Abstract
In vitro fermentation systems such as the rumen simulation technique (RUSITEC) are frequently used to assess dietary manipulations in livestock, thereby limiting the use of live animals. Despite being in use for nearly 40 years, improvements are continually sought in these systems to better reflect and mimic natural processes in ruminants. The aim of this study was to evaluate the effect of forage preparation, i.e., frozen minced (FM) and freeze-dried and ground (FDG), on the ruminal microbiota and on fermentation characteristics when included as a substrate in a RUSITEC system. A completely randomized design experiment was performed over a 15-day period, with 7 days of adaptation and an 8-day experimental period. Fermentation parameters (total gas, CH4, and volatile fatty acid production) were analyzed on a daily basis over the experimental period and the archaeal and bacterial microbiota (liquid-associated microbes [LAM] and solid-associated microbes [SAM] was assessed at 0, 5, 10, and 15 days using high-throughput sequencing of the 16S rRNA gene. Results from this study suggested a tendency (P = 0.09) of FM treatment to increase daily CH4 (mg/d) production by 16.7% when compared with FDG treatment. Of the major volatile fatty acids (acetate, propionate, and butyrate), only butyrate production was greater (P = 0.01) with FM treatment compared with FDG substrate. The archaeal and bacterial diversity and richness did not differ between the forage preparations, although feed particle size of the forage had a significant effect on microbial community structure in the SAM and LAM samples. The Bacteroidetes phylum was more relatively abundant in the FM substrate treatment, while Proteobacteria was enriched in the FDG treatment. At the genus-level, Butyrivibrio, Prevotella, and Roseburia were enriched in the FM substrate treatment and Campylobacter and Lactobacillus in the FDG substrate treatment. Evidence from this study suggests that forage preparation affects CH4 production, butyrate production, and the structure of the rumen microbiota during in vitro fermentation.
Keywords: RUSITEC, microbial ecology, rumen, methane, fermentation, forage, microbiota, cattle
Introduction
Reducing methane (CH4) emissions from anthropogenic activities is of considerable interest since enteric fermentation from ruminants accounts for 25% of the 40% derived from agriculture (Olivier et al., 1999; Steinfeld et al., 2006). Ruminants are considered economically important due to their capacity to digest low-quality forages (Flint, 1997) and their ability to convert these substrates into energy is largely dependent on the rumen microbiota (i.e., bacteria, anaerobic fungi, protozoa, and methanogenic archaea) which converts indigestible plant material into usable energy for the host. In addition, this allows ruminants to produce milk, meat, wool, and leather without competing directly with humans for food (Buddle et al., 2011).
Rumen microbial communities are known to respond to changes in diet, environment and to a lesser extent, the type of host (Henderson et al., 2015). Diet type (concentrate, forage), as well as the preparation, influences the fermentation process and the composition of the rumen microbiota (Henderson et al., 2015). In vitro fermentation systems such as the rumen simulation technique (RUSITEC) are frequently used to assess dietary manipulations in livestock, reducing the use of live animals for experiments. Although the RUSITEC system has been used in research for almost 40 years (Czerkawski and Breckenridge, 1977), improvements are sought in these systems that may better reflect and mimic natural processes in ruminants. For example, there is a requirement for appropriate methods that describe the preparation of fresh forage material to be used in in vitro systems. In recent years, it has been common practice to use forages in freeze-dried and ground (FDG) form as a substrate in these systems (Avila-Stagno et al., 2014), however, digestion products and kinetics are altered when fresh forage is used (Barrell et al., 2000).
The type of feed preparation has very significant effects on degradation kinetics, in terms of constituent disappearance, proteolysis, volatile fatty acids production, and microbial growth. Although conventional feed preparation involves freeze drying and grinding, this is not appropriate for fresh forages which have been both minced and freeze dried for comparison (McNabb et al., 1996). Barrell et al. (2000) and Cohen and Doyle (2001) demonstrated degradation kinetics of fresh chopped, minced and also freeze dried and ground material. They suggested the minced preparation to be most appropriate for fresh grasses and legumes. In vitro incubations should be carried out with feed prepared in a way that best mimics chewing by ruminants.
Presently, limited research has been conducted on the effect of the forage/substrate form on the rumen microbiota or on fermentation parameters in in vitro systems. The aim of this study was to evaluate the effect of two different forms of forage substrate, frozen minced (FM) and FDG, on fermentation parameters (including CH4) and the archaeal and bacterial microbiota, using a RUSITEC system. Our hypothesis was that by changing substrate preparation, digestibility, fermentation characteristics, and microbial population would be severely impacted.
Materials and Methods
Experimental Design and Treatments
This study was conducted using a RUSITEC system containing six fermentation vessels with samples taken over a 15-day period (7 days of adaptation followed by 8 days where samples were taken). The experiment was a completely randomized design with two treatments and three replicates per treatment. The fermentation substrate consisted of equal portions of concentrate and ryegrass that was included either as FM preparation (FM treatment) or FDG to 2 mm forage (FDG treatment). Fermentation parameters (total gas, CH4, and volatile fatty acid [VFA] production) were analyzed on a daily basis over the experimental period and the archaeal and bacterial microbiota (liquid-associated microbes [LAM] and solid-associated microbes [SAM]) were characterized on days 0, 5, 10, and 15.
Plant Material
Plant material of ryegrass (Lolium perenne) was collected on 15th January 2015 at the May Farm research site of the University of Sydney, Camden Campus, NSW, Australia (34°04′ S; 150°81 69′ E). The climate is warm-temperate with a mean annual minimum and maximum temperature of 10.7 and 23.3°C, respectively. The annual average rainfall is 738 mm (1900–2010). Multiple samples were randomly selected and harvested at grazing height ≥ 5 cm above ground level to mimic grazing by cattle. Plant material was processed immediately upon return to the laboratory (within 45 min of collection) and divided into two equal portions. One half of the plant material (FM sample) was prepared according to the methodology described by Chaves et al. (2006) as follows: material was frozen at -20°C and while still frozen, cut into 2–3 cm lengths using scissors and minced using a meat mincer (Rovtek MG-22SS, Commercial Meat Mincer, 800 W, 250 kg/h, blade speed 190 rotation/h, Sydney, NSW, Australia) fitted with a screen plate with 12 mm holes. The samples were stored at -20°C until the day of incubation. Preparation of FDG samples involved freeze-drying the ryegrass, grinding and then passing the ground ryegrass through a 2 mm screen. Samples were kept at room temperature until the day of incubation.
Chemical Analysis
Ryegrass and concentrate were analyzed for dry matter (DM) (method 967.03), ash (method 942), and ether extract (EE) content by extraction with diethyl ether using an Ankom XT10 Extraction System (Ankom® Technol. Corp., Fairport, NY, USA; method 920.39), following Association of Official Analytical Chemists [AOAC] (2006) methods. The concentration of neutral detergent fiber (NDF) was determined using procedures detailed by Van Soest et al. (1991) and modified for an Ankom 200/220 Fiber Analyzer (Ankom Technol. Corp., Fairport, NY, USA) using sodium sulfite and heat stable α-amylase. Crude Protein (CP) was analyzed by combustion [method 990.03 (Association of Official Analytical Chemists [AOAC], 2006)] using FP628 Food/Protein Analyzer (LECO, St Joseph, MI, USA) following the manufacturer’s guidelines. The feed chemical composition is presented in Supplementary Table S1.
Substrate and Rumen Inoculum
The rumen inoculum was collected on the initial day of the experiment from one ruminally fistulated (Soliva et al., 2011, 2015) Holstein dairy cow (9 years old, 750 kg). The donor animal was housed at The University of Sydney Corstorphine Dairy farm and was cared for in accordance with the guidelines of The University of Sydney Animal Ethics Committee (Project number 2015/835). The cow was fed the same diet (Supplementary Table S1) that was used as a substrate in the RUSITEC fermenters and the inoculum was collected 2 h after the morning feeding. The rumen digesta was filtered through four layers of cheesecloth to separate the solid and liquid portion to be used as an initial inoculum. The rumen inoculum was transported immediately to the laboratory using insulated containers.
Approximately 5 g (DM basis) of each of the treatments was weighed into pre-weighed nylon bags (70 mm × 140 mm; pore size = 150 μm) and approximately 5 g (DM basis) of concentrate feed was weighed into a separate nylon bag (70 mm × 100 mm; pore size = 150 μm). At the start of the experiment, 4.7 L of rumen fluid was evenly distributed into six fermentation vessels, and equal amounts (i.e., ∼50 g) of rumen solids were weighed into nylon bags.
RUSITEC Fermentation
The RUSITEC apparatus was equipped with six 800 mL fermentation vessels. Each vessel had an inlet for the infusion of buffer and an effluent output port. At the start of the experiment each fermentation vessel was filled with 780 mL of rumen fluid. The nylon bags (containing about 30 g of wet weight of rumen solids and the experimental diets) were placed inside each fermenter according to the randomized treatments. Fermenters were then submerged in a 39°C water bath and infused with McDougall’s buffer [3.69 g/L Na2HPO4 (anhydrous), NaHCO3 9.8 g/L, NaCl 0.47 g/L, KCl 0.57 g/L, MgCl26H2O 0.061 g/L, CaCl22H2O 0.0336 g/L] at a dilution rate of 30 mL/min.
After 24 h of incubation, the rumen solids bag was replaced with new nylon bags containing the experimental treatments, and from day 2 onward, the nylon bags were replaced each day, meaning that four bags were present at any given time (Narvaez et al., 2013; Avila-Stagno et al., 2014).
Total Gas and Methane Production
Total gas produced was collected on a daily basis in gas-tight bags (Plastigas, Linde AG, Munchen, Germany) connected to the effluent flasks. The total volume of gas was determined by water displacement. The gas bag was connected to a flask filled with water and the gas was evacuated by applying manual pressure to the flask. The evacuated water was then collected in a graduated cylinder and the volume collected was measured as daily gas production and expressed in mL/d.
From day 8 until the end of the experiment, 15 mL from each gas bag was removed with a syringe and transferred to an exetainer tube (Labco Ltd, Lampeter, UK) for analysis of methane concentration by gas chromatography (Bruker 450 GC, Bruker Technologies, Australia, with two packed columns and a Compass CDS data acquisition software (Bruker Technologies, Australia). Argon and helium were used as carrier gases at a flow rate of 30 mL/min. The oven, injector and detector temperatures were 50, 70, and 180°C, respectively.
Dry Matter Disappearance
Dry matter disappearance was determined using the residue remaining in each nylon bag after 48 h of fermentation. Nylon bags were removed, washed with cold water (tap water/distilled water) until the water was clear (Narvaez et al., 2013; Li et al., 2014) and then dried at 100°C for 24 h. The residue weight was recorded and used for the calculation of DM disappearance.
Fermentation Parameters and Volatile Fatty Acid (VFA) Production
Fermentation parameters, such as pH and volume outflow, were determined on a daily basis during the 15-day experimental period (i.e., 7 days of adaptation and 8 days of collection). Volume was measured daily at the time of feed bag exchange with a graduated cylinder. Starting at day 8, individual samples from each fermentation vessel were taken for quantification of VFA. Samples were preserved with metaphosphoric acid (25% w/v; 1:5 dilution) and stored at -20°C until analysis. Capillary gas chromatography was performed to analyze VFA (C:2 to C:6) according to the Biochemistry Laboratory Method NTR-7, Issue No. 5, 05/05/2015 (University of Western Australia). An Agilent 6892 Series GC with Agilent 7696 sample preparation station and HP 6890 injector with HP Chemstation software were used. The capillary column was a HP-FFAP, 30 m × 0.53 mm × 1.0 μm (HP Part. No. 199095F-123). Oven temperature was 240°C and hydrogen was used as carrier with a total flow of 55.7 mL/min at 3.5 psi. The HP Chemstation system calculated the concentration for the VFA in mmol/L. Concentration of volatile fatty acids production was calculated per day by multiplying concentration and volume of outflow.
Sample Collection for Microbiota Analysis
In order to investigate the effect of forage preparation on the rumen microbial community, samples were taken at the start of the experiment (day 0) from the original rumen fluid and solids, and on days 5, 10, and 15 for rumen fluid from the RUSITEC fermenters. To characterize LAM, a 10 mL sample from each fermenter was collected into a sterilized 25 mL falcon tube, immediately frozen in liquid nitrogen and kept at -80°C until DNA extraction. To assess SAM, on the last day of the experiment (i.e., day 15), the digesta in the nylon bags was frozen in liquid nitrogen and then subjected to freeze drying before DNA extraction.
DNA Extraction and Illumina Sequencing of the Archaeal and Bacterial 16S rRNA Gene
Total DNA was extracted using a QIAamp Fast DNA stool mini kit (Qiagen), according to manufacturer’s instructions and a lysis temperature of 80°C. The DNA yield and purity was assessed using a NanoDrop 2000 Spectrophotometer (Thermo Scientific) and extracted DNA was stored at -20°C. The 16S rRNA gene libraries were generated using a two-step PCR protocol. The first PCR step amplified the V4 region of the 16S rRNA gene using the universal bacterial and archaeal primers 515-F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806-R (5′-GGACTACVSGGGTATCTAAT-3′) (Caporaso et al., 2011). The second PCR step was used to add a unique 10-bp barcode at the 5′ end of the amplicon as well to add Illumina adapters. All PCR amplification and sequencing steps were carried out at Genome Quebec (Montreal, QC, Canada). Briefly, the 16S rRNA gene amplicons were quantified using a Quant-iT PicoGreen dsDNA assay kit (Invitrogen, Burlington, ON, Canada), pooled in equimolar ratios, and then purified with AMPure XP beads (Beckman Coulter, Mississauga, ON, Canada). The 16S rRNA gene amplicons were then sequenced using an Illumina MiSeq (2 × 250) and the MiSeq Reagent Kit v2 (500 cycles; Illumina, San Diego, CA, USA) according to manufacturer’s instructions.
16S rRNA Gene Sequence Analysis
The 16S rRNA gene sequences were processed and analyzed within the QIIME software package v. 1.9.1 (Caporaso et al., 2010b). Paired-end reads were joined using fastq-join with a minimum overlap of 35 bp and a maximum percent difference of 15 (Aronesty, 2013). Joined sequences were quality filtered with sequences being truncated following three consecutive base calls of a Phred score of less than 25. Sequences were retained only when 75% or more of the original sequence remained after truncation. Chimeric sequences were removed using the UCHIME algorithm (Edgar et al., 2011) implemented in USEARCH v. 6.1544 (Edgar, 2010). Sequences were then clustered into operational taxonomic units (OTUs) at 97% similarity using an open reference OTU picking method and the SILVA database v. 111 (Pruesse et al., 2007). Sequences that did not match OTUs in the SILVA database were clustered into OTUs using the de novo approach and USEARCH. The UCLUST consensus taxonomy assigner (Edgar, 2010) was used to assign taxonomy to OTUs using the SILVA database, with a minimum similarity of 0.8 and max accepts of 3. Representative sequences for the OTUs were aligned using PyNast (Caporaso et al., 2010a) and a phylogenetic tree was created using FastTree (Price et al., 2010). OTUs containing fewer than 10 sequences were excluded from further analysis as well as those OTUs classified as chloroplasts.
Each sample was randomly subsampled to 22,500 sequences per sample to account for uneven sequencing depth. The bacterial and archaeal diversity in each sample was calculated within QIIME using the Shannon index (Shannon, 1948) and phylogenetic diversity (PD whole tree) (Faith, 1992). Unweighted UniFrac distances (Lozupone and Knight, 2005) were used to assess the archaeal and bacterial community structure (beta-diversity) of each forage preparation type, sampling time, and sample type (solid vs. liquid). The subsequent distance matrices were visualized as principal coordinate analysis (PCoA) plots using Emperor (Vazquez-Baeza et al., 2013).
All 16S rRNA gene sequences were submitted to the NCBI Sequence Read Archive (SRA) under bio-project accession PRJNA3047651.
Statistical Analysis
Fermentation and alpha-diversity data were analyzed using the MIXED procedure of SAS (SAS, Inc., 2015; SAS Online Doc 9.1.3). The model included the fixed effects of forage preparation, day and forage preparation × day interaction. Therefore, the individual fermenter was used as the experimental unit for statistical analysis. The minimum values of Akaike’s information criterion were used to select the covariance structure. Significance was declared at P ≤ 0.05 and a trend was discussed when 0.05 < P ≤ 0.10.
Unweighted UniFrac distances were compared using ANOSIM (analysis of similarities) with 999 permutations. OTUs that were differentially abundant by sample type, forage preparation type and sampling time were identified using the G-test of independence with a false discovery rate (FDR) < 0.05. Linear discriminant analysis effect size (LEfSe) was used to determine which phyla and genera were enriched at each sampling time, forage preparation type, and sample type. LEfSe uses the Kruskal–Wallis test to identify different (P < 0.05) genera among sample groups and uses linear discriminant analysis (LDA) to estimate the effect size of each of these (Segata et al., 2011). A LDA score of 3.5 and an overall relative abundance of greater than 0.01% was used as the threshold for identifying differentially abundant genera.
Results
Fermentation Parameters
The type of forage substrate preparation did not affect total gas, CH4 (%, mg/g DM, and mg/g DDM) or digestibility of substrates (P ≥ 0.13; Table 1). Total VFA, propionate, BCVFA (branched-chain volatile fatty acids), acetate to propionate ratio and pH were also not affected by type of preparation, but there was a tendency of increase in daily CH4 (mg/d) production in FM compared to FDG (Table 2). An interaction between forage preparation and sampling time was observed for acetate, valerate and caproate (Figure 1; P ≤ 0.05). Butyrate production was greater (Table 1; P = 0.01) in FM compared to FDG treatments.
Table 1.
Effects of forage preparation on total gas, methane, and substrate disappearance in the RUSITEC fermentation.
|
P-value |
||||||
|---|---|---|---|---|---|---|
| Freeze-dried and ground | Frozen minced | SEM | Preparation | Day | Preparation × Day | |
| Total gas, mL/d | 1708.5 | 1758.6 | 144.59 | 0.82 | 0.05 | 0.66 |
| Total gas, mL/g DM | 170.0 | 179.0 | 10.13 | 0.55 | <0.01 | 0.21 |
| Total gas, mL/g DMD | 193.6 | 204.0 | 11.54 | 0.55 | <0.01 | 0.23 |
| CH4, % | 7.3 | 7.4 | 0.46 | 0.88 | 0.44 | 0.85 |
| CH4, mg/d | 90.8 | 106.0 | 5.54 | 0.09 | 0.10 | 0.46 |
| CH4, mg/g DM | 9.0 | 10.3 | 0.54 | 0.13 | 0.10 | 0.47 |
| CH4, mg/g DMD | 10.3 | 11.7 | 0.68 | 0.20 | 0.15 | 0.63 |
| Forage IVDMD, % | 91.1 | 92.3 | 0.67 | 0.24 | 0.02 | 0.12 |
| Concentrate IVDMD, % | 84.5 | 83.2 | 0.64 | 0.19 | 0.07 | 0.02 |
SEM, standard error of mean; DM, dry matter; DMD, dry matter disappearance; IVDMD, in vitro dry matter disappearance.
Table 2.
Effect of forage preparation on pH and volatile fatty acids (VFA) production.
|
P-value |
||||||
|---|---|---|---|---|---|---|
| Freeze-dried and ground | Frozen minced | SEM | Preparation | Day | Preparation × Day | |
| pH | 6.9 | 6.9 | 0.005 | 0.82 | 0.06 | 0.56 |
| Total VFA, mmol/d | 37.2 | 36.3 | 0.83 | 0.51 | <0.01 | 0.21 |
| Acetate, mmol/d | 17.4 | 16.1 | 0.41 | 0.07 | <0.01 | 0.05 |
| Propionate, mmol/d | 9.5 | 8.9 | 0.25 | 0.17 | 0.03 | 0.19 |
| Butyrate, mmol/d | 5.3b | 6.0a | 0.16 | 0.01 | 0.01 | 0.19 |
| Valerate, mmol/d | 2.7 | 2.9 | 0.06 | 0.05 | 0.27 | 0.03 |
| Caproate, mmol/d | 0.2b | 0.3a | 0.01 | 0.02 | <0.01 | 0.01 |
| BCVFA, mmol/d | 2.1 | 2.1 | 0.06 | 0.51 | 0.02 | 0.64 |
| Acetate:propionate | 1.8 | 1.8 | 0.05 | 0.71 | <0.01 | 0.42 |
Different lowercase letters indicate significant differences between freeze-dried and ground and frozen minced prepared forages (P < 0.05). BCVFA, branched-chain VFA; iso-butyrate + iso-valerate.
FIGURE 1.
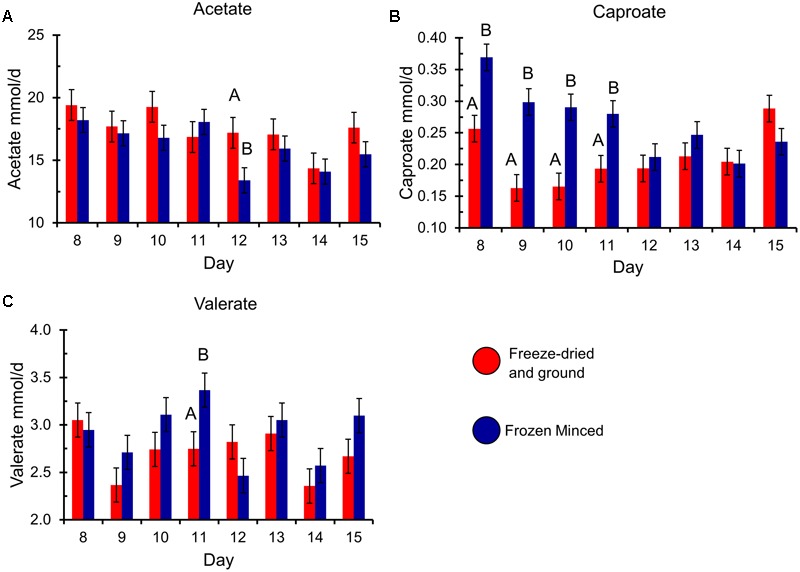
Production of the volatile fatty acids (A) acetate, (B) caproate, and (C) valerate, by experimental day and forage preparation type. Error bars represent standard error of the mean. Forage preparation types that significantly differ (P < 0.05) within each experimental day are indicated by different uppercase letters.
Archaeal and Bacterial Microbiota
A total of 858,881 sequences with an average length of 262 bp among all samples remained following quality filtering and removal of primer sequences. These sequences were clustered into 3,349 OTUs. Overall, the most relatively abundant phyla were: Bacteroidetes (60.1%), Firmicutes (24.3%), Spirochaetes (3.8%), Euryarchaeota (3.7%), Planctomycetes (2.6%), and Proteobacteria (1.6%) (Supplementary Figure S1). At the genus-level, Prevotella was the most relatively abundant (44.8%), while Dialister (8.4%), Succiniclasticum (3.5%), Lactobacillus (2.6%), and Treponema (2.1%) were the only other genera with an overall relative abundance > 2.0% (Figure 2).
FIGURE 2.
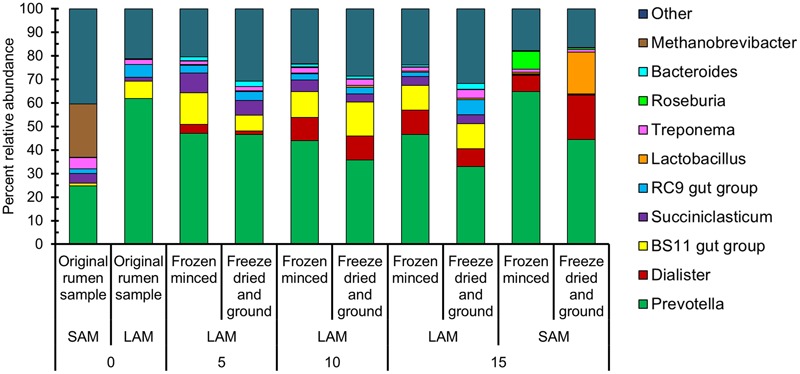
The 10 most relatively abundant genera among all samples for each sample type, forage preparation type, and sampling day. LAM, liquid-associated microbes; SAM, solid-associated microbes.
In regards to methanogens, only the solid fraction from the original rumen sample had a relatively high abundance of methanogens (classes Methanobacteria and Methanomicrobia; 22.9%). Nearly all of these methanogenic sequences were identified as Methanobrevibacter (22.6%). Excluding the original rumen samples, the relative abundance of the methanogens was 1.6% among all samples. Apart from Methanobrevibacter, the three other methanogenic genera identified among all samples were Methanosphaera, Methanomicrobium, and Methanimicrococcus. Overall, there were 28 OTUs that were classified into one of the four genera above.
Alpha-diversity (within-sample diversity) measures are presented in Table 3 for each sample preparation type. Archaeal and bacterial diversity and richness were not affected by forage preparation type (P > 0.05) in LAM samples, however, lower phylogenetic diversity was observed in LAM samples at day 5 compared with days 10 and 15 (68.84 vs. 74.8 and 74.2, respectively; P < 0.05). The archaeal and bacterial diversity and richness in SAM samples were also not affected by the forage preparation (P > 0.05; Table 3).
Table 3.
Archaeal and bacterial richness and diversity measures for liquid-associated microbes (LAM) and solid-associated microbes (SAM) samples for freeze-dried and ground and frozen minced forage types.
|
P-value |
||||||
|---|---|---|---|---|---|---|
| Freeze-dried and ground | Frozen minced | SEM | Preparation | Day | Preparation × Day | |
| LAM | ||||||
| Shannon index | 4.34 | 4.16 | 0.105 | 0.257 | 0.831 | 0.233 |
| PD whole tree | 72.2 | 73.1 | 1.06 | 0.541 | 0.022 | 0.172 |
| Number of OTUs | 1070 | 1088 | 26.0 | 0.643 | 0.243 | 0.135 |
| SAM | ||||||
| Shannon index | 3.61 | 3.87 | 0.101 | 0.141 | – | – |
| PD whole tree | 59.7 | 59.1 | 2.404 | 0.848 | – | – |
| Number of OTUs | 908 | 947 | 49.445 | 0.607 | – | – |
PD, phylogenetic diversity; OTU, operational taxonomic unit.
The effect of microbial sample type (LAM vs. SAM), forage preparation, and sampling time, on the structure of the archaeal and bacterial microbiota was analyzed using unweighted UniFrac distances (Figures 3–5). Visual inspection of the PCoA plots and ANOSIM of unweighted UniFrac distances demonstrated that sample type (LAM vs. SAM) was the most important factor in determining the structure of the microbiota (Figure 3, R-value = 0.982; P = 0.001). Similar to the within-sample diversity analysis, sampling time also had a significant effect on the structure of the microbiota in LAM samples. In particular, samples from day 5 clustered separately from those of days 10 and 15 (Figure 4, R-value = 0.423; P = 0.005). Samples also clustered together based on forage preparation type (Figure 5, R-value = 0.261; P = 0.002).
FIGURE 3.
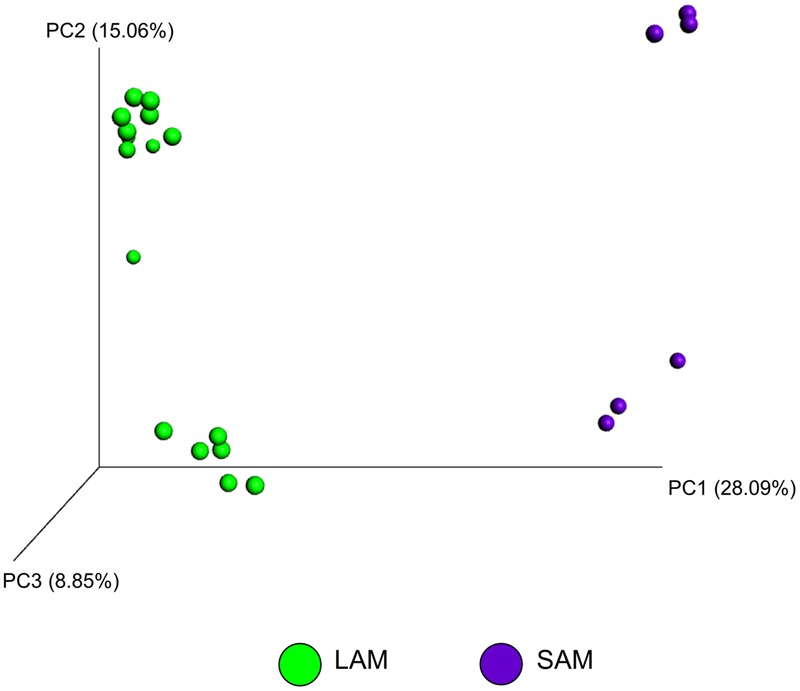
Principal coordinate analysis (PCoA) of the unweighted UniFrac distances for SAM and LAM samples. The percent variation explained by each principal coordinate is indicated on the axes. Original rumen samples are excluded. LAM, liquid-associated microbes; SAM, solid-associated microbes.
FIGURE 5.
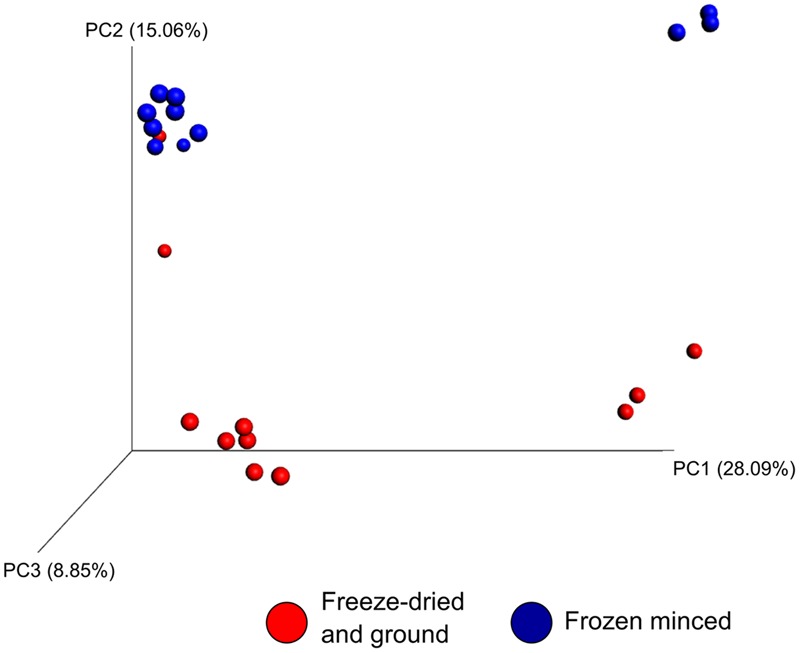
Principal coordinate analysis of the unweighted UniFrac distances for forage preparation for both LAM and SAM samples. The percent variation explained by each principal coordinate is indicated on the axes. No rumen samples included LAM, liquid-associated microbes; SAM, solid-associated microbes.
FIGURE 4.
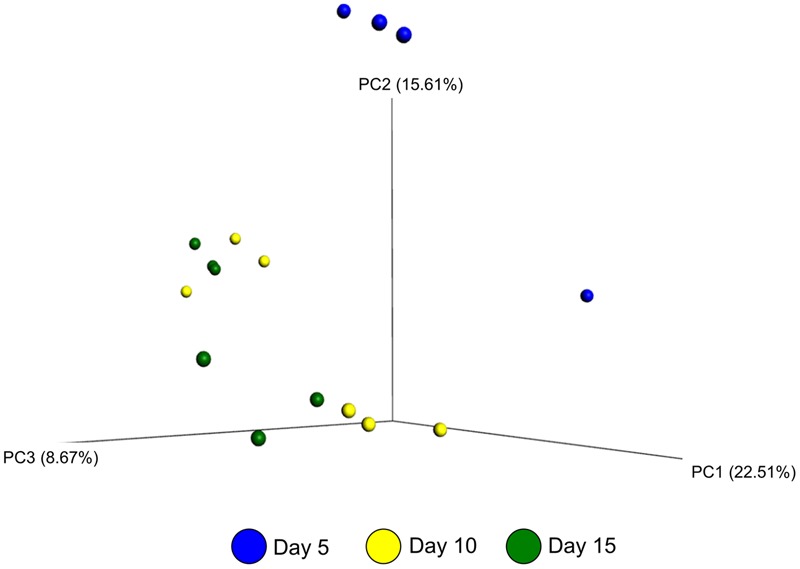
Principal coordinate analysis of the unweighted UniFrac distances for LAM samples at different sampling days. The percent variation explained by each principal coordinate is indicated on the axes. Original rumen samples are excluded. LAM, liquid-associated microbes.
Differentially abundant OTUs were identified using the G-test of independence. Excluding the original rumen samples, there were 244 OTUs that were differentially abundant (FDR < 0.05) between sample type (LAM vs. SAM) and 118 differentially abundant OTUs between the two forage preparations (both SAM and LAM samples included). Phyla enriched in one of the two sample types (LAM vs. SAM) were identified using LEfSe (Figure 6A; LDA [log10] score > 3.5). There was a total of six phyla enriched in the LAM samples and two in the SAM samples, with Firmicutes notably more relatively abundant in the SAM samples. At the genus-level, Succiniclasticum, Bacteroides, and Treponema were among the genera that were more relatively abundant in the LAM samples, while Lactobacillus and Roseburia were enriched in the SAM samples (Figure 6B).
FIGURE 6.
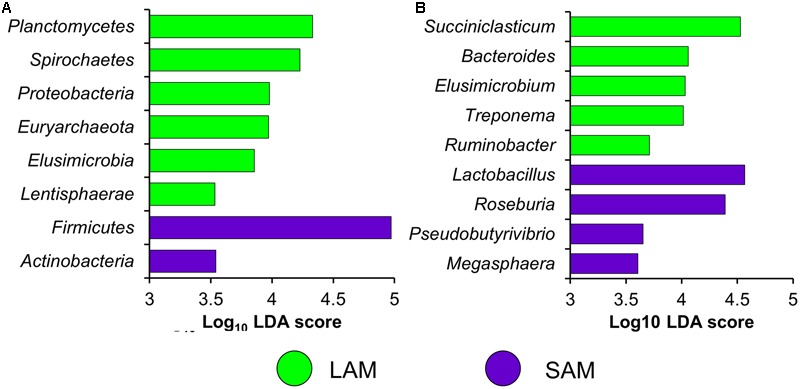
Differentially abundant (A) phyla and (B) genera between LAM and SAM samples as determined using linear discriminant analysis effect size (LEfSe) analysis. Only phyla and genera with a LDA score >3.5 and an overall relative abundance of >0.01% are included.
Only the Proteobacteria phylum was more relatively abundant in the FDG samples and Bacteroidetes was the only phylum enriched in the FM samples (LDA [log10] score > 4.0; P < 0.05). In the FDG samples, the genera Campylobacter and Lactobacillus were more relatively abundant while Butyrivibrio, Prevotella, and Roseburia were more relatively abundant in the FM samples (LDA [log10] score > 3.5; P < 0.05). Interestingly, Campylobacter was almost completely absent from the FM samples, as 9 of the 11 samples analyzed had no Campylobacter sequences, while all but two of the 11 FDG samples had greater than 0.1%. Among sampling times in the LAM samples, Firmicutes was also enriched at day 10 and Planctomycetes, Synergistetes, and Verrucomicrobia more relatively abundant at day 15. No phyla were enriched at day 5 compared to the other two sampling times. Similarly, among the genera with an overall relatively abundance greater than 0.01%, only Dialister was enriched at day 10 and Acidaminococcus and Megasphaera in day 15 samples (LDA [log10] score > 4.0; P < 0.05).
Discussion
Understanding and characterizing the rumen microbiota has become increasingly important in recent years as it has been demonstrated that the ruminal microbiota is influenced by and responds to changes in diet (Dehority, 2003; Henderson et al., 2015). In the present study, we examined the effect of FM vs. FDG forage preparation on CH4, total gas, and VFA production, and on the archaeal and bacterial microbiota, using a RUSITEC system.
Our findings indicated that forage substrate preparation had very little effect on fermentation parameters. FM preparation increased (P = 0.10) methane production (mg/d) by 16% compared to FDG. This is in agreement with previous reports from in vitro studies where authors had used freeze-dried plant samples for assessing anti-methanogenic compounds or alternative feed sources (García-González et al., 2008; Durmic et al., 2010; Li et al., 2014). However, care must be taken when interpreting these results, as some trends were observed in terms of methane production, and it may be possible that the freeze-drying is underestimating methane production. This is not unexpected, as grinding reduces particle size which in turn may result in better digestion (McAllister et al., 1994) and less methane production (Hales et al., 2012). However, FM preparation of forages achieves a forage particle distritution similar to what occurs in the chewing activity McNabb et al. (1996) and Barrell et al. (2000).
In terms of the molar proportion of VFA, a significant increase in butyrate production for FM forage preparation was found, a result that is in agreement with Mohammed et al. (2014) who reported a greater proportion of butyrate in fresh forage compared with conserved forage. However, our results did not show the same increasing trend for acetate, even though there was an interaction between time and forage preparation (Figure 1A). This interaction was only observed on day 12 of the experiment. Since propionate production and the acetate:propionate ratio was not affected by fresh or dried forage, differences in the VFA molar proportions could be related to the shifts in microbial communities. As Mohammed et al. (2014) reported, fresh or dried forage could result in differences based on the availability of specific nutrients.
The archaeal and bacterial microbiota was significantly affected by both microbial sample type (LAM vs. SAM) and the type of forage substrate preparation. Although archaeal and bacterial diversity and richness were unchanged, there were differences in various taxa between the two treatments (Figure 6). The changes observed in the microbial community structure (Figure 5) are noteworthy because to our knowledge, this is one of the first studies to use high-throughput sequencing to evaluate the rumen microbiota in response to different forage preparations. Although a number of studies have been conducted to evaluate the effect of diet on the rumen microbial community structure, most of these have investigated the effect of total mixed ration diets (Jami et al., 2014; Pitta et al., 2014; Veneman et al., 2015) or type of forages (Huws et al., 2010), rather than the forage preparation type.
de Menezes et al. (2011) used pyrosequencing of the 16S rRNA gene to compare the effect of feed pasture vs. total mixed ration diets on the rumen microbiota, noting several changes in the bacterial microbiota between diets and between the liquid and solid content. Our results are consistent with previous findings, as we found that the rumen microbiota was significantly different between the liquid and solid fractions, as demonstrated through the phylogenetic-based unweighted UniFrac distances (Figure 3). Mohammed et al. (2014) also found that bacterial communities clustered primarily by rumen digesta phase (solid vs. liquid) and that the microbiota of each fraction was dissimilar from the other.
In terms of archaeal community structure, the low relative abundance of the phylum Euryarchaeota limited our ability to perform an analysis based only on the methanogens. Methanogens were only present at a relative abundance of greater than 5.2% in the original rumen inoculum (SAM). Nonetheless, we found that Euryarchaeota, excluding the original rumen inoculum, was enriched in the LAM samples (Figure 6A). These findings indicate that methane production in the rumen may be influenced by the relative abundance of Archaea, rather than the microbial population structure as argued by Wallace et al. (2014) and Veneman et al. (2015), since the FM forage preparation showed an increased trend on methane production.
In the current study, the rumen microbiota was dominated by the Bacteroidetes and Firmicutes phyla, in both FM and freeze-dried preparation samples. Similarly, de Menezes et al. (2011) reported that Bacteroidetes and Firmicutes are the most relatively abundant bacterial phyla (>80%) in the rumen of dairy cattle on pasture and concentrate diets. However, these authors also noted a relatively high abundance of Fibrobacteres sequences in the solid phase of TMR and pasture diets (5–10%), although in the present study it only accounted for less than 1% of the sequences among all samples. However, this result to similar to a study of the rumen microbiota by Jami and Mizrahi (2012) where the relative abundance of Fibrobacteres was 0.02% in the rumen of dairy cattle fed 30% roughage and 70% concentrate.
We observed that Prevotella was the dominant genus among all samples, a result similar to that of in vivo studies of the rumen microbiota (Jami and Mizrahi, 2012; Jami et al., 2013; Thoetkiattikul et al., 2013; Paz et al., 2016). Jami et al. (2014) reported that certain genera in the Firmicutes phylum such as Dialister and Lactobacillus, were also negatively correlated with Prevotella. In the current study, Prevotella was more relatively abundant in the FM samples and Lactobacillus in the FDG samples. The enrichment of Bacteroidetes in the FM forage and Firmicutes in the SAM samples concurs with the findings of de Menezes et al. (2011) where a pasture diet vs. TMR diet was compared in vivo. In terms of forage preparation, the fact that Butyrivibrio, Prevotella, and Roseburia were enriched in the FM samples is consistent with the increased concentration of butyrate in these samples, as all of these genera contain major producers of butyrate (Flint et al., 2012; Sun et al., 2015). Caproate, the concentration of which was also greater (P < 0.05) in the FM samples, has also been found to be positively associated with Prevotella spp. (Tap et al., 2015). Therefore, the higher relative abundance of Prevotella in the FM samples may indicate that this forage preparation method results in a microbiota that is more similar to the natural rumen.
Conclusion
We can partially reject our initial hypothesis since this study demonstrated that fermentation parameters are generally not affected by the type of forage preparation with the exception of butyrate and a small trend in methane production. Prevotella and the butyrate producers, Butyrivibrio and Roseburia, were more relatively abundant in the FM prepared forage treatment, suggesting that this forage preparation method yields a microbiota that is more similar to that of the natural rumen. The diversity and richness of the microbiota was also not influenced by forage treatment, however, the structure of the rumen microbiota was significantly different between the LAM and SAM samples, as well as between forage substrate treatments.
Author Contributions
AD and AC designed the study. AD, AC, DH, TA, and ZD conducted the experimental procedures and laboratory analyses. AD, AC, DH, and TA analyzed and interpreted the data. AD drafted the manuscript. AD, AC, DH, TA, ZD, and PV critically revised the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Joy Vadhanabhuti (The University of Western Australia) for the CH4 gas analysis and Dr. Ajantha Horadagoda (MC Franklin lab – The University of Sydney) for the grass and rumen contents sampling.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00704/full#supplementary-material
The eight most relatively abundant phyla among all samples for each sample type, forage preparation type, and sampling day. LAM, liquid-associated microbes; SAM, solid-associated microbes.
References
- Aronesty E. (2013). Comparison of sequencing utility programs. Open Bioinform. J. 7 1–8. 10.2174/1875036201307010001 [DOI] [Google Scholar]
- Association of Official Analytical Chemists [AOAC] (2006). Official Methods of Analysis. Arlington, VA: Association of Official Analytical Chemists. [Google Scholar]
- Avila-Stagno J., Chaves A. V., Ribeiro G. O., Jr., Ungerfeld E. M., Mcallister T. A. (2014). Inclusion of glycerol in forage diets increases methane production in a rumen simulation technique system. Br. J. Nutr. 111 829–835. 10.1017/S0007114513003206 [DOI] [PubMed] [Google Scholar]
- Barrell L., Burke J., Waghorn G., Attwood G., Brookes I., Peterson S. (2000). Preparation of fresh forages for incubation and prediction of nutritive value. Proc. N. Z. Soc. Anim. Prod. 60 5–8. [Google Scholar]
- Buddle B. M., Denis M., Attwood G. T., Altermann E., Janssen P. H., Ronimus R. S., et al. (2011). Strategies to reduce methane emissions from farmed ruminants grazing on pasture. Vet. J. 188 11–17. 10.1016/j.tvjl.2010.02.019 [DOI] [PubMed] [Google Scholar]
- Caporaso J. G., Bittinger K., Bushman F. D., Desantis T. Z., Andersen G. L., Knight R. (2010a). PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26 266–267. 10.1093/bioinformatics/btp636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., et al. (2010b). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7 335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G., Lauber C. L., Walters W. A., Berg-Lyons D., Lozupone C. A., Turnbaugh P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Nat. Acad. Sci. Proc. 108 4516–4522. 10.1073/pnas.1000080107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves A. V., Waghorn G. C., Brookes I. M., Woodfield D. R. (2006). Effect of maturation and initial harvest dates on the nutritive characteristics of ryegrass (Lolium perenne L.). Anim. Feed Sci. Technol. 127 293–318. 10.1016/j.anifeedsci.2005.08.015 [DOI] [Google Scholar]
- Cohen D., Doyle P. (2001). Effect of sample preparation on in situ estimates of protein degradability for white clover herbages. Anim. Prod. Sci. 41 619–624. 10.1071/EA00168 [DOI] [Google Scholar]
- Czerkawski J. W., Breckenridge G. (1977). Design and development of a long-term rumen simulation technique (Rusitec). Br. J. Nutr. 38 371–384. 10.1079/BJN19770102 [DOI] [PubMed] [Google Scholar]
- de Menezes A. B., Lewis E., O’donovan M., O’neill B. F., Clipson N., Doyle E. M. (2011). Microbiome analysis of dairy cows fed pasture or total mixed ration diets. FEMS Microbiol. Ecol. 78 256–265. 10.1111/j.1574-6941.2011.01151.x [DOI] [PubMed] [Google Scholar]
- Dehority B. (2003). “Numbers, factors affecting the population and distribution of rumen bacteria,” in Rumen Microbiology ed. Dehority B. A. (Nottingham: Nottingham University Press; ) 265–294. [Google Scholar]
- Durmic Z., Hutton P., Revell D., Emms J., Hughes S., Vercoe P. (2010). In vitro fermentative traits of Australian woody perennial plant species that may be considered as potential sources of feed for grazing ruminants. Anim. Feed Sci. Technol. 160 98–109. 10.1016/j.anifeedsci.2010.07.006 [DOI] [Google Scholar]
- Edgar R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26 2460–2461. 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- Edgar R. C., Haas B. J., Clemente J. C., Quince C., Knight R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27 2194–2200. 10.1093/bioinformatics/btr381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith D. P. (1992). Conservation evaluation and phylogenetic diversity. Biol. Conserv. 61 1–10. 10.1016/0006-3207(92)91201-3 [DOI] [Google Scholar]
- Flint H. J. (1997). The rumen microbial ecosystem—some recent developments. Trends Microbiol. 5 483–488. 10.1016/S0966-842X(97)01159-1 [DOI] [PubMed] [Google Scholar]
- Flint H. J., Scott K. P., Duncan S. H., Louis P., Forano E. (2012). Microbial degradation of complex carbohydrates in the gut. Gut microbes 3 289–306. 10.4161/gmic.19897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-González R., López S., Fernandez M., Bodas R., González J. S. (2008). Screening the activity of plants and spices for decreasing ruminal methane production in vitro. Anim. Feed Sci. Technol. 147 36–52. 10.1016/j.anifeedsci.2007.09.008 [DOI] [Google Scholar]
- Hales K. E., Cole N. A., Macdonald J. C. (2012). Effects of corn processing method and dietary inclusion of wet distillers grains with solubles on energy metabolism, carbon-nitrogen balance, and methane emissions of cattle. J. Anim. Sci. 90 3174–3185. 10.2527/jas.2011-4441 [DOI] [PubMed] [Google Scholar]
- Henderson G., Cox F., Ganesh S., Jonker A., Young W., Collaborators G. R. C., et al. (2015). Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci. Rep. 5:14567 10.1038/srep14567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huws S. A., Lee M. R., Muetzel S. M., Scott M. B., Wallace R. J., Scollan N. D. (2010). Forage type and fish oil cause shifts in rumen bacterial diversity. FEMS Microbiol. Ecol. 73 396–407. 10.1111/j.1574-6941.2010.00892.x [DOI] [PubMed] [Google Scholar]
- Jami E., Israel A., Kotser A., Mizrahi I. (2013). Exploring the bovine rumen bacterial community from birth to adulthood. ISME J. 7 1069–1079. 10.1038/ismej.2013.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jami E., Mizrahi I. (2012). Composition and similarity of bovine rumen microbiota across individual animals. PLoS ONE 7:e33306 10.1371/journal.pone.0033306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jami E., White B. A., Mizrahi I. (2014). Potential role of the bovine rumen microbiome in modulating milk composition and feed efficiency. PLoS ONE 9:e85423 10.1371/journal.pone.0085423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Durmic Z., Liu S., Mcsweeney C. S., Vercoe P. E. (2014). Eremophila glabra reduces methane production and methanogen populations when fermented in a rusitec. Anaerobe 29 100–107. 10.1016/j.anaerobe.2013.10.008 [DOI] [PubMed] [Google Scholar]
- Lozupone C., Knight R. (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71 8228–8235. 10.1128/AEM.71.12.8228-8235.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister T. A., Bae H. D., Jones G. A., Cheng K. J. (1994). Microbial attachment and feed digestion in the rumen. J. Anim. Sci. 72 3004–3018. [DOI] [PubMed] [Google Scholar]
- McNabb W., Waghorn G., Peters J., Barry T. (1996). The effect of condensed tannins in Lotus pedunculatus on the solubilization and degradation of ribulose-1, 5-bis phosphate carboxylase (EC 4.1. 1.39; Rubisco) protein in the rumen and the sites of Rubisco digestion. Br. J. Nutr. 76 535–549. 10.3389/fmicb.2014.00689 [DOI] [PubMed] [Google Scholar]
- Mohammed R., Brink G. E., Stevenson D. M., Neumann A. P., Beauchemin K. A., Suen G., et al. (2014). Bacterial communities in the rumen of Holstein heifers differ when fed orchardgrass as pasture vs. hay. Front. Microbiol. 5:689 10.3389/fmicb.2014.00689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narvaez N., Wang Y., Xu Z., Alexander T., Garden S., Mcallister T. (2013). Effects of hop varieties on ruminal fermentation and bacterial community in an artificial rumen (rusitec). J. Sci. Food Agric. 93 45–52. 10.1002/jsfa.5725 [DOI] [PubMed] [Google Scholar]
- Olivier J., Bouwman A., Berdowski J., Veldt C., Bloos J., Visschedijk A., et al. (1999). Sectoral emission inventories of greenhouse gases for 1990 on a per country basis as well as on 1°× 1°. Environ. Sci. Policy 2 241–263. 10.3389/fmicb.2016.01206 [DOI] [Google Scholar]
- Paz H. A., Anderson C. L., Muller M. J., Kononoff P. J., Fernando S. C. (2016). Rumen bacterial community composition in Holstein and Jersey cows is different under same dietary condition and is not affected by sampling method. Front. Microbiol. 7:1206 10.3389/fmicb.2016.01206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitta D., Kumar S., Veiccharelli B., Parmar N., Reddy B., Joshi C. (2014). Bacterial diversity associated with feeding dry forage at different dietary concentrations in the rumen contents of Mehshana buffalo (Bubalus bubalis) using 16S pyrotags. Anaerobe 25 31–41. 10.1016/j.anaerobe.2013.11.008 [DOI] [PubMed] [Google Scholar]
- Price M. N., Dehal P. S., Arkin A. P. (2010). FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE 5:e9490 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E., Quast C., Knittel K., Fuchs B. M., Ludwig W., Peplies J., et al. (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35 7188–7196. 10.1093/nar/gkm864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60 10.1186/gb-2011-12-6-r60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon C. E. (1948). A mathematical theory of communication. Bell Syst. Tech. J. 27 379–423. 10.1017/S0007114510005684 [DOI] [Google Scholar]
- Soliva C., Amelchanka S., Duval S., Kreuzer M. (2011). Ruminal methane inhibition potential of various pure compounds in comparison with garlic oil as determined with a rumen simulation technique (Rusitec). Br. J. Nutr. 106 114–122. 10.1017/S0007114510005684 [DOI] [PubMed] [Google Scholar]
- Soliva C. R., Amelchanka S. L., Kreuzer M. (2015). The requirements for rumen-degradable protein per unit of fermentable organic matter differ between fibrous feed sources. Front. Microbiol. 6:715 10.3389/fmicb.2015.00715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinfeld H., Gerber P., Wassenaar T., Castel V., De Haan C. (2006). Livestock’s Long Shadow: Environmental Issues and Options. Rome: Food and Agriculture Organization of the United Nations. 10.1371/journal.pone.0119697 [DOI] [Google Scholar]
- Sun X., Henderson G., Cox F., Molano G., Harrison S. J., Luo D., et al. (2015). Lambs fed fresh winter forage rape (Brassica napus L.) emit less methane than those fed perennial ryegrass (Lolium perenne L.), and possible mechanisms behind the difference. PLoS ONE 10:e0119697 10.1371/journal.pone.0119697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tap J., Furet J. P., Bensaada M., Philippe C., Roth H., Rabot S., et al. (2015). Gut microbiota richness promotes its stability upon increased dietary fibre intake in healthy adults. Environ. Microbiol. 17 4954–4964. 10.1111/1462-2920.13006 [DOI] [PubMed] [Google Scholar]
- Thoetkiattikul H., Mhuantong W., Laothanachareon T., Tangphatsornruang S., Pattarajinda V., Eurwilaichitr L., et al. (2013). Comparative analysis of microbial profiles in cow rumen fed with different dietary fiber by tagged 16S rRNA gene pyrosequencing. Curr. Microbiol. 67 130–137. 10.1007/s00284-013-0336-3 [DOI] [PubMed] [Google Scholar]
- Van Soest P., Roberston J., Lewis B. (1991). Methods for dietary fibre, neutral detergent fibre and non starch polysaccharides in relation to animal nutrition. J. Dairy. Sci. 74 3583–3593. 10.1186/2047-217X-2-16 [DOI] [PubMed] [Google Scholar]
- Vazquez-Baeza Y., Pirrung M., Gonzalez A., Knight R. (2013). EMPeror: a tool for visualizing high-throughput microbial community data. Gigascience 2 16 10.1186/2047-217X-2-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veneman J. B., Muetzel S., Hart K. J., Faulkner C. L., Moorby J. M., Perdok H. B., et al. (2015). Does dietary mitigation of enteric methane production affect rumen function and animal productivity in dairy cows? PLoS ONE 10:e0140282 10.1371/journal.pone.0140282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace R. J., Rooke J. A., Duthie C.-A., Hyslop J. J., Ross D. W., Mckain N., et al. (2014). Archaeal abundance in post-mortem ruminal digesta may help predict methane emissions from beef cattle. Sci. Rep. 4:5892 10.1038/srep05892 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The eight most relatively abundant phyla among all samples for each sample type, forage preparation type, and sampling day. LAM, liquid-associated microbes; SAM, solid-associated microbes.


