Abstract
Actin, one of the most highly conserved and abundant eukaryotic proteins, is constantly being polymerized and depolymerized within cells as part of cellular motility, tissue formation and repair, and embryonic development. Many proteins exist that bind to monomeric or filamentous (F) forms of actin to regulate the polymerization state. It has become increasingly apparent that the ability of different proteins to bind to and regulate actin filament dynamics depends on the ability of the filament to exist in altered conformations. Yet, little is known about how these conformational changes occur at the molecular level. We have destabilized F-actin filaments by forming a disulfide that locks the “hydrophobic plug” to the body of the actin subunit or by altering the C terminus of actin with a tetramethylrhodamine label. We also examined F-actin filaments at short times after the initiation of polymerization. In all three cases, a substantial fraction of protomers can be found in a “tilted” state that also is induced by actin depolymerizing factor/cofilin proteins. These observations suggest that F-actin filaments are annealed over time into a stable filament and that actin-depolymerizing proteins can effect a time reversal of polymerization.
Keywords: cytoskeleton, EM, image analysis
Actin is one of the most highly conserved and abundant eukaryotic proteins. Actin is very dynamic, and its filaments are constantly rearranged within cells (1). Many proteins exist that bind to monomeric (G) or filamentous (F) forms of actin to regulate the polymerization state. Whereas actin was first isolated and studied as part of the contractile apparatus of vertebrate striated muscle (2), we now understand that actin is ubiquitous and plays an important role in many nonmuscle tissues. Within muscle, the traditional notion that actin is a passive cable on which myosin “walks” has been challenged by many observations (3–6). Structural studies of actin filaments have revealed a surprising degree of internal plasticity, such as a variability in the twist (7, 8) and tilt (9) of the component protomers, and these internal dynamics could play an important role in the interactions between actin and many other proteins. Within nonmuscle cells, proteins in the actin depolymerizing factor (ADF)/cofilin family (10) play a crucial role in the rapid depolymerization and repolymerization of actin filaments that is needed for cell motility. Actin-containing structures that were assumed to be static, such as the dense core of the stereocilia of inner ear hair cells, have now been shown to be quite dynamic (11), with a treadmilling of actin protomers proceeding through these tightly packed filaments by an unknown mechanism (12).
Materials and Methods
Sample Preparation. F-Mg2+-actin was prepared from rabbit skeletal muscle as described (13). Polymerization time was either 2 min or 2 h at room temperature. Wild-type yeast actin was prepared as described (14), and G-actin was polymerized in 50 mM NaCl/3 mM MgCl2/10 mM 4-morpholinepropanesulfonic acid (Mops), pH 7.6/0.5 mM ATP for 2 h. Tetramethylrhodamine (TMR)-labeled actin was prepared as described (15). A mixture of 10 μM G-TMR-actin/10 μM unlabeled G-actin/10 mM Hepes, pH 7.5/0.5 mM ATP was converted to Mg2+ form by incubation with EGTA and MgCl2 and then polymerized by 50 mM KCl/2 mM MgCl2 over 3 h. F-Mg2+-TMR copolymers were diluted to 1.5 μM before application to EM grids. The yeast triple mutant (LC)2CA (L180C/L269C/C374A) was prepared as described (16). The mutant G-actin (10 μM) in 50 mM KCl/2 mM MgCl2/10 mM Hepes, pH 7.4/0.5 mM ATP was polymerized at room temperature in the presence of equimolar phalloidin over 3 h. For the disulfide formation between Cys-180 and Cys-269 in solution, mutant F-actin (10 μM) in 10 mM Mops, pH 7.2/0.2 mM CaCl2/0.2 mM ATP/100 mM KCl/3 mM MgCl2 was oxidized with 5 μM CuSO4 at room temperature for either 3 or 15 min. For the EM experiments, F-actin was diluted to 5 μM, applied to an EM grid, and washed with F-buffer before oxidation. Disulfide formation within actin filaments was catalyzed by incubation on the grid for 2–3.5 min in one drop of 25 μM CuSO4/2 mM MgCl2.
Light Scattering. Actin (either TMR-labeled or unlabeled) in G-buffer (5 mM Hepes, pH 7.5/0.2 mM CaCl2/0.2 mM ATP/1 mM DTT) was supplemented with 0.3 mM EGTA/0.1 mM MgCl2 and incubated for 10 min at room temperature to replace Ca2+ with Mg2+ in the high-affinity divalent cation binding site on G-actin. Then, 2.0 mM MgCl2 was added to initiate polymerization. The increase in light scattering upon actin polymerization was monitored by using a spectrofluorometer (PTI, South Brunswick, NJ) with the emission and excitation wavelengths set at 350 nm.
EM. Filaments were applied to glow-discharged carbon-coated EM grids, followed by negative staining with 2% (wt/vol) uranyl acetate. A Tecnai-12 EM (FEI, Hillsboro, OR) was used at an accelerating voltage of 80 kV and a nominal magnification of ×30,000. Densitometry of negatives was performed by using a Leaf 45 (Scitex, Tel Aviv) scanner with a raster of 3.9 Å per pixel.
Image Analysis. The spider software package (17) was used for most of the image processing. Four structural states were isolated from complexes of F-actin with actin-binding proteins (18). A twist from 146° to 176° per subunit, with a step of 2°, was imposed on these structures, and the resultant 64 volumes (4 × 16) were projected onto two-dimensional images with different azimuthal orientations from 0° to 360°, with an increment of 4°. This procedure generated 5,760 reference projections (64 × 90). Filament segments (100 × 100 pixels) from EM images of actin filaments were cross-correlated with the reference projections to sort these segments by both symmetry (twist) and structural state. Classes were then reconstructed by using the iterative helical real space reconstruction (IHRSR) method (19). For the experiments involving time after initiation of polymerization, 11,550 segments were analyzed from the 2-h control, and 4,500 segments were analyzed from the 2-min sample. For the TMR-labeled actin copolymers with unlabeled actin, 9,650 segments were analyzed. The experiments with the (LC)2CA yeast involved 8,930 segments under oxidizing conditions when cross-links were formed and 10,040 segments under reducing conditions as a control. The control yeast actin filaments involved analysis of 14,610 segments. The initial sorting thus involved >341 million cross-correlations (5,760 × 59,280).
Results
We have used two different modifications of actin subunits to destabilize F-actin filaments. The original Holmes model for F-actin involved a postulated “hydrophobic plug” between the two strands, with a loop (residues 263–274) that swung out from the body of the monomer to make a contact with subunits on the opposing strand (20). Subsequent experiments have partially supported this model (21–23), and it has been shown that locking this loop to the body of the actin subunit by engineered disulfide bonds can prevent normal polymerization (16). We have used the yeast actin triple mutant (L180C/L269C/C374A), which places two cysteine residues in positions allowing for the loop to be locked to the body of actin and eliminates the reactive cysteine at position 374 (16). Remarkably, we have been able to form actin filaments first under reducing conditions and then allow the disulfide to form within filaments by oxidation. The disulfide formation can be monitored on SDS/PAGE (Fig. 1a) by taking advantage of the different mobilities of oxidized and reduced actin. The rate of disulfide formation depends strongly on the presence and concentration of oxidation catalysts (Cu2+), temperature, and pH.
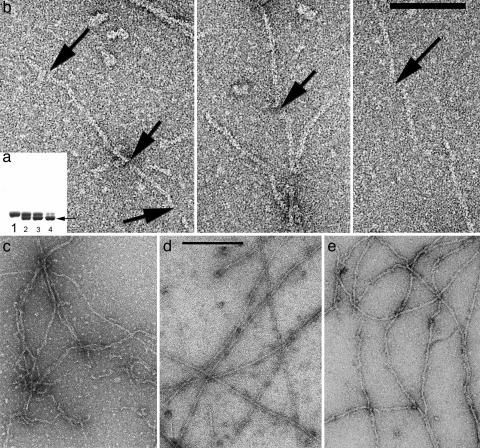
Fig. 1.
Under conditions where almost all actin is polymerized into filaments, intramolecular disulfide bonds can be introduced into actin subunits containing a triple mutation (L180C/L269C/C374A). These intramolecular disulfides lock the hydrophobic plug of actin (residues 263–274) to the body of the subunit and prevent it from forming a bond with subunits in the opposing strand. (a) SDS/PAGE can show this disulfide formation because the cross-linked monomer runs with higher mobility (lower band, arrow) than the un-cross-linked molecule. Lane 1 is the control, under reducing conditions. After 1 min of oxidation (lane 2), almost half the actin molecules have an intramolecular disulfide. Lanes 3 and 4 show actin after 2 and 15 min of oxidation, respectively. (b) Electron micrographs show the fragmentation of filaments that occurs after disulfide bonds are introduced into filaments that have been first adsorbed to an EM grid. The arrows indicate the breaks that can be seen within filaments. (Scale bar: 1,000 Å.) (c–e) Copolymers of TMR-labeled actin and unlabeled actin (c) are typically shorter than control actin filaments (d), and many kinks and sharp bends are present. Filaments that are examined 2 min after the initiation of polymerization (e) have a much less uniform appearance than the control filaments (d) that are examined 2 h after the initiation of polymerization.
If the loop was locked into the proposed filament conformation (20), we would expect that this disulfide would never form in an existing filament. There are two explanations for how this extensive disulfide formation might occur. One is that the loop is dynamic, so that it is exploring many different conformations within the filament, including one that is monomer-like. Another possibility is that the extended conformation of the loop exists for only a subset of the protomers, which may be all that is required to hold the two strands together. The observation that the two strands can actually separate within F-actin (24) is consistent with both possibilities.
When filaments containing the triple mutant are first polymerized in solution and placed on an EM grid, and disulfides are then formed, breaking of the filaments occurs extensively (Fig. 1b). To examine this process further, we have analyzed the segments between the observed breaks. By using the IHRSR method (19), we can generate three-dimensional reconstructions from short lengths of F-actin. More importantly, we do not need to average over long filaments and can sort segments into different classes based on such parameters as twist and conformation. Based on the analysis of >100,000 segments from filaments formed under different conditions, we used four different conformational states for sorting (Fig. 2). If the disulfides occurred only at the breaks, these segments should look like normal F-actin. In fact, they do not, and the distribution of observed states (Fig. 3c) is very different from what is observed for wild-type yeast actin filaments (Fig. 3 a and b). The possibility that the difference is caused by the triple mutation, not the disulfide, can be excluded, because the distribution of states for the triple-mutant filament under reducing conditions is very similar to that of the wild type (data not shown). We interpret these results as showing that the disulfide formation destabilizes an existing actin filament so that it becomes fragile and spontaneously breaks. Within these fragile filaments, the probability of observing segments in a tilted state becomes significantly elevated.
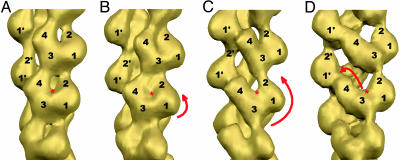
Fig. 2.
Four structural states of F-actin were isolated from complexes of F-actin with either cofilin (32) or Abl-related gene (Arg) kinase. The subdomains (SDs) of actin are numbered, and the nucleotide-binding cleft is marked with a red asterisk. The difference between “regular” state (A) and “intermediate” state (B) is a slight rotation of the whole protomer (short red arrow) that might still allow actin to maintain similar protomer–protomer contacts. The larger rotation of protomers (long red arrow), which we call “tilted state I” (C), does not allow SD2 to interact with SD1 and establishes a new contact between SD2 and SD3 of adjacent protomers in the same long-pitch helix strand. Propeller rotation of SD4 toward the opposite strand (shown by red arrow in D) is called “tilted state II.” In this state, SD4 establishes a contact with SD1 from a protomer on the opposite strand, and this contact is absent in the three other states.
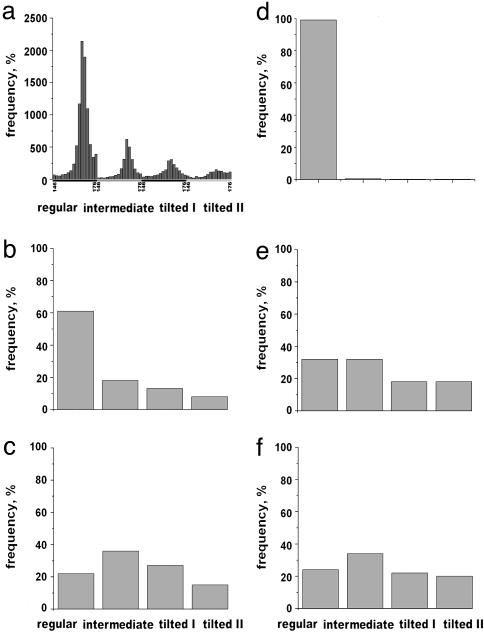
Fig. 3.
The distribution of states found among filament segments extracted from different populations of F-actin. The four structural states are those shown in Fig. 2. The full sorting, by both twist and structural state, is for the control wild-type yeast actin (a). For simplicity, different twists within the same structural state are grouped together in b–f. Wild-type yeast F-actin (b) as well as the (LC)2CA mutant under reducing conditions (data not shown) have >60% of segments in the regular state. Under oxidizing conditions (when an intramolecular disulfide is formed), only 20% of segments are in the regular state (c), with a significant increase in the number of segments assigned to the tilted state. Almost all segments of rabbit F-actin after2hof polymerization exhibit regular structure (d), compared with early stages of polymerization (e), where only ∼30% of segments are assigned to this class and >35% are in the tilted state. A similar reduction in the fraction of regular actin and increase in the tilted states occurs when copolymers of unlabeled actin and TMR-labeled actin are formed (f).
The second method of destabilization of F-actin involves a modification at position 374. The covalent attachment of the fluorescent dye TMR to Cys-374 in actin prevents normal polymerization (25). However, copolymers containing up to 50% TMR-labeled actin can be formed with unmodified actin subunits (26). Electron micrographs of these copolymers (Fig. 1c) show filaments that are shorter and more kinked than control filaments (Fig. 1d). When these copolymers are reconstructed, they display a similar distribution of states (Fig. 3f) as seen for the disulfide cross-linked F-actin (Fig. 3c). Of course, one needs to prove that the distribution of states found actually corresponds to the structural differences illustrated in Fig. 2 and is not an artifact. The IHRSR method lends itself to such a proof, and this distribution is shown in Fig. 4 for the filaments formed from a mixture of TMR-labeled and unlabeled actin. By starting with the same filament structure for each of the four states in which the segments have been classified, we can show that the resulting reconstructions diverge, and the final reconstructions from each set (after 60 iterations) look very similar to the corresponding reference volumes (Fig. 2) used for the initial sorting. This procedure breaks any potential circularity, arising when a reference volume serves as a template for alignment, leading to a reconstruction resembling the reference volume that has been used. The exact same test has been performed for all of the sorting shown in Fig. 3, with the result that the segments sorted as being in one particular state can actually be shown to be in that state when they are reconstructed starting from a completely different filament structure. In fact, we have demonstrated that actin filaments converge to the same reconstruction starting with filament models made from either cofilin or RecA (27).
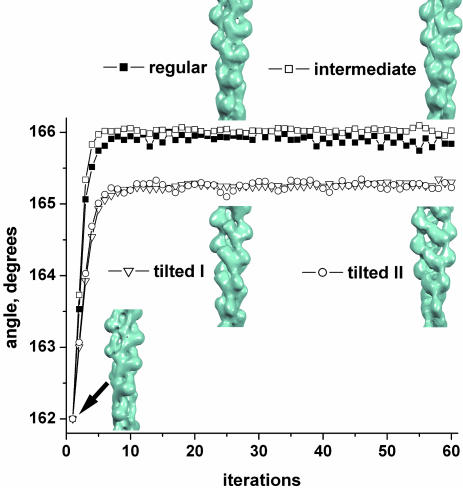
Fig. 4.
The validity of the classification method is illustrated for F-actin formed from a mixture of unlabeled actin and TMR-labeled actin. Segments have been classified as being in one of four states based on cross-correlations with four different reference structures. These segments are then reconstructed by using the IHRSR approach (19), with a modified Holmes et al. model for F-actin (20) used as an initial reference. The modification involves changing the twist of this volume to 162° per subunit, and this initial reference volume is shown at the bottom left (iteration 1). After 60 cycles, the resulting reconstructions are shown for the four different sets. Because these reconstructions correspond quite well to the references used for purposes of classification but have been reconstructed using a very different structure as an initial model, the sorting is shown to be reliable.
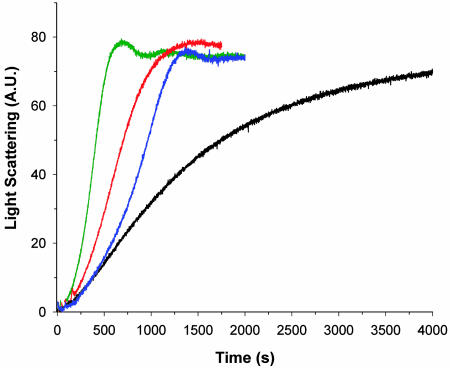
The destabilization of F-actin in these copolymers of TMR-labeled and unlabeled actin can be seen by the light scattering observed as a function of time after the initiation of polymerization (Fig. 5). Copolymers containing the TMR modification behave in a manner similar to polymerization in the presence of cofilin, where filaments are being severed and depolymerized at the same time that a net addition of subunits to the polymerized state is taking place. Because filament-severing increases the number of filament ends, the net result is acceleration of actin polymerization by cofilin and TMR-labeled actin and a synergistic effect of the two factors together (Fig. 5). Whereas quantitative analysis of the light-scattering data is difficult, due to the fact that the scattered intensity will be a function of both the filament length distribution and the amount of material polymerized, a consistent description of actin polymerization in the presence of cofilin does emerge based on combining the light-scattering observations with pyrene fluorescence data (28) and EM observations (29). Together, these methods yield a picture that the total monomer pool is being depleted at the same time that more and more short filaments are being created. The formation of short filaments during polymerization (presumably from fragmentation) also has been seen in mixtures of TMR-labeled and unlabeled actin (26).
Fig. 5.
The effect on the polymerization kinetics of adding TMR-labeled actin to unlabeled actin is similar to the effect of cofilin, as judged by a light-scattering assay of filament polymerization growth. A.U., arbitrary units. Unlabeled actin (5 μM) has the most gradual slope (black trace), resulting from the kinetics of limited nucleation and few filament ends. In the presence of 0.083 μM cofilin (blue trace), extensive fragmentation of filaments occurs, which leads to a much greater increase in the rate of polymerization. The incorporation of TMR-labeled actin into copolymers with unlabeled actin has an effect similar to that of cofilin, as seen (red trace) when 0.5 μM TMR-labeled actin is mixed with 4.5 μM unlabeled actin. The addition of 0.083 μM cofilin to this same 9:1 unlabeled actin/TMR-labeled actin mixture (green trace) leads to an even further enhancement in the overall rate of polymerization.
Last, we used the IHRSR method to look at filaments that were formed only 2 min after the initiation of polymerization (Fig. 1e). A previous report has described that such filaments have a “ragged” morphology, which disappears after longer incubation times (30). As with the TMR-labeled copolymer and the disulfide cross-linked filaments, the segments examined at such an early time contain a significant proportion of the tilted state (Fig. 3e). Because the TMR modification has been made to rabbit skeletal muscle actin and not yeast actin, we have examined F-actin polymers 2 h after the initiation of polymerization as a control (Fig. 3d). Rabbit F-actin polymers show a distribution of states similar to that seen with wild-type yeast actin (Fig. 3b).
Discussion
We have shown that actin filaments are structurally quite heterogeneous a short time after the initiation of polymerization. After several hours, these filaments appear much more homogeneous by EM. The polymerization process for actin thus appears to involve an annealing mechanism, where a multiplicity of states present at short times after polymers are formed is reduced to one dominant state. This dominant state is the basis for current models of F-actin structure (20). Interestingly, the multiplicity of states present initially includes a significant number of subunits in a tilted conformation (T-actin), which involves very different subunit–subunit contacts than those that exist in F-actin (9). This tilted state was initially seen under conditions when actin filaments were being actively depolymerized by ADF (18), and we have now shown that this tilted state can exist when filaments are destabilized either by the formation of an internal disulfide bond within protomers or by modification of Cys-374 with TMR.
What is the relationship between these observations and the standard model for an actin filament at steady state (31), where subunits can add on at the barbed end and dissociate at the pointed end? The main point is that actin filaments in the cell do not, in general, depolymerize by the slow dissociation of subunits from a pointed end. Proteins such as ADF/cofilin are engaged in the rapid depolymerization of actin filaments (1). The structural heterogeneity that we find in “young” actin filaments, involving tilted actin, is not simply related to the structural differences between the barbed and pointed ends of “mature” actin filaments at steady state, because we showed recently (32) that whereas the two ends are different, a substantial amount of tilted actin is found at neither end. We suggest, therefore, that the destabilization of the filament that we observe (such as by ADF/cofilin) may be more relevant biologically than that which occurs in vitro at the pointed ends of F-actin. Our findings also may be relevant to the observation that, even in vitro, single actin filaments cannot be described at steady state “by the simple association and dissociation of monomers at both ends of the filaments” (33).
We suggest that proteins such as ADF/cofilin exert their action in depolymerizing F-actin not by inducing a novel structure but rather by driving filaments back to a less stable state that exists at early stages of polymerization. This model provides insight into how other actin-binding proteins, such as myosin (4, 34), may take advantage of intrinsic multiple conformational states within F-actin.
Acknowledgments
We thank Mai Phan and Martin Phillips for technical assistance. This work was supported by grants from the National Institutes of Health (to P.A.R., E.H.E., and E.R.) and the National Science Foundation (to E.R.).
Author contributions: P.A.R., E.H.E., and E.R. designed research; A.O., A.S., V.E.G., D.S.K., and E.H.E. performed research; V.E.G., P.A.R., and E.H.E. contributed new reagents/analytic tools; V.E.G. and E.R. analyzed data; and E.H.E. and E.R. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: G-actin, monomeric actin; F-actin, filamentous actin; TMR, tetramethylrhodamine; ADF, actin depolymerizing factor; IHRSR, iterative helical real space reconstruction.
References
- 1.Pollard, T. D., Blanchoin, L. & Mullins, R. D. (2000) Annu. Rev. Biophys. Biomol. Struct. 29, 545–576. [DOI] [PubMed] [Google Scholar]
- 2.Feuer, G., Molnar, F., Pettko, E. & Straub, F. B. (1948) Hung. Acta Physiol. 1, 150–163. [PubMed] [Google Scholar]
- 3.Schwyter, D. H., Kron, S. J., Toyoshima, Y. Y., Spudich, J. A. & Reisler, E. (1990) J. Cell Biol. 111, 465–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borovikov, Y. S., Dedova, I. V., dos Remedios, C. G., Vikhoreva, N. N., Vikhorev, P. G., Avrova, S. V., Hazlett, T. L. & Van Der Meer, B. W. (2004) Biophys. J. 86, 3020–3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prochniewicz, E. & Yanagida, T. (1990) J. Mol. Biol. 216, 761–772. [DOI] [PubMed] [Google Scholar]
- 6.Drummond, D. R., Peckham, M., Sparrow, J. C. & White, D. C. (1990) Nature 348, 440–442. [DOI] [PubMed] [Google Scholar]
- 7.Egelman, E. H., Francis, N. & DeRosier, D. J. (1982) Nature 298, 131–135. [DOI] [PubMed] [Google Scholar]
- 8.McGough, A., Pope, B., Chiu, W. & Weeds, A. (1997) J. Cell Biol. 138, 771–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galkin, V. E., Van Loock, M. S., Orlova, A. & Egelman, E. H. (2002) Curr. Biol. 12, 570–575. [DOI] [PubMed] [Google Scholar]
- 10.Chen, H., Bernstein, B. W. & Bamburg, J. R. (2000) Trends Biochem. Sci. 25, 19–23. [DOI] [PubMed] [Google Scholar]
- 11.Schneider, M. E., Belyantseva, I. A., Azevedo, R. B. & Kachar, B. (2002) Nature 418, 837–838. [DOI] [PubMed] [Google Scholar]
- 12.Rzadzinska, A. K., Schneider, M. E., Davies, C., Riordan, G. P. & Kachar, B. (2004) J. Cell Biol. 164, 887–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orlova, A. & Egelman, E. H. (1995) J. Mol. Biol. 245, 582–597. [DOI] [PubMed] [Google Scholar]
- 14.Kim, E., Wriggers, W., Phillips, M., Kokabi, K., Rubenstein, P. A. & Reisler, E. (2000) J. Mol. Biol. 299, 421–429. [DOI] [PubMed] [Google Scholar]
- 15.Kudryashov, D. S. & Reisler, E. (2003) Biophys. J. 85, 2466–2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shvetsov, A., Musib, R., Phillips, M., Rubenstein, P. A. & Reisler, E. (2002) Biochemistry 41, 10787–10793. [DOI] [PubMed] [Google Scholar]
- 17.Frank, J., Radermacher, M., Penczek, P., Zhu, J., Li, Y., Ladjadj, M. & Leith, A. (1996) J. Struct. Biol. 116, 190–199. [DOI] [PubMed] [Google Scholar]
- 18.Galkin, V. E., Orlova, A., Lukoyanova, N., Wriggers, W. & Egelman, E. H. (2001) J. Cell Biol. 153, 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Egelman, E. H. (2000) Ultramicroscopy 85, 225–234. [DOI] [PubMed] [Google Scholar]
- 20.Holmes, K. C., Popp, D., Gebhard, W. & Kabsch, W. (1990) Nature 347, 44–49. [DOI] [PubMed] [Google Scholar]
- 21.Chen, X., Cook, R. K. & Rubenstein, P. A. (1993) J. Cell Biol. 123, 1185–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuang, B. & Rubenstein, P. A. (1997) J. Biol. Chem. 272, 4412–4418. [DOI] [PubMed] [Google Scholar]
- 23.Musib, R., Wang, G., Geng, L. & Rubenstein, P. A. (2002) J. Biol. Chem. 277, 22699–22709. [DOI] [PubMed] [Google Scholar]
- 24.Bremer, A., Millonig, R. C., Sutterlin, R., Engel, A., Pollard, T. D. & Aebi, U. (1991) J. Cell Biol. 115, 689–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Otterbein, L. R., Graceffa, P. & Dominguez, R. (2001) Science 293, 708–711. [DOI] [PubMed] [Google Scholar]
- 26.Kudryashov, D. S., Phillips, M. & Reisler, E. (2004) Biophys. J. 87, 1136–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang, S., Yu, X., Galkin, V. E. & Egelman, E. H. (2003) J. Struct. Biol. 144, 162–171. [DOI] [PubMed] [Google Scholar]
- 28.Du, J. & Frieden, C. (1998) Biochemistry 37, 13276–13284. [DOI] [PubMed] [Google Scholar]
- 29.Ono, S. & Benian, G. M. (1998) J. Biol. Chem. 273, 3778–3783. [DOI] [PubMed] [Google Scholar]
- 30.Schoenenberger, C. A., Bischler, N., Fahrenkrog, B. & Aebi, U. (2002) FEBS Lett. 529, 27–33. [DOI] [PubMed] [Google Scholar]
- 31.Wegner, A. (1976) J. Mol. Biol. 108, 139–150. [DOI] [PubMed] [Google Scholar]
- 32.Galkin, V. E., Orlova, A., Van Loock, M. S., Shvetsov, A., Reisler, E. & Egelman, E. H. (2003) J. Cell Biol. 163, 1057–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fujiwara, I., Takahashi, S., Tadakuma, H., Funatsu, T. & Ishiwata, S. (2002) Nat. Cell Biol. 4, 666–673. [DOI] [PubMed] [Google Scholar]
- 34.Kim, E., Miller, C. J., Motoki, M., Seguro, K., Muhlrad, A. & Reisler, E. (1996) Biophys. J. 70, 1439–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]