabolic-noness. | 395 | 4.6127 | 4.3158 | 21 | 0.0240 | 3.3 | 70.4 |
| (P < 0.3) | (P < 0.8) | (P < 0.85) | (P < 6 × 10-4) | (P < 1.2 × 10-3) | (P < 0.4) |
Values in bold indicate that the measured value is smaller than the one obtained by using the randomized networks; values in italics indicate that the measured value is greater than the one obtained by using the radomized networks. Noness., Nonessential.
Robustness of the Network Results. All network measures so far relied on protein–protein interactions obtained from the Database of Interacting Proteins (26), which includes high-throughput, genome-wide data, such as yeast two-hybrid (2, 5, 7) and mass spectrometric analyses of protein complexes (1, 31) as well as interactions collected from small-scale screens in hundreds of individual research papers. We performed computations on an exhaustive yeast interactome that includes all reported protein–protein interactions to evaluate the network characteristics of all our identified toxicity-modulating proteins. However, false-positive protein–protein interactions might affect our observed trends, so it is important to assess the robustness of the results reported here. We have recomputed all network measures by using an additional smaller yeast protein–protein interaction network: the core yeast interactome obtained from the Database of Interacting Proteins as of October 2004 (2,628 proteins and 6,337 interactions) (26, 32) (see Supporting Methods). Results are shown in Table 12 and are in agreement with those obtained by using the complete yeast interactome, indicating that the results are robust to false-positive protein–protein interactions.
We have also identified the party and date hubs [as recently defined by Han et al. (23)] in the toxicity-modulating networks (Table 13). Although the majority of these hubs are essential, most of the remaining nonessential party and date hubs are involved in modulating toxicity. These results further emphasize the topological similarities between essential and toxicity-modulating proteins.
Discussion
We have presented a systematic investigation of global protein networks in a phenotypic context. Recovery from exposure to DNA-damaging agents was chosen because of the wide range of cellular activities required to prevent cell death and because of the association of many toxicity-modulating pathways with human diseases, such as cancer, aging, and other degenerative diseases. Our findings suggest that toxicity-modulating proteins have attributes somewhat similar to essential proteins. All of the measures reported here lead us to the same conclusion. Specifically, toxicity-modulating proteins have greater direct interactions, smaller shortest paths, are more connected, and are significantly more clustered than the average yeast protein, suggesting that there exists a higher-order organization for these toxicity-modulating networks. These results reflect two underlying phenomena: the toxicity-modulating proteins have more hubs, which allow them to be more connected and to exhibit shorter path lengths, and the network composed of these proteins is more clustered, which indicates the existence of many protein complexes and dense signaling pathways. We were also able to identify, by using global measures, targeted pathways and complexes essential for modulating toxicity.
Because of their phenotypic role in cell survival, toxicity-modulating proteins might represent a middle ground between essential and no-phenotype proteins. Essential proteins dictate cell viability under all conditions of life and their place in the network makes them the most centralized. The centrality of essential proteins may serve to provide facile communication between the processes vital for maintaining proper cellular function and homeostasis. Toxicity-modulating proteins are less centralized compared with essential proteins, perhaps because they are only required for cell viability some of the time (i.e., during stress). It may be that toxicity-modulating proteins are more centralized in the network than no-phenotype proteins, because, under stressful conditions, toxicity-modulating proteins need to rapidly coordinate a wide variety of cellular processes that ultimately dictate cellular viability (25). For MMS, it has been postulated that extensive damage occurs to DNA, RNA, lipids, and proteins (24, 25, 33, 34); it thus seems likely that a highly coordinated response to carry out repair, removal, and replacement of a multitude of damaged molecules is required for survival. Short path lengths by means of access to a number of highly connected nodes might serve to provide toxicity-modulating proteins with a means of optimizing cellular responses that together prevent damage-induced cell death.
Our metabolic network results indicate that other cellular networks do not necessarily share similar quantitative features. As can be seen in Table 2, the metabolic network has lower average degree than the corresponding randomized network, similar to the nophenotype network. We can speculate why the individual network metrics are so different for the metabolic network and how the toxicity-modulating network metrics provide a biological advantage for achieving an effective toxicity-modulating response. One possibility is that small diffusible metabolites important in metabolism may be crucial components for keeping the network connected, thereby contrasting with direct protein–protein interactions that we presume are more important in signaling pathways. This contrast would be consistent with a view that signaling and other regulatory networks may be more complex in organizational structure than those devoted to core functions such as metabolism and energy generation (35).
Finally, toxicity-modulating pathways may be highly conserved across evolution (certainly this is true for DNA repair pathways) and as a result, it is expected that the pathway characteristics unraveled for S. cerevisiae will parallel those in higher organisms. Protein–DNA interactions represent an additional form of connectivity for this regulatory network, as for many others. We have restricted our attention to protein–protein interactions in this present study to focus on a relatively well-defined time frame for network operation; this restriction also permitted us to make an important, clear distinction by comparison to the metabolic pathway protein–protein interaction network. Nonetheless, expanding our scope to the protein–DNA interactions in the toxicity-modulating network will be useful and we are underway with a corresponding effort.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants RO1-CA-55042, U19-ES11399, and P30-ES02109 (to L.D.S.); National Research Service Award F32-ES11733; a Merck–Massachusetts Institute of Technology Computational and Systems Biology Fellowship (to T.J.B.); the Defense Advanced Research Projects Agency Bio:Info:Micro Program; the Army Institute for Collaborative Biotechnologies; the National Institute of General Medical Sciences Cell Decision Processes Center (D.A.L. and A.V.O.); and a Merck–Massachusetts Institute of Technology Fellowship in Bioinformatics (to M.R.S.). L.D.S. is an Ellison American Cancer Society Research Professor.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: MMS, methyl methanesulfonate; 4NQO, 4-nitroquinoline-N-oxide; t-BuOOH, tert-butyl hydroperoxide.
References
- 1.Ho, Y., Gruhler, A., Heilbut, A., Bader, G. D., Moore, L., Adams, S. L., Millar, A., Taylor, P., Bennett, K., Boutilier, K., et al. (2002) Nature 415, 180–183. [DOI] [PubMed] [Google Scholar]
- 2.Uetz, P., Giot, L., Cagney, G., Mansfield, T. A., Judson, R. S., Knight, J. R., Lockshon, D., Narayan, V., Srinivasan, M., Pochart, P., et al. (2000) Nature 403, 623–627. [DOI] [PubMed] [Google Scholar]
- 3.Tong, A. H., Evangelista, M., Parsons, A. B., Xu, H., Bader, G. D., Page, N., Robinson, M., Raghibizadeh, S., Hogue, C. W., Bussey, H., et al. (2001) Science 294, 2364–2368. [DOI] [PubMed] [Google Scholar]
- 4.Tong, A. H., Lesage, G., Bader, G. D., Ding, H., Xu, H., Xin, X., Young, J., Berriz, G. F., Brost, R. L., Chang, M., et al. (2004) Science 303, 808–813. [DOI] [PubMed] [Google Scholar]
- 5.Fromont-Racine, M., Rain, J. C. & Legrain, P. (1997) Nat. Genet. 16, 277–282. [DOI] [PubMed] [Google Scholar]
- 6.Ito, T., Chiba, T., Ozawa, R., Yoshida, M., Hattori, M. & Sakaki, Y. (2001) Proc. Natl. Acad. Sci. USA 98, 4569–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rain, J. C., Selig, L., De Reuse, H., Battaglia, V., Reverdy, C., Simon, S., Lenzen, G., Petel, F., Wojcik, J., Schachter, V., et al. (2001) Nature 409, 211–215. [DOI] [PubMed] [Google Scholar]
- 8.Giot, L., Bader, J. S., Brouwer, C., Chaudhuri, A., Kuang, B., Li, Y., Hao, Y. L., Ooi, C. E., Godwin, B., Vitols, E., et al. (2003) Science 302, 1727–1736. [DOI] [PubMed] [Google Scholar]
- 9.Li, S., Armstrong, C. M., Bertin, N., Ge, H., Milstein, S., Boxem, M., Vidalain, P. O., Han, J. D., Chesneau, A., Hao, T., et al. (2004) Science 303, 540–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walhout, A. J., Sordella, R., Lu, X., Hartley, J. L., Temple, G. F., Brasch, M. A., Thierry-Mieg, N. & Vidal, M. (2000) Science 287, 116–122. [DOI] [PubMed] [Google Scholar]
- 11.Achacoso, T. & Yamamoto, W. (1992) Neuroanatomy of C. elegans for Computation (CRC, Boca Raton, FL).
- 12.Watts, D. J. & Strogatz, S. H. (1998) Nature 393, 440–442. [DOI] [PubMed] [Google Scholar]
- 13.Jeong, H., Tombor, B., Albert, R., Oltvai, Z. N. & Barabasi, A. L. (2000) Nature 407, 651–654. [DOI] [PubMed] [Google Scholar]
- 14.Wagner, A. & Fell, D. A. (2001) Proc. R. Soc. London Ser. B 268, 1803–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeong, H., Mason, S. P., Barabasi, A. L. & Oltvai, Z. N. (2001) Nature 411, 41–42. [DOI] [PubMed] [Google Scholar]
- 16.Maslov, S. & Sneppen, K. (2002) FEBS. Lett. 530, 255–256. [DOI] [PubMed] [Google Scholar]
- 17.Maslov, S. & Sneppen, K. (2002) Science 296, 910–913. [DOI] [PubMed] [Google Scholar]
- 18.Guelzim, N., Bottani, S., Bourgine, P. & Kepes, F. (2002) Nat. Genet. 31, 60–63. [DOI] [PubMed] [Google Scholar]
- 19.Lee, T. I., Rinaldi, N. J., Robert, F., Odom, D. T., Bar-Joseph, Z., Gerber, G. K., Hannett, N. M., Harbison, C. T., Thompson, C. M., Simon, I., et al. (2002) Science 298, 799–804. [DOI] [PubMed] [Google Scholar]
- 20.Ren, B., Robert, F., Wyrick, J. J., Aparicio, O., Jennings, E. G., Simon, I., Zeitlinger, J., Schreiber, J., Hannett, N., Kanin, E., et al. (2000) Science 290, 2306–2309. [DOI] [PubMed] [Google Scholar]
- 21.Barabasi, A.-L. & Oltvai, Z. N. (2004), Nat. Rev. Genet. 5, 101–113. [DOI] [PubMed] [Google Scholar]
- 22.Dezso, Z., Oltvai, Z. N. & Barabasi, A. L. (2003) Genome. Res. 11, 2450–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han, J-D. J., Bertin, N., Hao, T., Goldberg, D. S., Berriz, G. F., Zhang, L. V., Dupuy, D., Walhout, A. J. M., Cusick. M. E., Roth, F. P. & Vidal, M. (2004) Nature 430, 88–93. [DOI] [PubMed] [Google Scholar]
- 24.Begley, T. J., Rosenbach, A. S., Ideker, T. & Samson, L. D. (2002) Mol. Cancer Res. 1, 103–112. [PubMed] [Google Scholar]
- 25.Begley, T. J., Rosenbach, A. S., Ideker, T. & Samson, L. D. (2004) Mol. Cell., 16, 117–125. [DOI] [PubMed] [Google Scholar]
- 26.Xenarios, I., Salwinski, L., Duan, X. J., Higney, P., Kim, S. M. & Eisenberg, D. (2002) Nucleic. Acids Res. 30, 303–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Devore, J. L. (2004) Probability and Statistics (Brooks/Cole–Thomason, Belmont, CA), 6th Ed.
- 28.Spirin, V. & Mirny, L. A. (2003) Proc. Natl. Acad. Sci. USA 100, 12123–12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., Amin, N., Schwikowski, B. & Ideker, T. (2003) Genome. Res. 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Forster, J., Famili, I., Fu, P., Palsson, B. & Nielsen, J. (2003) Gen. Res. 13, 244–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gavin, A.-C., Bosche, M., Krause, R., Grandi, P., Marzioch, M., Bauer, A., Schultz, J., Rick, J. M., Michon, A.-M., Cruciat, C.-M., et al. (2002) Nature 415, 141–147. [DOI] [PubMed] [Google Scholar]
- 32.Deane, C. M., Salwinski, L., Xenarios, I. & Eisenberg, D. (2002). Mol. Cell. Proteomics 1, 349–356. [DOI] [PubMed] [Google Scholar]
- 33.Jelinsky, S. A. & Samson, L. D. (1999) Proc. Natl. Acad. Sci. USA 96, 1486–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jelinsky, S. A., Estep, P., Church, G. M. & Samson, L. D. (2000) Mol. Cell. Biol. 20, 8157–8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lauffenburger, D. A. (2000) Proc. Natl. Acad. Sci. USA 97, 5031–5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.