Abstract
It is becoming increasingly clear that single cortical neurons encode complex and behaviorally relevant signals, but efficient means to study gene functions in small networks and single neurons in vivo are still lacking. Here, we establish a method for genetic manipulation and subsequent phenotypic analysis of individual cortical neurons in vivo. First, lentiviral vectors are used for neuron-specific gene delivery from α-calcium/calmodulin-dependent protein kinase II or Synapsin I promoters, optionally in combination with gene knockdown by means of U6 promoter-driven expression of short-interfering RNAs. Second, the phenotypic analysis at the level of single cortical cells is carried out by using two-photon microscopy-based techniques: high-resolution two-photon time-lapse imaging is used to monitor structural dynamics of dendritic spines and axonal projections, whereas cellular response properties are analyzed electrophysiologically by two-photon microscopydirected whole-cell recordings. This approach is ideally suited for analysis of gene functions in individual neurons in the intact brain.
Keywords: patch-clamp recording, two-photon imaging
Altering expression of specific genes in living organisms has become an invaluable approach to the study of gene functions in systems biology. In mammalian neurobiology, the typical approach is the generation of transgenic mice or of mice with a targeted gene disruption (knockout), sometimes in combination with means to developmentally and/or regionally regulate the onset of the genetic manipulation and the cell type that is affected (1–3). However, even the most specific genetic designs affect entire populations of neurons, typically in multiple brain regions. The current transgenic/knockout technologies thus do not allow targeting of small neuronal networks or individual neurons in the intact brain. Yet, individual neurons are believed to critically contribute to network functions in the mammalian cortex.
The manifold and complex response properties of single neurons have been a focus of cortical physiology ever since the groundbreaking work of Hubel and Wiesel (4) in the cat visual cortex. The emphasis on the analysis of single-neuron activity has been enforced by studies demonstrating a tight link between psychophysical judgments and spiking activity of single cortical neurons in distinct brain regions of primates (5, 6). In addition, a cause-and-effect evidence for functional significance of individual neurons has been provided by a recent study (7) that showed that intracellular stimulation of single pyramidal neurons in the rat vibrissae motor cortex can drive detectable movements. Taken together, these experiments demonstrate that individual cortical neurons encode meaningful and behaviorally significant information. The development of methods for genetic manipulations in individual cortical neurons in vivo is thus highly desirable.
Recombinant retroviral vectors have been used for stable gene expression in many cell types (8, 9). One of the most promising systems for gene delivery to neurons is based on lentiviruses that infect postmitotic nondividing cells (10, 11). The most commonly used so-called self-inactivating lentiviral vector contains an internally placed recombinant promoter that determines the efficiency of gene expression in different cell types (12).
Here, we present a method that includes a sparse lentiviral infection of a small number of cortical neurons in vivo, and their subsequent functional analysis by targeted optical and electrophysiological methods in anesthetized animals. Because the temporal and spatial control of the gene manipulations is reliably achieved by stereotactic delivery of the viral particles, this method can be used to study gene functions in individual neurons of defined cortical systems at a postnatal stage of choice.
Methods
All animal experiments were carried out according to the animal welfare guidelines of the Max Planck Society.
Cloning of Lentiviral Vectors. Flip, ubiquitin promoter, GFP, and WRE (FUGW) vector (13) cut by PacI/BamHI yields a backbone for the following: Cytomegalovirus (CMV)-IE/chicken β-actin, 1.7-kb fragment cut SalI/EcoRI from pCAGGS-lacZ (14); mouse Thy1.2, ≈4-kb fragment cut EcoRI/XhoI from puc18-Thy1 (15); mouse α-calcium/calmodulin-dependent protein kinase II (α-CaMKII), 0.4-kb PCR fragment amplified by primers prCK0.4/5′ (CCCTTAATTAACTTGTGGACTAAGTTTGTTCACATCCC) and prCK/3′ (GCTCTAGAGCTGCCCCCAGAACTAGGGGCCACTCG) from pMM403 template (16); α-CaMKII, 1.2-kb PCR fragment amplified by prCK1.2/5′ (CCCTTAATTAACATTATGGCCTTAGGTCACTT) and prCK/3′ from pMM403 template; α-CaMKII, 2.4-kb fragment cut BamHI from pMM403; rat Synapsin I 1.1-kb PCR fragment amplified by primers prSy1.1/5′ (CCCTTAATTAAGGGTTTTGGCTACGTCCAGAG) and prSy1.1/3′ (CGCGGATCCAAGGGGCAGTGGGTCGGTGGG) from pBL4.3SynCAT template (17); rat Synapsin I 1.1-kb PCR fragment amplified by primers prSy0.5/5′ (CCCTTAATTAACAAGTATCTGGGAAGGGTAAC) and prSy1.1/3′; and neuron-restrictive silencor element/simian virus (SV)40, 0.3-kb fragment cut by KpnI/HindIII from pSyNRSE2/SV40luc (18).
Lentivirus Production/Enhanced GFP (EGFP) Expression. Lentiviruses were produced as described (9, 12). Human embryonic kidney 293FT cells (Invitrogen) were transfected by using the calciumphosphate method with the expression and two helper, ▵8.9 and vesicular stomatitis virus G protein (19, 20), plasmids at 1, 7.5, and 5.5 μg of DNA per 10-cm plate. After 48 h, the supernatants of four plates were pooled, spun at 780 × g for 5 min, filtered at a 0.45-μm pore size, spun at 83,000 × g for 1.5 h, and the pellet was resuspended in 100 μl of PBS.
Hippocampal cultures (21) were infected a day after plating and fixed with 4% paraformaldehyde (PFA) at 14 days in vitro. For analysis, the cells were imaged with a Leica TCS-NT confocal microscope, 0.5-μm z step, at constant laser intensity and photomultiplier tube settings. Square regions of interest (ROIs) were selected over individual cell nuclei (to standardize the region selection) in maximal projection images; mean fluorescence intensity per area was measured by using imagej-1.3 software (National Institutes of Health, Bethesda). The in vivo injections were with similar titer and volume for all viruses, resulting in an uniform 500- to 600-μm diameter infected area. After an expression period, animals were anesthetized with an overdose of Halothane and transcardially perfused with 4% PFA in PBS. Coronal brain sections (100 μm) were imaged (0.5- or 1.0-μm z step, with stacks up to 60 μm). EGFP expression analysis was done as for the cultured neurons, with the exception that ROIs were selected from single z sections taken through the midregion of individual neuronal cell bodies. Care was taken to analyze cells from the edges of injection sites to avoid bias for cells with multiple infection at the injection center.
Lentivirus-Based Short-Interfering (siRNA) Expression. The U6 promoter was generated by PCR from mouse genomic DNA with primers NheI/5′U6; CCCGCTAGCATCCGACGCCGCCATCTCTA and XhoI/BbsI 3′U6; CCGCTCGAGGAAGACCACAAACAAGGCTTTTCTCCAA (22), and cloned into pBudCE4.1 (Invitrogen) by means of NheI/XhoI, creating a vector pCMV-U6. The double-stranded hairpin oligo GFPsiRNA (23) was cloned in pCMV-U6 through BbsI/BstBI sites (22). The U6- GFPsiRNA cassette was recloned into FlkURW through NheI/BstBI (FlkURW was made from FUGW by adding NheI/BstBI linker in the PacI site of FUGW, and replacing EGFP with dsRed2, Clontech), creating a vector FU6siGURW. For testing GFPsiRNA-based silencing in neurons, cultures were infected at 1 day in vitro with a high titer of FUGW and at 3 days in vitro with a low titer of FU6siGURW virus. After 7 days, cells were fixed in 4% PFA. EGFP fluorescence in the soma region of infected cells was quantified with imagej (above).
In Vivo Infection of Cortical Neurons. Wistar rats [aged postnatal day (P)8–P28] or C57BL6 or NMR1 mice (P10 to P48) were anesthetized with an i.p. injection of ketamine (100 mg/kg)/xylazine (10 mg/kg), in some cases, with addition of atropine (0.02 mg/kg). Animals were kept deeply anesthetized as assessed by monitoring pinch withdrawal, eyelid reflex, corneal reflex, respiration rate, and vibrissae movements. Body temperature was maintained at 37°C by using a heating blanket (Watlow). One to three craniotomies, ≈300–400 μm in diameter, were drilled above the somatosensory cortex. Viral stock, ≈30 nl, was slowly injected, by using an ultraprecise small animal stereotaxic apparatus (Kopf Instruments, Tujunga, CA) at target depths of 300–500 μm below the pia mater with pulled glass pipettes (Blaubrand intraMARK, tips broken to an outer diameter of ≈30 μm).
In Vivo Two-Photon Imaging. Four to eight days after infections, animals were anesthetized with urethane (2 g/kg of body weight). A metal plate was attached to the skull with dental acrylic cement as described (24), and a large craniotomy (1.5–3 mm diameter) was opened. The dura was removed to improve optical access in rats (this procedure was not necessary in mice). To dampen heartbeat/breathing-induced brain motion, the craniotomy was filled with agarose (1.5%, type III-A, Sigma) in normal rat Ringer and covered with a coverslip. In vivo images were acquired with a custom-built two-photon laser-scanning microscope with a 40× water-immersion objective lens (0.8 numerical aperture, Zeiss), by using custom-written software (R. Stepnoski, Lucent Technologies, Murray Hill, NJ, and M. Müller, Max Planck Institute for Medical Research, Heidelberg). No apparent changes in morphology resulted from laser illumination during time-lapse recordings (stacks of 10–34 images, 45- to 300-sec stack interval, 0.5- to 1.5-μm z spacing, and 20–350 min total acquisition time). For overview fluorescence image stack, a maximum-intensity side projection was created in the z direction. Consecutive projection images of time-lapse recordings were aligned based on the position shift of the crosscorrelation peak by using custom-written macros in nih image.
In Vivo Two-Photon Targeted-Patch Recording. Ten to 14 days after viral delivery, a craniotomy was prepared as for two-photon imaging (above), but without agar and a glass coverslip. Images were acquired on a custom-made laser-scanning microscope, and a two-channel whole-area detector (using Hamamatsu R6357 photomultipliers) was used to collect emitted light selected by interference filters (Chroma Technology, Rockingham, VT; 520/BP30 for monitoring EGFP and 610/BP40 for Alexa 594 signals). Patch pipettes were fabricated by using nonfilamented borosilicate glass (OD at 2.0 mm, ID at 1.5 mm, Hilgenberg, Malsfeld, Germany). The standard intracellular solution was as follows: 130 mM potassium gluconate, 10 mM sodium gluconate, 10 mM Hepes, 10 mM phosphocreatine, 4 mM MgATP, 0.3 mM GTP, 4 mM NaCl, 0.3–1% biocytin, and Alexa 594 0.05 (pH 7.2). The liquid junction potential of y7 mV was corrected. The surface of the cortex was covered with normal rat Ringer. Recordings were obtained by using an Axoclamp-2B amplifier (Axon Instruments, Foster City, CA), filtered at 3–10 kHz, and digitized at 5–20 kHz (ITC-16, Instrutech, Mineola, NY) by using heka software. For whisker stimulation, mechanical stimuli (≈6° deflection angle) were delivered by a glass capillary attached to a piezo apparatus (Physik Instrumente, Karlsruhe, Germany) for 200 ms at a frequency of 1 Hz (25). After each successful two-photon microscopy targeted-patch (TPTP) recording, the animals were perfused with 4% PFA in PBS, and the brains were sectioned tangentially at 100 μm, and processed by cytochrome c oxidase staining (26) and the avidin-biotinperoxidase method (27). Cell reconstructions were generated with neurolucida (Microbrightfield, Williston, VT).
Statistics given are for mean ± SEM unless otherwise stated.
Results
To establish efficient pyramidal neuron-specific expression in the rodent cortex in vivo, we constructed eight lentiviral vectors that expressed EGFP from different internal promoter sequences: a hybrid promoter containing a human CMV enhancer element in front of a chicken β-actin promoter (14), a mouse Thy1.2 promoter (15), mouse α-CaMKII promoters (16) in three different sizes, rat Synapsin I promoters (17) in two different sizes, and a hybrid promoter containing two Synapsin I neuron-restrictive silencer elements in front of an SV40 promoter (18) (Table 1, and Fig. 6, which is published as supporting information on the PNAS web site). The efficiency of expression was compared with a previously described vector containing the human ubiquitin C promoter, a construct termed FUGW, which drives high EGFP expression in various cell types (13).
Table 1. Recombinant promoters used for generation of lentiviral vectors.
| Promoter | Size, kb | Lentiviral vector |
|---|---|---|
| CMV-IE/β-Actin | 1.7 | FCbAGW |
| Thy1.2 promoter | 4.1 | FThGW |
| α-CaMKII | 0.4 | FCK(0.4)GW |
| α-CaMKII | 1.3 | FCK(1.3)GW |
| α-CaMKII | 2.4 | FCK(2.4)GW |
| Synapsin I | 0.5 | FSy(0.5)GW |
| Synapsin I | 1.1 | FSy(1.1)GW |
| Syn. I NRSE/SV40 | 0.3 | FNRSE/SV40GW |
The names of the genes and viral promoter components are indicated in the left column, the sizes of the recombinant promoter fragments (in kb) in the middle column, and the abbreviations for the corresponding vectors in the right column. The names are from the original FUGW promoter. See ref. 13 for more details.
As a first step to test the efficiency of the promoters, we compared EGFP expression in hippocampal primary cultures and organotypic slice cultures infected with the lentiviruses at normalized titers (Fig. 7, which is published as supporting information on the PNAS web site). In cultured neurons, the FUGW vector drove the highest expression, whereas the other vectors expressed EGFP at slightly lower levels and the FNRSE/SV40GW vector expressed very weakly (Fig. 7c).
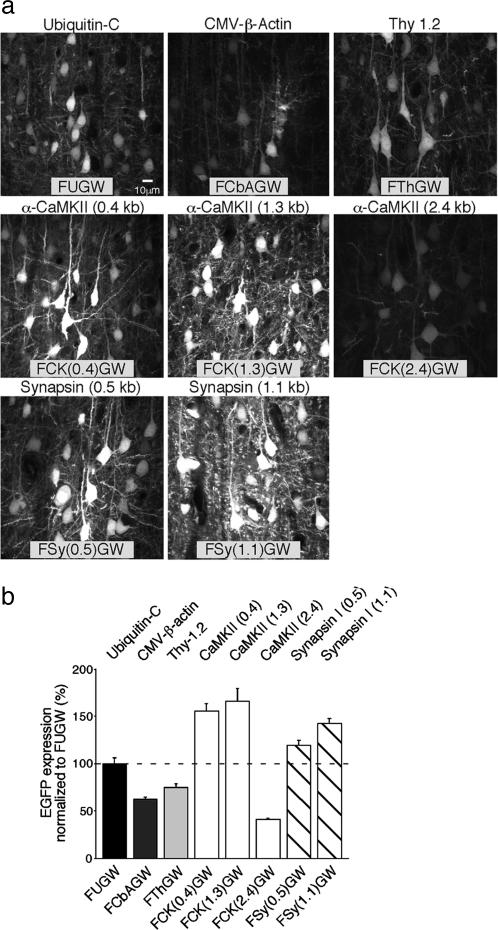
Analysis of the Recombinant Promoters in Cortical Neurons in Vivo. To test the efficiency of the vectors in vivo, we injected viruses into layer 2/3 somatosensory barrel cortex in P21–P24 animals. After 7 days, animals were killed, and EGFP fluorescence from individual neurons was quantified in fixed brain sections (see Methods). The analysis was restricted to neurons at the periphery of the injection sites to avoid measurements from superinfected cells in the center of the injections. As shown in Fig. 1, the α-CaMKII promoter-based vectors FCK(0.4)GW and FCK(1.3)GW drove the strongest level of EGFP expression, followed by the Synapsin I promoter-based vectors FSy(0.5)GW and FSy(1.1)GW. These data are consistent with developmental up-regulation of endogenous α-CaMKII expression in pyramidal neurons in the second postnatal week (28). Interestingly, the 2.4-kb α-CaMKII promoter, vector FCK(2.4)GW, drove very weak expression (Fig. 1). Hence, inclusion of the extra 1.1 kb of 5′ promoter sequence down-regulates the promoter activity in vivo in comparison with the shorter constructs. This finding underscores the importance of testing different-length promoter fragments in the lentiviral vectors.
Fig. 1.
Lentivirus-based EGFP expression in cortical neurons in vivo: P21–P28 infection. (a) Examples of lentivirus infected layer 2/3 neurons in the somatosensory cortex. The internal promoter types and corresponding vector names are indicated at the top and bottom of each image, respectively. The images are maximum-intensity projections of five z sections separated by 1.0 μm, collected by confocal microscopy from the outer region of the injection sites (within 100 μm from the periphery of an injection area of a 500- to 600-μm diameter) in brain sections. [Scale bar, 10 μm (Left Upper) is valid for all images.] (b) Quantification of EGFP expression based on fluorescent signal within nuclei of individual infected neurons (n = number of cells analyzed). The promoter types and corresponding vector names are indicated above and below the bar graphs, respectively. The values (percent mean ± SEM) are normalized to FUGW expression: FUGW = 100 ± 6.9, n = 69; FCbAGW = 62.7 ±1.8, n = 13; FThGW = 74.8 ± 3.9, n = 19; FCK(0.4)GW = 155.1 ± 7.9, n = 20; FCK(1.3)GW = 165.7 ± 13.5, n = 41; FCK(2.4)GW = 41.1 ± 1.0, n = 30; FSy(0.5)GW = 142.5 ± 4.7, n = 20; FSy(1.1)GW = 119.4 ± 5.2, n = 20.
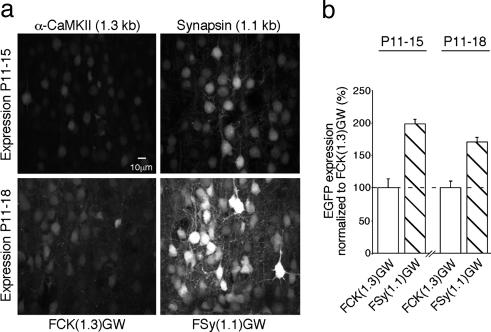
Because Synapsin I expression peaks between the second and third postnatal week (17), the Synapsin I-based vectors may be suitable for early postnatal expression. To evaluate this possibility, we compared EGFP expression from α-CaMKII (1.3 kb) and Synapsin (1.1 kb) promoters during the second postnatal week. As expected, the Synapsin I promoter drove approximately 2-fold higher EGFP expression compared with the α-CaMKII promoter in animals injected at P11 and analyzed at P15 or P18 (Fig. 2). Regarding the neuron specificity of expression, at all developmental stages, the α-CaMKII promoter-based expression was restricted to cortical pyramidal neurons identified by a typical apical dendrite and dendritic branching in the layer 1 (Fig. 3; see also Movie 1, which is published as supporting information on the PNAS web site). The Synapsin I promoter also caused the strongest expression in pyramidal neurons, with weaker expression in cortical interneurons (Fig. 3 and data not shown).
Fig. 2.
Lentivirus-based EGFP expression in cortical neurons in vivo: P11 to P15/18 infection. (a) Examples of infected layer 2/3 neurons in the somatosensory cortex, as in Fig. 1a. The infection period is indicated on the left. (b) Quantification of EGFP expression normalized to FCK(1.3)GW expression, as in Fig. 1. The infection period is indicated above the bar graphs. P11–P15: FCK(1.3)GW = 100 ± 13.6, n = 10; FSy(1.1)GW = 197.9 ± 7.1, n = 10; P11-P18: FCK(1.3)GW = 100 ± 10.3, n = 8; FSy(1.1)GW = 170.4 ± 6.9, n = 13.
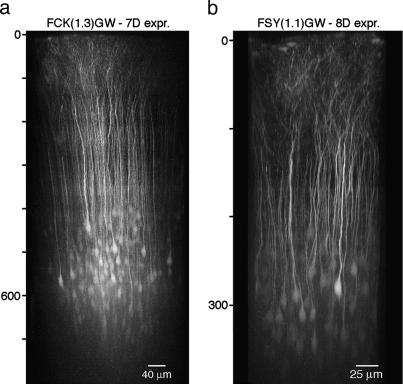
Fig. 3.
In vivo expression pattern of cortical layer 2/3 infected neurons. Each image is a maximum-intensity side projection from an overview stack of fluorescence images recorded by using in vivo two-photon microscopy. Note the different depth scaling (indicated on the left of each image) and the different scale bars. (a) FCK(1.3)GW-infected neurons in P28 rat after 7 days of expression. (b) FSy(1.1)GW-infected neurons in P48 mouse after 8 days of expression.
To test whether infected cells remain healthy, we examined the intrinsic biophysical properties of regular spiking layer 2/3 FCK(1.3)GW-infected neurons by whole-cell patch recordings in acute slices prepared from P31–P33 rats, 18–20 days after in vivo injections. All parameters (resting membrane potential, steady-state input resistance, firing threshold, spike width, and amplitude) examined in the infected neurons were indistinguishable from control, uninfected neurons (Table 2, which is published as supporting information on the PNAS web site). We conclude that the 1.1-kb Synapsin I and 1.3-kb α-CaMKII promoter-based vectors drive the highest level of expression in early postnatal and mature cortical pyramidal neurons in vivo, respectively.
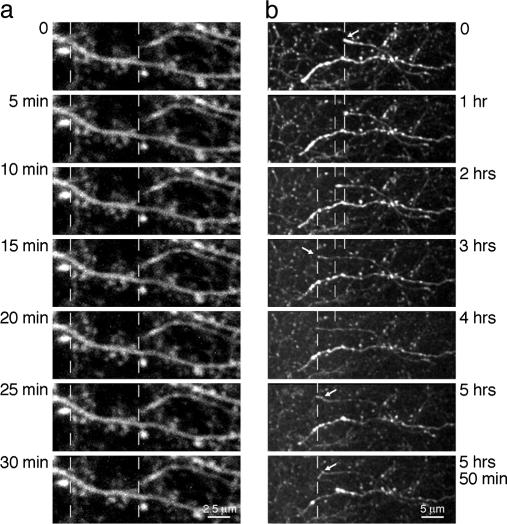
In Vivo Two-Photon Imaging. Two-photon imaging of EGFP-expressing cells allows visualization of morphological changes, such as structural dynamics of dendritic spines, in individual pyramidal neurons in the intact brain (29–31). We tested whether EGFP expression from the FCK(1.3)GW or FSy(1.1)GW vectors is sufficient for in vivo two-photon imaging, at the resolution of dendritic spines and axonal branches. After 7 days of expression, both FCK(1.3)GW- and FSy(1.1)GW-infected pyramidal neurons could be imaged routinely down to a depth of 500 μm below the pial surface and down to 800 μm under favorable conditions (e.g., removed dura, and lack of large blood vessels in the field of view) (Fig. 3 and Movie 1). High-resolution time-lapse imaging of dendritic spines in cortical layer 1 of P31–P48 mice, after 7–8 days of EGFP expression, showed negligible amounts of photobleaching (Fig. 4a, and Movie 2, which is published as supporting information on the PNAS web site; ≈97.5% of fluorescence intensity remained after 30 min imaging at a 1-min sampling interval). Similarly, EGFP-labeled axons in layer 1 region adjacent to the injection site could be imaged for extended time, >1 h when sampled at 45-sec intervals, or close to 6 h at 5-min intervals (Fig. 4b, and Movie 3, which is published as supporting information on the PNAS web site; ≈66% of fluorescence intensity remained after 300 min of imaging of stable structures at a 5-min sampling interval, in P15–P17 mice after 5–6 days of expression; note the clear outgrowth of an axonal fiber ending in the time-lapse movie).
Fig. 4.
Two-photon time-lapse imaging of dendritic spines and axonal projections in the mouse cortex. Acquisition times are indicated on the sides of each image. Each image is a maximum-intensity projection of image stacks (z step, 1 μm) collected in cortical layer 1 in vivo (infected neurons were located in layer 2/3). Dashed vertical lines serve as a guide for comparison of structures over time. (a) Imaging of dendritic spines caused no photobleaching after 30 min at a 1-min sampling interval, with ▵F(t = 30 min) ≈0.97·▵F(t = 0min). (b) Imaging of axons in a layer 1 region adjacent to the injection site for >5 h at a 5-min sampling interval, with ▵F(t = 300 min) ≈0.66·▵F(t = 0 min). Some axonal projection endings, an example indicated by an arrow, showed directed outgrowth for several micrometers on a time scale of several hours in young animals (P15–P17 mice). Note the different scale bars in a and b.
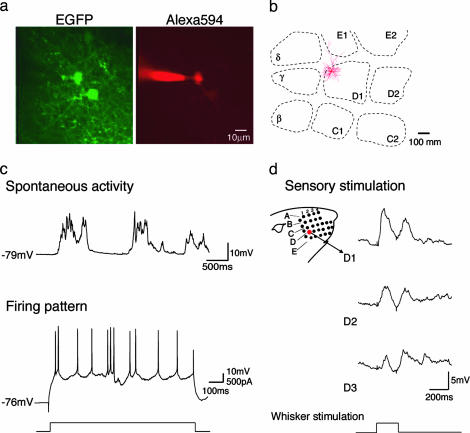
TPTP Recording. In the TPTP technique, two-photon imaging is used to simultaneously visualize EGFP-expressing neurons and a patch pipette containing a fluorescent dye to visually guide the pipette toward the labeled neurons and obtain targeted recording in vivo (32). We applied TPTP recording to layer 2/3 pyramidal neurons in the rat barrel cortex (Fig. 5). Animals were infected with the FCK(1.3)GW virus between P8 and P11, and the recordings were obtained between P21 and P24. After establishing the technique, the frequency of successful targeted recordings was, on average, one cell per five penetration attempts. Typically, we recorded from one neuron per animal, which allowed us to compare the sensory-evoked responses to the position of the neuron in the barrel cortex, as revealed by subsequent reconstruction of the cell's morphology (27) and cytochrome c staining of layer four barrels (26) (Fig. 5b). Recordings lasted, on average, for 30–40 min, and the infected neurons had normal passive membrane properties, somatic current injection-evoked firing patterns, and whisker deflection-evoked sensory responses (Fig. 5 c and d), as compared with previously reported data obtained by using blind-patch techniques (25, 33).
Fig. 5.
Physiological analysis of infected neurons by TPTP recording. (a) An example of an infected in vivo-patched neuron. The EGFP image shows FCK(1.3)GW-infected layer 2/3 cells, at a depth of 182 μm below pia; The Alexa 594 image shows a patch pipette in a whole-cell configuration on the infected cell shown in the center on the left. (b) Horizontal projection of dendritic arbors relative to position of layer 4 barrels of the patched infected neuron shown in a. The cell was reconstructed from tangential sections. (c) Spontaneous activity and firing pattern of FCK(1.3)GW-infected layer 2/3 neuron shown in a. Top trace shows typical two-state membrane fluctuations (up and down states) observed in cortical neurons in anesthetized animals; bottom trace shows spiking in the same cell, elicited with DC current injection. In total, nine regular spiking FCK(1.3)GW-infected neurons were recorded, with average resting membrane potential, Vrest (mV) = 69.3 ± 2.0; steady-state input resistance, Rin (MΩ) = 65.3 ± 9.3; depolarization required for AP initiation (mV) = 30.7 ± 1.7.(d) Examples of whisker deflection-evoked responses in the same cell. Deflection of the D1, D2, and D3 whiskers, as illustrated in the schema on the left, evoked the corresponding responses shown on the right. The time course of the whisker deflection is shown at the bottom.
The main objective of TPTP is to record from genetically modified cells in vivo. For this aim, the combination of TPTP recording with RNA interference, a method for gene silencing through expression of double-stranded siRNAs (34) may be particularly useful. Recently, the self-inactivating lentiviral vector was adapted for siRNA-based gene silencing by inserting a polymerase III promoter (either U6 or H1), which is used to express siRNA as a fold-back stem–loop structure (23, 35), and several studies have reported successful gene silencing with similar retroviral vectors in neurons (36–39). To quantitatively assess the efficiency of lentiviral vector-based gene silencing in neurons, we tested siRNA-based knockdown of EGFP expression in hippocampal primary cultures. To this end, neurons were infected first with the FUGW virus by using a high titer, leading to superinfection of all cells, and 3 days later, they were infected at a low titer with virus coexpressing a hairpin siRNA sequence against EGFP mRNA, GFPsiRNA (23), and a red fluorescent protein, dsRed2 (vector FU6siGURW, Fig. 8, which is published as supporting information on the PNAS web site). Even though basal EGFP expression in the infected neurons was high, both due to the efficient transcription driven from the ubiquitin C promoter in cultured neurons (Fig. 7) and multiple FUGW integrations per infected cells due to the high titer of the virus, we observed a strong (≈70%) decrease of EGFP fluorescence in GFPsiRNA-expressing neurons (Fig. 8). We conclude that siRNA driven from the U6 promoter in lentiviral vectors can cause a significant knockdown of gene expression in neurons, and, in combination with expression of EGFP, can be used to study gene functions in single neurons in vivo by two-photon imaging or TPTP recordings.
Discussion
Lentivirus-Based Gene Expression in Cortical Pyramidal Neurons in Vivo. The promoters tested here were chosen based on three criteria: pyramidal neuron-specificity of expression, high transcriptional rate, and a preference for compact promoter size. The first criterion is important because the viral particles were pseudotyped with vesicular stomatitis virus (VSV) glycoproteins that bind nonselectively to membrane phospholipids of all mammalian cells (40). Consequently, the CMV-IE/β-Actin and the ubiquitin C promoters drove considerable expression in glia, as well as neurons. Three promoters, Thy1.2, α-CaMKII, and Synapsin I, were found to be neuron-specific, with the Thy1.2 and α-CaMKII promoters labeling selectively pyramidal neurons, as determined from the infected cells' morphology. This result confirms that using lentiviruses pseudotyped with the VSV glycoprotein coat requires the selection of an internal promoter tailored to the cell type of choice.
In terms of the transcriptional strength, the ubiquitin C promoter was found to be best suited for expression in cultured neurons. The Synapsin I (1.1 kb) and α-CaMKII (1.3 kb) promoters drove neuron-specific expression, with the onset of efficient expression matching transcriptional regulation of endogenous Synapsin I and α-CaMKII genes in vivo. The rather low expression from the Thy1.2 promoter was surprising, because this promoter drove high expression in transgenic mice (15, 41). It is possible that we compromised the promoter efficiency to accommodate for the size limitation of the lentiviral vectors. The maximum combined size for the promoter and gene of interest cassette in these vectors is ≈10 kb (13). Because the original Thy1.2 transgenic cassette was 6.5 kb (15), we used only the 4-kb region upstream of the transcriptional start.
Several lentiviral vector types were previously described for expression of recombinant genes in neurons by using other promoters, e.g., CMV (12, 19, 20), phosphoglycerate kinase 1 (42), or neuron-specific enolase (43). The Synapsin I (1.1 kb) and α-CaMKII (1.3 kb) promoter-based vectors presented here provide a highly efficient expression in principal cortical neurons in vivo, and their compact size indicates that they can be used to express large genes, perhaps up to ≈8 kb. This size gives the vectors an additional advantage over other viral systems used for gene delivery to neurons, where the insert size is often more limited: for example, the broadly used adenoassociated viruses have promoter and gene of interest cassette limit of only ≈4.7 kb (8).
Application of Lentiviral Vectors to Study Gene Functions in Cortical Neurons in Vivo. Genetic manipulations described here offer several unique advantages for studying gene functions in cortical neurons in vivo. First, stereotactic injection makes it easy to control the delivery of the recombinant genetic material to a discreet region and to produce expression in a defined time window. Second, the fact that only a small number of neurons is affected offers additional benefits. A small population of neurons in the intact brain can be altered in a way that would result in a lethal phenotype or in an activation of compensatory mechanisms if the entire brain or whole-brain subregions were affected. This possibility may allow one to study in vivo functions of genes that otherwise would be possible to study only by the use of in vitro methods. Furthermore, fluorescent labeling of sparse neurons offers a high signal-to-noise resolution (bright labeling in an otherwise unlabeled tissue) for structural studies by two-photon imaging.
In summary, we show that the experimental approach described here can be used for efficient gene expression or gene knockdown in a small population of cortical neurons in vivo. Combination of the lentivirus-based genetic manipulations with high-resolution two-photon time-lapse imaging or TPTP recording can be applied to study a specific gene product for a function in single neurons in the intact cortex, with the option to selectively examine its role(s), either during early postnatal development or in the adult in a cortical region of choice.
Supplementary Material
Acknowledgments
We thank Peter H. Seeburg for support and interest, and Carlos Lois for FUGW, ▵8.9, and vesicular stomatitis virus G protein plasmids, as well as for assistance with setting up the lentivirus production. This work was supported in part by Deutsche Forschungsgemeinschaft Grant Os 191/1-1 (to P.O.) and by Boehringer Ingelheim Fonds (to A.N.).
Author contributions: T.W.M., F.H., W.D., M.B., and P.O. designed research; T.D., A.N., S.K., P.L., J.W., T.W.M., F.H., M.B., and P.O. performed research; T.D., A.N., S.K., and J.W. analyzed data; and P.O. wrote the paper.
Abbreviations: EGFP, enhanced GFP; α-CaMKII, α-calcium/calmodulin-dependent protein kinase II; CMV, cytomegalovirus; siRNA, short-interfering RNA; PFA, paraformaldehyde; Pn, postnatal day n; TPTP, two-photon microscopy targeted-patch; SV40, simian virus 40; FUGW, Flip, ubiquitin promoter,GFP, and WRE.
References
- 1.Mayford, M., Mansuy, I. M., Muller, R. U. & Kandel, E. R. (1997) Curr. Biol. 7, R580–R589. [DOI] [PubMed] [Google Scholar]
- 2.Sauer, B. (1998) Methods 14, 381–392. [DOI] [PubMed] [Google Scholar]
- 3.Gossen, M. & Bujard, H. (2002) Annu. Rev. Genet. 36, 153–173. [DOI] [PubMed] [Google Scholar]
- 4.Hubel, D. H. & Wiesel, T. N. (1962) J. Physiol. 160, 106–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Romo, R. & Salinas, E. (2003) Nat. Rev. Neurosci. 4, 203–218. [DOI] [PubMed] [Google Scholar]
- 6.Cohen, M. R. & Newsome, W. T. (2004) Curr. Opin. Neurobiol. 14, 169–177. [DOI] [PubMed] [Google Scholar]
- 7.Brecht, M., Schneider, M., Sakmann, B. & Margrie, T. W. (2004) Nature 427, 704–710. [DOI] [PubMed] [Google Scholar]
- 8.Kootstra, N. A. & Verma, I. M. (2003) Annu. Rev. Pharmacol. Toxicol. 43, 413–439. [DOI] [PubMed] [Google Scholar]
- 9.Lois, C., Refaeli, Y., Qin, X. F. & Van Parijs, L. (2001) Curr. Opin. Immunol. 13, 496–504. [DOI] [PubMed] [Google Scholar]
- 10.Ailles, L. E. & Naldini, L. (2002) Curr. Top. Microbiol. Immunol. 261, 31–52. [DOI] [PubMed] [Google Scholar]
- 11.Trono, D. (2000) Gene Ther. 7, 20–23. [DOI] [PubMed] [Google Scholar]
- 12.Miyoshi, H., Blomer, U., Takahashi, M., Gage, F. H. & Verma, I. M. (1998) J. Virol. 72, 8150–8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lois, C., Hong, E. J., Pease, S., Brown, E. J. & Baltimore, D. (2002) Science 295, 868–872. [DOI] [PubMed] [Google Scholar]
- 14.Niwa, H., Yamamura, K. & Miyazaki, J. (1991) Gene 108, 193–199. [DOI] [PubMed] [Google Scholar]
- 15.Caroni, P. (1997) J. Neurosci. Methods 71, 3–9. [DOI] [PubMed] [Google Scholar]
- 16.Mayford, M., Bach, M. E., Huang, Y. Y., Wang, L., Hawkins, R. D. & Kandel, E. R. (1996) Science 274, 1678–1683. [DOI] [PubMed] [Google Scholar]
- 17.Hoesche, C., Sauerwald, A., Veh, R. W., Krippl, B. & Kilimann, M. W. (1993) J. Biol. Chem. 268, 26494–26502. [PubMed] [Google Scholar]
- 18.Thiel, G., Lietz, M. & Cramer, M. (1998) J. Biol. Chem. 273, 26891–26899. [DOI] [PubMed] [Google Scholar]
- 19.Naldini, L., Blomer, U., Gallay, P., Ory, D., Mulligan, R., Gage, F. H., Verma, I. M. & Trono, D. (1996) Science 272, 263–267. [DOI] [PubMed] [Google Scholar]
- 20.Zufferey, R., Nagy, D., Mandel, R. J., Naldini, L. & Trono, D. (1997) Nat. Biotechnol. 15, 871–875. [DOI] [PubMed] [Google Scholar]
- 21.Brewer, G. J., Torricelli, J. R., Evege, E. K. & Price, P. J. (1993) J. Neurosci. Res. 35, 567–576. [DOI] [PubMed] [Google Scholar]
- 22.Yu, J. Y., DeRuiter, S. L. & Turner, D. L. (2002) Proc. Natl. Acad. Sci. USA 99, 6047–6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tiscornia, G., Singer, O., Ikawa, M. & Verma, I. M. (2003) Proc. Natl. Acad. Sci. USA 100, 1844–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kleinfeld, D. & Denk, W. (1999) in Imaging Neurons: A Laboratory Manual, eds. Yuste, R., Lanni, F. & Konnerth, A. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 23.1–23.15.
- 25.Brecht, M., Roth, A. & Sakmann, B. (2003) J. Physiol. 553, 243–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wong-Riley, M. (1979) Brain Res. 171, 11–28. [DOI] [PubMed] [Google Scholar]
- 27.Horikawa, K. & Armstrong, W. E. (1988) J. Neurosci. Methods 25, 1–11. [DOI] [PubMed] [Google Scholar]
- 28.Burgin, K. E., Waxham, M. N., Rickling, S., Westgate, S. A., Mobley, W. C. & Kelly, P. T. (1990) J. Neurosci. 10, 1788–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lendvai, B., Stern, E. A., Chen, B. & Svoboda, K. (2000) Nature 404, 876–881. [DOI] [PubMed] [Google Scholar]
- 30.Trachtenberg, J. T., Chen, B. E., Knott, G. W., Feng, G., Sanes, J. R., Welker, E. & Svoboda, K. (2002) Nature 420, 788–794. [DOI] [PubMed] [Google Scholar]
- 31.Grutzendler, J., Kasthuri, N. & Gan, W. B. (2002) Nature 420, 812–816. [DOI] [PubMed] [Google Scholar]
- 32.Margrie, T. W., Meyer, A. H., Caputi, A., Monyer, H., Hasan, M. T., Schaefer, A. T., Denk, W. & Brecht, M. (2003) Neuron 39, 911–918. [DOI] [PubMed] [Google Scholar]
- 33.Margrie, T. W., Brecht, M. & Sakmann, B. (2002) Pflügers Arch. 444, 491–498. [DOI] [PubMed] [Google Scholar]
- 34.Elbashir, S. M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K. & Tuschl, T. (2001) Nature 411, 494–498. [DOI] [PubMed] [Google Scholar]
- 35.Qin, X. F., An, D. S., Chen, I. S. & Baltimore, D. (2003) Proc. Natl. Acad. Sci. USA 100, 183–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xia, H., Mao, Q., Paulson, H. L. & Davidson, B. L. (2002) Nat. Biotechnol. 20, 1006–1010. [DOI] [PubMed] [Google Scholar]
- 37.Hommel, J. D., Sears, R. M., Georgescu, D., Simmons, D. L. & DiLeone, R. J. (2003) Nat. Med. 9, 1539–1544. [DOI] [PubMed] [Google Scholar]
- 38.Van den Haute, C., Eggermont, K., Nuttin, B., Debyser, Z. & Baekelandt, V. (2003) Hum. Gene Ther. 14, 1799–1807. [DOI] [PubMed] [Google Scholar]
- 39.Rubinson, D. A., Dillon, C. P., Kwiatkowski, A. V., Sievers, C., Yang, L., Kopinja, J., Rooney, D. L., Ihrig, M. M., McManus, M. T., Gertler, F. B., et al. (2003) Nat. Genet. 33, 401–406. [DOI] [PubMed] [Google Scholar]
- 40.Burns, J. C., Friedmann, T., Driever, W., Burrascano, M. & Yee, J. K. (1993) Proc. Natl. Acad. Sci. USA 90, 8033–8037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feng, G., Mellor, R. H., Bernstein, M., Keller-Peck, C., Nguyen, Q. T., Wallace, M., Nerbonne, J. M., Lichtman, J. W. & Sanes, J. R. (2000) Neuron 28, 41–51. [DOI] [PubMed] [Google Scholar]
- 42.Kordower, J. H., Bloch, J., Ma, S. Y., Chu, Y., Palfi, S., Roitberg, B. Z., Emborg, M., Hantraye, P., Deglon, N. & Aebischer, P. (1999) Exp. Neurol. 160, 1–16. [DOI] [PubMed] [Google Scholar]
- 43.Lai, Z. & Brady, R. O. (2002) J. Neurosci. Res. 67, 363–371. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.