ABSTRACT
Genomic analysis has placed the origins of two human-pathogenic fungi, the Cryptococcus gattii species complex and the Cryptococcus neoformans species complex, in South America and Africa, respectively. Molecular clock calculations suggest that the two species separated ~80 to 100 million years ago. This time closely approximates the breakup of the supercontinent Pangea, which gave rise to South America and Africa. On the basis of the geographic distribution of these two species complexes and the coincidence of the evolutionary divergence and Pangea breakup times, we propose that a spatial separation caused by continental drift resulted in the emergence of the C. gattii and C. neoformans species complexes from a Pangean ancestor. We note that, despite the spatial and temporal separation that occurred approximately 100 million years ago, these two species complexes are morphologically similar, share virulence factors, and cause very similar diseases. Continuation of these phenotypic characteristics despite ancient separation suggests the maintenance of similar selection pressures throughout geologic ages.
KEYWORDS: Cryptococcus neoformans, evolution, fungus
INTRODUCTION
Cryptococcosis is a disease of humans and animals that is caused by Cryptococcus spp. The disease is most frequent in individuals with impaired immunity, and there are currently over a million cases worldwide (1). In recent years, most pathogenic cryptococcal strains have been grouped within two species known as Cryptococcus neoformans and C. gattii, but genomic analysis reveals a complex taxonomy such that each of these taxa almost certainly includes numerous individual species (2). Given the rapidly accumulating genomic information and concerns about nomenclature instability, it was recently proposed that species complex nomenclature be used such that the broad taxa C. neoformans and C. gattii will be referred to as the C. neoformans species complex and the C. gattii species complex (3), a temporary expedient that we use in this essay. Each of these species complexes potentially includes numerous species, but those in each complex are more closely related to each other than to those across the two complexes.
The same genomic studies that have shown great taxonomic diversity have also provided important new insights into the evolution of the C. gattii and C. neoformans species complexes. The research effort launched to understand the origins of the C. gattii strains causing outbreaks among otherwise healthy humans in the Pacific Northwest of Canada and the United States revealed that the best-studied lineage of the C. gattii species complex has a center of genetic diversity that includes individuals of both the a and α mating types in the rainforest of Northern Brazil (4–6). Subsequent studies of additional Brazilian isolates suggest that an even more diverse population of the C. gattii species complex may be found in more arid areas of northwest Brazil (7). In contrast, in the C. neoformans species complex, the lineage known as VNB, or VNII-AFLP1A, has a center of genetic diversity that again includes individuals of both the a and α mating types in the Botswana region of southern Africa (8). Consistent with an African origin of C. neoformans var. grubii is an analysis of isolates in Southeast Asia that reveals low genetic diversity (9). Consequently, the available genomic data place the origins of lineages of the C. gattii and C. neoformans species complexes on two different continents. Previously, molecular clock calculations based on an estimated 32% substitution of synonymous nucleotide positions in orthologous coding regions and a neutral mutation rate of 2 × 10−9 substitutions per nucleotide per year suggested that the two cryptococcal species complexes separated about 80 million years ago (mya), with a range of 16 to 160 mya (10). We aligned 13.6 × 106 nucleotides of the genomes of C. neoformans species complex B-3501A and C. gattii species complex WM276, finding that 15.54% were polymorphic, a value that grew to 17.4% after multiple substitutions were accounted for (11). Using the full range of mutation rates estimated for coding regions in ascomycete filamentous fungi (0.9 × 10−9 to 16.7 × 10−9) (12), the divergence time would lie between 5.2 ×106 and 96.7 ×106 years ago, which encompasses the previous estimate. If the recently published mutation rate measured experimentally in Saccharomyces cerevisiae, 1.6 × 10−10, were used, the divergence would be pushed back to 544 × 106 years before the present, which seems far too long ago (13, 14). Realizing that no substitution rate has been estimated for Basidiomycota and cognizant of the strong difference between the substitution rates estimated for filamentous Ascomycota and yeast, we have no basis on which to dispute a divergence time of 80 × 106 to 100 × 106 years, which would lie roughly in the middle of the Cretaceous period (145 to 65 mya).
Reviewing the geography of planet Earth at the time that the cryptococcal species complexes separated places us at a geologic time dominated by the breakup of the supercontinent Pangea. This breakup formed the minor supercontinent of west Gondwana, which subsequently broke up to generate the current continents of South America and Africa. The breakup of west Gondwana began in the Early Cretaceous, about 130 mya, and Africa was separate from South America by approximately 100 mya. However, dinosaur fossil data suggest that some land connections between Africa and South America existed as late as 95 mya (15). The details of the breakup of west Gondwana leading to the formation of the South Atlantic are lost in time, but the process is thought to have occurred gradually over tens of millions of years. Although the time estimates for the divergence of the cryptococcal species complexes and the separation of the South American and African continents both have tremendous uncertainty, we are struck by the coincidence of the time scales of both processes and the ancient geographic juxtaposition of the regions now posited as regions of origin. On this basis, we hypothesize that the C. gattii and C. neoformans species complexes emerged following continental drift events that resulted in the physical separation of a Pangean cryptococcal ancestor population that occupied a discrete region of the supercontinent Pangea, a region that subsequently became parts of South America and Africa (Fig. 1). Given the modern association of Cryptococcus species with birds, particularly invasive rock doves (16), it might be thought that continental rafting would have scant effect on the distribution of the fungus given the potential for the aerial transport of fungal isolates across great distances. Although birds evolved from dinosaurs in the Early Cretaceous, the major radiation of modern birds occurred after the end of the Cretaceous at 65 mya, and on the basis of the phylogeny of birds, the radiation of the clade that includes the rock dove occurred after 25 mya (17). Nevertheless, it is possible that there was an association between the primordial Cryptococcus ancestor and early birds, which could have allowed the intercontinental transport of isolates during the early phases of the Pangean breakup, when the continents were much closer than they are today and thus prevented species isolation until more recent epochs. Such intercontinental transport of isolates by birds, wind, ocean currents, or even another animal would account for estimates of a more recent separation of the Cryptococcus complex species. One can imagine scenarios where intercontinental transport mechanisms prevented isolation of the primordial Cryptococcus ancestor for millions of years following continental separation until increasing continental distance created conditions for spatial isolation. Hence, the wide difference between estimates of the separation time of the C. gattii and C. neoformans species complexes, which include significantly more recent dates than the separation of Africa and South America, could still be reconciled with a precipitating event associated with the breakup of Pangea. Furthermore, we note that the suggested speciation event is very different from the current situation, where lineages of both complexes occur simultaneously in diverse geographic regions throughout the globe. We propose that continental drift was the initial trigger of speciation and that the current geographic distribution of the two cryptococcal species complexes is the result of subsequent introduction and dispersal events, including anthropomorphic causes, such as the global dispersal of Columbia livia (rock dove or pigeon) from its Mediterranean origin in recent centuries (16).
FIG 1 .
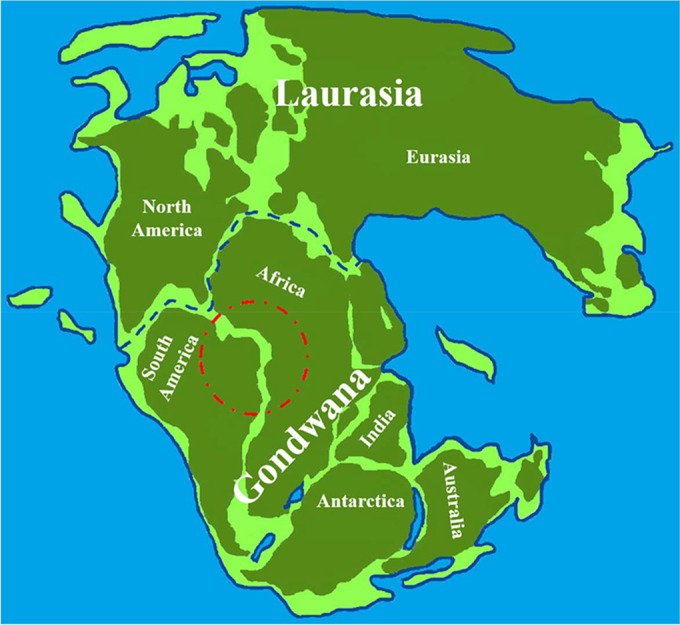
Representation of the supercontinent of Pangea with outlines of the present continents. The red circle denotes the proposed biogeography of the Pangean ancestor of both the C. gattii and C. neoformans species complexes. Also shown are the two supercontinents that came together to form Pangea, Laurasia and Gondwana (separated by a dashed blue line). This map was designed by tracing an outline of the supercontinent of Pangea via a Google Image search. The organization of subcontinents (Laurasia and Gondwana) is based on reference 47.
One of the remarkable aspects of the two cryptococcal species complexes is how similar they are with regard to virulence factors despite their distant separation in time. For example, the two cryptococcal complex species complexes share such virulence-associated phenotypes as polysaccharide capsules (18), thermotolerance of mammalian temperatures (18), melanin production (18), urease (19, 20) and phospholipase (21, 22) activities, intracellular replication (23, 24), nonlytic exocytosis (25, 26), and inositol production (27). Although it is possible that these phenotypes evolved independently, the fact that they are not shared, or rarely shared, by other closely related species supports the view that they evolved in the common ancestor of both the C. neoformans and C. gattii species complexes. For example, a survey of heterobasidiomycetous yeasts found that only Cryptococcus podzolicus had a capsule and was capable of making melanin (28). Both cryptococcal species complexes have been isolated from tree hollows (29), and we note that their current and past locations are near forested equatorial regions. Strains of both cryptococcal species complexes have been shown to be facultative intracellular pathogens capable of replicating in mammalian macrophages. For C. neoformans, the intracellular pathogenic strategy in macrophages and amoebae has been shown to be uncannily similar, which led to the proposal that the capacity for animal virulence arose from selection pressures in the environment that included predaceous phagocytic cells such as those now found in amoebae and slime molds (30, 31). Strains of both cryptococcal species complexes have been shown to interact with amoebae, which they can exploit for growth under certain circumstances (30, 32). Given that the origins of both fungi and amoebae occur in deep Earth time and that the closest outgroup to the kingdom Fungi is that of nucleariid amoebae (33), the ancestors of these two groups of eukaryotes could have been interacting even before the emergence of multicellular life forms. Consequently, we hypothesize that the remarkable similarities between the two cryptococcal species complexes with regard to their distinctive encapsulated morphology and shared virulence factors is a consequence of similar selection pressures, which continued after the breakup of West Gondwana.
The facts that the C. neoformans and C. gattii species complexes are likely to have diverged in the Cretaceous and share so many phenotypes associated with virulence have interesting implications for their Pangean ancestor and for the origin of virulence in fungi. First, it must have shared the virulence phenotypes now found in C. gattii and C. neoformans, which implies that it was encapsulated, made melanin, possessed numerous enzymes that can damage host cells, and had the capacity for intracellular replication in animal and environmental phagocytic cells. Second, since cryptococcosis can occur in reptiles (34) and the Cretaceous was remarkable for its reptilian megafauna, the Pangean ancestor could have been pathogenic for some animals at the time. Third, the capacity of cryptococcal species for virulence must have existed in the ancient past. Since fungal and amoebal evolutionary lineages predate the appearance of animals and interactions between these organisms, such as predation of fungi by amoebae, may have selected for traits that accidentally enabled a capacity for mammalian virulence, it is conceivable that fungi with the potential to be pathogenic in metazoans existed before the latter appeared. A thorough phylogenetic and phenotypic study of C. gattii and C. neoformans and close relatives in the Filobasidella and Kwoniella clades, many of which had been isolated from insect guts or frass, concluded that ancestors of the human pathogens had the abilities to grow at 30°C, produce a capsule, and produce melanin (35). It seems likely, therefore, that the ancestors of the pathogenic Cryptococcus species had the capacity for animal virulence, which was passed on to the descendant species. An origin of virulence in Cryptococcus complex species in deep time could help explain the remarkable nonspecificity of their pathogenic potential, given their capacity for virulence to vertebrates (18), insects (36, 37), nematodes (38), amoebae (30), and plants (39). This scenario differs from the situation with two groups of Ascomycota, Onygenales (40) and Clavicipitales (41), where virulence emerged independently. Interestingly, in the case of Clavicipitales, the animal-parasitic species may have evolved from mycoparasitic ancestors. Relatives of the pathogenic Cryptococcus species in the Tremella clade are also mycoparasitic, but it appears likely that this trait was not exhibited by the ancestor of the C. gattii and C. neoformans species complexes (35).
Coming back to continental drift and its ability to change the continuity of land masses and isolate species or bring them together, in addition to Cryptococcus species, this phenomenon has been used to explain the biogeography of many land species, including animals and plants and their associated microbes. Continental drift has been used to interpret the biogeography of many pathogenic microbes, including geminiviruses (42), trypanosomes (43), and the conifer root rot fungus Heterobasidion annosum (44). Here we extend that reasoning to the human-pathogenic cryptococcal species complexes and note with great interest the congruence of molecular and geologic scales and the glimpse they provide into the origins of fungal virulence. We are fully aware that we are just beginning to sample the genetic diversity of the fungal world and that as additional data are accrued, the current hypothesis may be further supported, require modification, or be abandoned. In associating cryptococcal speciation with continental drift, the most uncertain estimates are those of the divergence of the C. neoformans and C. gattii complex lineages. To improve the estimate of the time of divergence of the two Cryptococcus lineages would require either a measured mutation rate for Cryptococcus spp. or a solid geologic calibration for at least one node on a phylogenetic tree that includes the two Cryptococcus species, neither of which is available at the present time.
Our goal in formulating this hypothesis is to stimulate thought and discussion about the mechanisms driving fungal speciation and the origins of fungal virulence that will, we hope, promote further experimental work. In this regard, we note that the hypothesis already suggests new lines of inquiry. For example, if continental drift did indeed trigger the speciation of the C. neoformans and C. gattii complexes, then there may be a great level of spatial difference between individual cryptococcal lineages among different continents, which could be revealed by careful geographic environmental sampling. Also, the hypothesis suggests that the current distribution of C. neoformans and C. gattii complex strains should be viewed as resulting from a layering of strains descending from ancient ancestors with more recent dispersal events. In this regard, the occurrence of C. neoformans and C. gattii hybrids (45) could represent descendants of ancient mating events or recent crosses following the introduction of strains that retained the capacity for sexual reproduction. In this regard, the hypothesis provides a new conceptual approach to understanding their origin. For example, if these hybrids are the result of ancient mating events, these strains should be more prevalent in areas of Africa and South America that remained connected or physically related for the longest time. Finally, we hope that the hypothesis stimulates more research into the mutation rates of the cryptococcal complex species and into the nascent field of paleomycology. With regard to the latter, fungi from the Cretaceous have been found in amber (46) and a search for yeast related to Cryptococcus in amber combined with advanced DNA and protein sequencing techniques could provide important information to support, modify, or refute the proposed hypothesis.
ACKNOWLEDGMENTS
Arturo Casadevall is supported by National Heart, Lung, and Blood Institute (NHLBI) grant HL059842 and National Institute of Allergy and Infectious Diseases (NIAID) grants AI033142 and AI052733. John Taylor is supported by University of California Office of the President grant MRP-17-454959.
REFERENCES
- 1.Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, Chiller TM. 2009. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS 23:525–530. doi: 10.1097/QAD.0b013e328322ffac. [DOI] [PubMed] [Google Scholar]
- 2.Hagen F, Khayhan K, Theelen B, Kolecka A, Polacheck I, Sionov E, Falk R, Parnmen S, Lumbsch HT, Boekhout T. 2015. Recognition of seven species in the Cryptococcus gattii/Cryptococcus neoformans species complex. Fungal Genet Biol 78:16–48. doi: 10.1016/j.fgb.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 3.Kwon-Chung KJ, Bennett JE, Wickes BL, Meyer W, Cuomo CA, Wollenburg KR, Bicanic TA, Castaneda E, Chang YC, Chen J, Cogliati M, Dromer F, Ellis D, Filler SG, Fisher MC, Harrison TS, Holland SM, Kohno S, Kronstad JW, Lazera M, Levitz SM, Lionakis MS, May RC, Ngamskulrongroj P, Pappas PG, Perfect JR, Rickerts V, Sorrell TC, Walsh TJ, Williamson PR, Xu J, Zelazny AM, Casadevall A. 2017. The case for adopting the “species complex” nomenclature for the etiologic agents of cryptococcosis. mSphere 2:e00357. doi: 10.1128/mSphere.00357-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hagen F, Ceresini PC, Polacheck I, Ma H, van Nieuwerburgh F, Gabaldón T, Kagan S, Pursall ER, Hoogveld HL, van Iersel LJ, Klau GW, Kelk SM, Stougie L, Bartlett KH, Voelz K, Pryszcz LP, Castañeda E, Lazera M, Meyer W, Deforce D, Meis JF, May RC, Klaassen CH, Boekhout T. 2013. Ancient dispersal of the human fungal pathogen Cryptococcus gattii from the Amazon rainforest. PLoS One 8:e71148. doi: 10.1371/journal.pone.0071148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Engelthaler DM, Hicks ND, Gillece JD, Roe CC, Schupp JM, Driebe EM, Gilgado F, Carriconde F, Trilles L, Firacative C, Ngamskulrungroj P, Castañeda E, Lazera MS, Melhem MS, Pérez-Bercoff A, Huttley G, Sorrell TC, Voelz K, May RC, Fisher MC, Thompson GR III, Lockhart SR, Keim P, Meyer W. 2014. Cryptococcus gattii in North American Pacific Northwest: whole-population genome analysis provides insights into species evolution and dispersal. mBio 5:e01464-14. doi: 10.1128/mBio.01464-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Billmyre RB, Croll D, Li W, Mieczkowski P, Carter DA, Cuomo CA, Kronstad JW, Heitman J. 2014. Highly recombinant VGII Cryptococcus gattii population develops clonal outbreak clusters through both sexual macroevolution and asexual microevolution. mBio 5:e01494-14. doi: 10.1128/mBio.01494-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Souto AC, Bonfietti LX, Ferreira-Paim K, Trilles L, Martins M, Ribeiro-Alves M, Pham CD, Martins L, Dos Santos W, Chang M, Brito-Santos F, Santos DC, Fortes S, Lockhart SR, Wanke B, Melhem MS, Lazéra MS, Meyer W. 2016. Population genetic analysis reveals a high genetic diversity in the Brazilian Cryptococcus gattii VGII population and shifts the global origin from the Amazon rainforest to the semi-arid desert in the northeast of Brazil. PLoS Negl Trop Dis 10:e0004885. doi: 10.1371/journal.pntd.0004885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Litvintseva AP, Carbone I, Rossouw J, Thakur R, Govender NP, Mitchell TG. 2011. Evidence that the human pathogenic fungus Cryptococcus neoformans var. grubii may have evolved in Africa. PLoS One 6:e19688. doi: 10.1371/journal.pone.0019688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simwami SP, Khayhan K, Henk DA, Aanensen DM, Boekhout T, Hagen F, Brouwer AE, Harrison TS, Donnelly CA, Fisher MC. 2011. Low diversity Cryptococcus neoformans variety grubii multilocus sequence types from Thailand are consistent with an ancestral African origin. PLoS Pathog 7:e1001343. doi: 10.1371/journal.ppat.1001343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharpton TJ, Neafsey DE, Galagan JE, Taylor JW. 2008. Mechanisms of intron gain and loss in Cryptococcus. Genome Biol 9:R24. doi: 10.1186/gb-2008-9-1-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tajima F, Nei M. 1984. Estimation of evolutionary distance between nucleotide sequences. Mol Biol Evol 1:269–285. [DOI] [PubMed] [Google Scholar]
- 12.Kasuga T, White TJ, Taylor JW. 2002. Estimation of nucleotide substitution rates in Eurotiomycete fungi. Mol Biol Evol 19:2318–2324. doi: 10.1093/oxfordjournals.molbev.a004056. [DOI] [PubMed] [Google Scholar]
- 13.Taylor JW, Berbee ML. 2006. Dating divergences in the fungal tree of life: review and new analyses. Mycologia 98:838–849. doi: 10.3852/mycologia.98.6.838. [DOI] [PubMed] [Google Scholar]
- 14.Chang Y, Wang S, Sekimoto S, Aerts AL, Choi C, Clum A, LaButti KM, Lindquist EA, Yee Ngan C, Ohm RA, Salamov AA, Grigoriev IV, Spatafora JW, Berbee ML. 2015. Phylogenomic analyses indicate that early fungi evolved digesting cell walls of algal ancestors of land plants. Genome Biol Evol 7:1590–1601. doi: 10.1093/gbe/evv090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sereno PC, Wilson JA, Conrad JL. 2004. New dinosaurs link southern landmasses in the mid-Cretaceous. Proc Biol Sci 271:1325–1330. doi: 10.1098/rspb.2004.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Litvintseva AP, Mitchell TG. 2012. Population genetic analyses reveal the African origin and strain variation of Cryptococcus neoformans var. grubii. PLoS Pathog 8:e1002495. doi: 10.1371/journal.ppat.1002495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prum RO, Berv JS, Dornburg A, Field DJ, Townsend JP, Lemmon EM, Lemmon AR. 2015. A comprehensive phylogeny of birds (Aves) using targeted next-generation DNA sequencing. Nature 526:569–573. doi: 10.1038/nature15697. [DOI] [PubMed] [Google Scholar]
- 18.Casadevall A, Perfect JR. 1998. Cryptococcus neoformans. American Society for Microbiology, Washington, DC. [Google Scholar]
- 19.Feder V, Kmetzsch L, Staats CC, Vidal-Figueiredo N, Ligabue-Braun R, Carlini CR, Vainstein MH. 2015. Cryptococcus gattii urease as a virulence factor and the relevance of enzymatic activity in cryptococcosis pathogenesis. FEBS J 282:1406–1418. doi: 10.1111/febs.13229. [DOI] [PubMed] [Google Scholar]
- 20.Cox GM, Mukherjee J, Cole GT, Casadevall A, Perfect JR. 2000. Urease as a virulence factor in experimental cryptococcosis. Infect Immun 68:443–448. doi: 10.1128/IAI.68.2.443-448.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Latouche GN, Sorrell TC, Meyer W. 2002. Isolation and characterisation of the phospholipase B gene of Cryptococcus neoformans var. gattii. FEMS Yeast Res 2:551–561. doi: 10.1111/j.1567-1364.2002.tb00122.x. [DOI] [PubMed] [Google Scholar]
- 22.Cox GM, McDade HC, Chen SC, Tucker SC, Gottfredsson M, Wright LC, Sorrell TC, Leidich SD, Casadevall A, Ghannoum MA, Perfect JR. 2001. Extracellular phospholipase activity is a virulence factor for Cryptococcus neoformans. Mol Microbiol 39:166–175. doi: 10.1046/j.1365-2958.2001.02236.x. [DOI] [PubMed] [Google Scholar]
- 23.Feldmesser M, Kress Y, Novikoff P, Casadevall A. 2000. Cryptococcus neoformans is a facultative intracellular pathogen in murine pulmonary infection. Infect Immun 68:4225–4237. doi: 10.1128/IAI.68.7.4225-4237.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma H, Hagen F, Stekel DJ, Johnston SA, Sionov E, Falk R, Polacheck I, Boekhout T, May RC. 2009. The fatal fungal outbreak on Vancouver Island is characterized by enhanced intracellular parasitism driven by mitochondrial regulation. Proc Natl Acad Sci U S A 106:12980–12985. doi: 10.1073/pnas.0902963106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alvarez M, Casadevall A. 2006. Phagosome extrusion and host-cell survival after Cryptococcus neoformans phagocytosis by macrophages. Curr Biol 16:2161–2165. doi: 10.1016/j.cub.2006.09.061. [DOI] [PubMed] [Google Scholar]
- 26.Ma H, Croudace JE, Lammas DA, May RC. 2006. Expulsion of live pathogenic yeast by macrophages. Curr Biol 16:2156–2160. doi: 10.1016/j.cub.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 27.Xue C, Liu T, Chen L, Li W, Liu I, Kronstad JW, Seyfang A, Heitman J. 2010. Role of an expanded inositol transporter repertoire in Cryptococcus neoformans sexual reproduction and virulence. mBio 1:e00084. doi: 10.1128/mBio.00084-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petter R, Kang BS, Boekhout T, Davis BJ, Kwon-Chung KJ. 2001. A survey of heterobasidiomycetous yeasts for the presence of the genes homologous to virulence factors of Filobasidiella neoformans, CNLAC1 and CAP59. Microbiology 147:2029–2036. doi: 10.1099/00221287-147-8-2029. [DOI] [PubMed] [Google Scholar]
- 29.Refojo N, Perrotta D, Brudny M, Abrantes R, Hevia AI, Davel G. 2009. Isolation of Cryptococcus neoformans and Cryptococcus gattii from trunk hollows of living trees in Buenos Aires City, Argentina. Med Mycol 47:177–184. doi: 10.1080/13693780802227290. [DOI] [PubMed] [Google Scholar]
- 30.Steenbergen JN, Shuman HA, Casadevall A. 2001. Cryptococcus neoformans interactions with amoebae suggest an explanation for its virulence and intracellular pathogenic strategy in macrophages. Proc Natl Acad Sci U S A 98:15245–15250. doi: 10.1073/pnas.261418798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steenbergen JN, Nosanchuk JD, Malliaris SD, Casadevall A. 2003. Cryptococcus neoformans virulence is enhanced after growth in the genetically malleable host Dictyostelium discoideum. Infect Immun 71:4862–4872. doi: 10.1128/IAI.71.9.4862-4872.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malliaris SD, Steenbergen JN, Casadevall A. 2004. Cryptococcus neoformans var. gattii can exploit Acanthamoeba castellanii for growth. Med Mycol 42:149–158. doi: 10.1080/13693786310001616500. [DOI] [PubMed] [Google Scholar]
- 33.Steenkamp ET, Wright J, Baldauf SL. 2006. The protistan origins of animals and fungi. Mol Biol Evol 23:93–106. doi: 10.1093/molbev/msj011. [DOI] [PubMed] [Google Scholar]
- 34.Hough I. 1998. Cryptococcosis in an eastern water skink. Aust Vet J 76:471–472. doi: 10.1111/j.1751-0813.1998.tb10183.x. [DOI] [PubMed] [Google Scholar]
- 35.Findley K, Rodriguez-Carres M, Metin B, Kroiss J, Fonseca A, Vilgalys R, Heitman J. 2009. Phylogeny and phenotypic characterization of pathogenic Cryptococcus species and closely related saprobic taxa in the Tremellales. Eukaryot Cell 8:353–361. doi: 10.1128/EC.00373-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Apidianakis Y, Rahme LG, Heitman J, Ausubel FM, Calderwood SB, Mylonakis E. 2004. Challenge of Drosophila melanogaster with Cryptococcus neoformans and role of the innate immune response. Eukaryot Cell 3:413–419. doi: 10.1128/EC.3.2.413-419.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mylonakis E, Moreno R, El Khoury JB, Idnurm A, Heitman J, Calderwood SB, Ausubel FM, Diener A. 2005. Galleria mellonella as a model system to study Cryptococcus neoformans pathogenesis. Infect Immun 73:3842–3850. doi: 10.1128/IAI.73.7.3842-3850.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mylonakis E, Ausubel FM, Perfect JR, Heitman J, Calderwood SB. 2002. Killing of Caenorhabditis elegans by Cryptococcus neoformans as a model of yeast pathogenesis. Proc Natl Acad Sci U S A 99:15675–15680. doi: 10.1073/pnas.232568599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Warpeha KM, Park YD, Williamson PR. 2013. Susceptibility of intact germinating Arabidopsis thaliana to human fungal pathogens Cryptococcus neoformans and C. gattii. Appl Environ Microbiol 79:2979–2988. doi: 10.1128/AEM.03697-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharpton TJ, Stajich JE, Rounsley SD, Gardner MJ, Wortman JR, Jordar VS, Maiti R, Kodira CD, Neafsey DE, Zeng Q, Hung CY, McMahan C, Muszewska A, Grynberg M, Mandel MA, Kellner EM, Barker BM, Galgiani JN, Orbach MJ, Kirkland TN, Cole GT, Henn MR, Birren BW, Taylor JW. 2009. Comparative genomic analyses of the human fungal pathogens Coccidioides and their relatives. Genome Res 19:1722–1731. doi: 10.1101/gr.087551.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spatafora JW, Sung GH, Sung JM, Hywel-Jones NL, White JF Jr.. 2007. Phylogenetic evidence for an animal pathogen origin of ergot and the grass endophytes. Mol Ecol 16:1701–1711. doi: 10.1111/j.1365-294X.2007.03225.x. [DOI] [PubMed] [Google Scholar]
- 42.Nawaz-ul-Rehman MS, Fauquet CM. 2009. Evolution of geminiviruses and their satellites. FEBS Lett 583:1825–1832. doi: 10.1016/j.febslet.2009.05.045. [DOI] [PubMed] [Google Scholar]
- 43.Stevens JR, Gibson W. 1999. The molecular evolution of trypanosomes. Parasitol Today 15:432–437. doi: 10.1016/S0169-4758(99)01532-X. [DOI] [PubMed] [Google Scholar]
- 44.Dalman K, Olson A, Stenlid J. 2010. Evolutionary history of the conifer root rot fungus Heterobasidion annosum sensu lato. Mol Ecol 19:4979–4993. doi: 10.1111/j.1365-294X.2010.04873.x. [DOI] [PubMed] [Google Scholar]
- 45.Aminnejad M, Diaz M, Arabatzis M, Castañeda E, Lazera M, Velegraki A, Marriott D, Sorrell TC, Meyer W. 2012. Identification of novel hybrids between Cryptococcus neoformans var. grubii VNI and Cryptococcus gattii VGII. Mycopathologia 173:337–346. doi: 10.1007/s11046-011-9491-x. [DOI] [PubMed] [Google Scholar]
- 46.Poinar GO Jr., Buckley R. 2007. Evidence of mycoparasitism and hypermycoparasitism in Early Cretaceous amber. Mycol Res 111:503–506. doi: 10.1016/j.mycres.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 47.Rogers JJW, Santosh M. 2003. Supercontinents in Earth history. Gondwana Res 6:357–368. doi: 10.1016/S1342-937X(05)70993-X. [DOI] [Google Scholar]


