Since its initial identification in epidemic multidrug-resistant Salmonella enterica serovar Typhimurium DT104 strains, several SGI1 variants, SGI1 lineages, and SGI1-related elements (SGI2, PGI1, and AGI1) have been described in many bacterial genera (Salmonella, Proteus, Morganella, Vibrio, Shewanella, etc.). They constitute a family of multidrug resistance site-specific integrative elements acquired by horizontal gene transfer, SGI1 being the best-characterized element. The horizontal transfer of SGI1/PGI1 elements into other genera is of public health concern, notably with regard to the spread of critically important resistance genes such as ESBL and carbapenemase genes. The identification of SGI1 in Morganella morganii raises the issue of (i) the potential for SGI1 to emerge in other human pathogens and (ii) its bacterial host range. Further surveillance and research are needed to understand the epidemiology, the spread, and the importance of the members of this SGI1 family of integrative elements in contributing to antibiotic resistance development.
KEYWORDS: Salmonella genomic island 1, integrative mobilizable element, integrons, multidrug resistance
ABSTRACT
Salmonella genomic island 1 (SGI1) is a multidrug resistance integrative mobilizable element that harbors a great diversity of antimicrobial resistance gene clusters described in numerous Salmonella enterica serovars and also in Proteus mirabilis. A serious threat to public health was revealed in the recent description in P. mirabilis of a SGI1-derivative multidrug resistance island named PGI1 (Proteus genomic island 1) carrying extended-spectrum-β-lactamase (ESBL) and metallo-β-lactamase resistance genes, blaVEB-6 and blaNDM-1, respectively. Here, we report the first description of Salmonella genomic island 1 (SGI1) in a multidrug-resistant clinical Morganella morganii subsp. morganii strain isolated from a patient in France in 2013. Complete-genome sequencing of the strain revealed SGI1 variant SGI1-L carrying resistance genes dfrA15, floR, tetA(G), blaPSE-1 (now referred to as blaCARB-2), and sul1, conferring resistance to trimethoprim, phenicols, tetracyclines, amoxicillin, and sulfonamides, respectively. The SGI1-L variant was integrated into the usual chromosome-specific integration site at the 3′ end of the trmE gene. Beyond Salmonella enterica and Proteus mirabilis, the SGI1 integrative mobilizable element may thus also disseminate its multidrug resistance phenotype in another genus belonging to the Proteae tribe of the family Enterobacteriaceae.
IMPORTANCE Since its initial identification in epidemic multidrug-resistant Salmonella enterica serovar Typhimurium DT104 strains, several SGI1 variants, SGI1 lineages, and SGI1-related elements (SGI2, PGI1, and AGI1) have been described in many bacterial genera (Salmonella, Proteus, Morganella, Vibrio, Shewanella, etc.). They constitute a family of multidrug resistance site-specific integrative elements acquired by horizontal gene transfer, SGI1 being the best-characterized element. The horizontal transfer of SGI1/PGI1 elements into other genera is of public health concern, notably with regard to the spread of critically important resistance genes such as ESBL and carbapenemase genes. The identification of SGI1 in Morganella morganii raises the issue of (i) the potential for SGI1 to emerge in other human pathogens and (ii) its bacterial host range. Further surveillance and research are needed to understand the epidemiology, the spread, and the importance of the members of this SGI1 family of integrative elements in contributing to antibiotic resistance development.
OBSERVATION
Salmonella genomic island 1 (SGI1) is a multidrug resistance (MDR) site-specific integrative mobilizable element (IME) initially described in Salmonella that integrates into the last 18 bp of the conserved chromosomal trmE gene (formerly thdF) (1, 2). Among the most prevalent incompatibility groups of plasmids, only the conjugative plasmids of the IncA/C family have been shown to specifically mobilize SGI1 in trans (3). Recently, the major IncA/C-encoded transcriptional activator complex, AcaCD, was shown to trigger SGI1 excision and in trans conjugative mobilization (4). SGI1 contains a complex class 1 integron, named In104 in accordance with its initial host strain (1, 5, 6). Since the identification of SGI1 in Salmonella enterica serovar Typhimurium DT104, the high genetic plasticity of its MDR region, highlighted by the diversity of the class 1 integron resistance gene cassettes and by the presence of recombination and insertion sequence (IS) element-mediated rearrangements, has led to the description of more than 30 different MDR regions of SGI1 in many S. enterica serovars (7–14). In addition, genetic variations are observed also in the backbone of the island, i.e., IS-mediated insertion/deletion, single nucleotide polymorphism (SNP), and transpositional insertion of the complex class 1 integron structure at another position (8, 10, 12, 14).
In 2006, SGI1 was identified in a clinical Proteus mirabilis strain from a diabetic patient from Palestine (15). Since then, the number of reported cases of SGI1 variants of this bacterial species in isolates from humans, food-producing animals, foodstuffs, and companion animals in China and France has been increasing (16–23). Recently, a novel SGI1 derivative MDR genomic island named Proteus genomic island 1 (PGI1) has been described in human and animal P. mirabilis strains in France (20, 21, 24, 25). PGI1 showed gene synteny similar to that of SGI1 and was also found integrated into the last 18 bp of the conserved chromosomal trmE gene. The recent emergence of P. mirabilis strains carrying SGI1 or PGI1 islands with extended-spectrum-β-lactamase and/or metallo-β-lactamase resistance genes, blaVEB-6 and blaNDM-1, respectively, representing the first description of the latter gene in an MDR genomic island, is a serious threat to public health (25). In this study, we analyzed the first SGI1-positive Morganella morganii subsp. morganii (here M. morganii) strain isolated from a human case in France.
A hepatitis C virus (HCV)- and human immunodeficiency virus (HIV)-positive 52-year-old man was hospitalized with high blood pressure and cirrhosis complicated by a hepatocellular carcinoma in January 2013 at the Limoges University Hospital center in France. The patient mentioned having had prostatitis in December 2012. MDR M. morganii strain LIM90 was isolated from urine sample during his stay at the hospital in January 2013. The strain was screened for antibiotic susceptibility by the disc diffusion method according to the guidelines of the EUCAST committee (26). Besides intrinsic resistance to several β-lactam antibiotics (amoxicillin ± clavulanic acid, cephalotin, cefuroxim), tetracycline, nitrofurantoin, fosfomycin, and colistin, M. morganii strain LIM90 was resistant to chloramphenicol, florfenicol, streptomycin, spectinomycin, sulfonamides, trimethoprim, and ticarcillin, which suggested the possible presence of SGI1. The result of PCR performed using primers corresponding to the integrase genes of SGI1 and related islands (FwintSGI1HR [5′-ATGTTGCGTCAGGCYGAGGC-3′] and RvintSGI1HR [5′-GAGTGYCCAAGAAGSCGAGAG-3′]) was positive, suggesting the presence of a SGI1-related island in M. morganii strain LIM90.
To identify the SGI1-related island, its chromosomal location, and its resistance gene content, the whole genome of LIM90 was sequenced using an IonProton system (99-fold average read depth). The reads were assembled using MIRA software. The ResFinder and PlasmidFinder tools available at the Center for Genomic Epidemiology were used for identification of acquired resistance genes and plasmid detection, respectively (27). The complete sequence of SGI1 was assembled using the relevant contigs detected by BLAST searches, PCR gap closure, and PCR product sequencing. The sequence of SGI1 was annotated using the Microbial Genome Annotation and Analysis Platform MicroScope (Genoscope, France) and deposited in the European Nucleotide Archive (ENA) under accession number LT630458 (28).
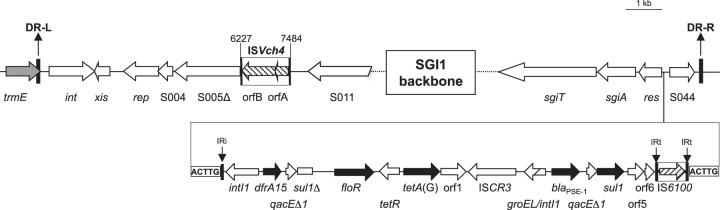
M. morganii strain LIM90 harbored the SGI1-L variant shown in Fig. 1. SGI1-L-related islands have been previously described in S. enterica serovars and P. mirabilis strains but were never fully sequenced (10, 14). This SGI1-L variant carried the dfrA15 and blaCARB-2 (previously named blaPSE-1) resistance gene cassettes inserted at the two SGI1 integron attI recombination sites, conferring resistance to trimethoprim and ticarcillin, respectively (Fig. 1). The floR gene, which confers resistance to chloramphenicol and florfenicol, and tetracycline resistance genes tetR(G) and tetA(G) were found to be flanked by these two integron structures (Fig. 1). The presence of ISVch4 (also called IS1359) at position 6227 to position 7484 of the SGI1 backbone sequence (ENA accession number LT630458) was found as previously described in a few SGI1 variants (SGI1-H, SGI1-K, and SGI1-L derivatives) in S. enterica serovars and P. mirabilis. An adjacent 2,779-bp deletion removed the region extending from within open reading frame (ORF) S005 to within ORF S009 (Fig. 1). The absence of target site duplication created upon ISVch4 insertion suggested that additional recombinational events may have occurred after the transposition of this ISVch4 copy, i.e., replicative transposition of ISVch4 into ORF S009 and subsequent recombination between the two copies of ISVch4 creating the 2,779-bp deletion.
FIG 1 .
Schematic view of SGI1-L and its specific features encountered in Morganella morganii strain LIM90. The gray arrow corresponds to chromosomal gene trmE into which SGI1 is integrated into the last 18 bp. DR-L and DR-R are the 18-bp left and right direct repeats, respectively, bracketing SGI1. The insertion points of complex class 1 integron InSGI1-L between the res gene and ORF S044 of the SGI1 backbone and the 5-bp target site duplication are indicated. IRi and IRt are 25-bp imperfect inverted repeats defining the left and right end of the complex class 1 integron. Black arrows correspond to SGI1 antibiotic resistance genes. IS elements are indicated by boxes containing black hatched arrows representing the transposase genes. Base pair coordinates are from the complete SGI1-L sequence of M. morganii strain LIM90 (ENA accession no. LT630458).
The backbone of SGI1-L (24,093 bp, excluding mobile elements like ISVch4 and the complex class 1 integron structure) was identical to that of SGI1 variants harboring the insertion/deletion of ISVch4 and previously described in P. mirabilis and S. Kentucky ST198 (Table 1) (13, 18). In addition to the ISVch4 insertion/deletion, the backbone of this group of SGI1 variants showed two single-base differences from other fully sequenced variants (29). One SNP was identified compared with ISVch4-negative SGI1 of epidemic S. Typhimurium DT104 strains, resulting in an amino acid (AA) change in toxin SgiT (ORF S025) of the sgiAT toxin-antitoxin addiction system (Table 1) (28, 29). The putative role of this AA change in the SgiAT toxin-antitoxin system and in the stability of SGI1 remains to be determined. Concomitantly with the first SNP position described above, a second one was found in antitoxin SgiA (ORF S026, no AA change) of ISVch4-negative variants described in other S. enterica serovars and P. mirabilis strains (Table 1) (29, 30). Interestingly, the original SGI1 sequence of S. Typhimurium (GenBank accession number AF261825), the recently released SGI1-F sequence of S. Cerro (GenBank accession no. KU847976), and the SGI2 sequence of S. Emek showed 7, 7, and 95 different base pair substitutions relative to the ISVch4-positive SGI1 backbone, respectively (Table 1). All these SGI1 backbone characteristics suggest different lineages that evolve and horizontally spread among Enterobacteriaceae.
TABLE 1 .
Characteristics of complete SGI1 sequences and backbone SNP analysis
| Host strain | SGI1 variant | ISVch4 indel | SNP positiona |
GenBank accession no. | |
|---|---|---|---|---|---|
| 22001 (in sgiT) | 24286 (in sgiA) | ||||
| M. morganii | SGI1-L | + | C | G | LT630458 |
| S. Kentucky | SGI1-K | + | C | G | AY463797 |
| P. mirabilis | SGI1-PmMAT | +b | C | G | JX089583 |
| P. mirabilis | SGI1-PmABB | + | C | G | KP313760 |
| P. mirabilis | SGI1-PmGUE | + | C | G | JX121641 |
| P. mirabilis | SGI1-PmVER | + | C | G | JX121640 |
| P. mirabilis | SGI1-PmSCO | + | C | G | JX121639 |
| P. mirabilis | SGI1-PmABB | + | C | G | JX121638 |
| S. Typhimurium | SGI1 | − | A | G | KU499918 |
| S. Typhimurium | SGI1 | − | A | G | CP014979 |
| S. Typhimurium | SGI1 | − | A | G | CP014975 |
| S. Typhimurium | SGI1 | − | A | G | CP014969 |
| S. Typhimurium | SGI1 | − | A | G | CP014967 |
| S. Typhimurium | SGI1 | − | A | G | CP012985 |
| S. Typhimurium | SGI1 | − | A | G | CP014358 |
| S. Typhimurium | SGI1 | − | A | G | CP007581 |
| S. Typhimurium | SGI1 | − | A | G | HF937208 |
| S. Infantis | SGI1-D | − | A | G | KU854986 |
| S. Derby | SGI1-I | − | A | T | KU563154 |
| S. Rissen | SGI1-I | − | A | T | KM234279 |
| P. mirabilis | SGI1-B0616 | − | A | T | KU987432 |
| P. mirabilis | SGI1-O | − | A | T | KU987431 |
| P. mirabilis | SGI1-B | − | A | T | KU987430 |
| P. mirabilis | SGI1-Z | − | A | T | KP662516 |
| P. mirabilis | SGI1-X | − | A | T | KJ186154 |
| P. mirabilis | SGI1 | − | A | T | KJ186153 |
| P. mirabilis | SGI1-I | − | A | T | KJ186152 |
| P. mirabilis | SGI1-W | − | A | T | KJ186151 |
| P. mirabilis | SGI1-O | − | A | T | KJ186150 |
| P. mirabilis | SGI1-Y | − | A | T | KJ186149 |
| P. mirabilis | SGI1-PmBRI | − | A | T | JX089582 |
| P. mirabilis | SGI1-PmCAU | − | A | T | JX089581 |
| P. mirabilis | SGI1-B2 | − | A | T | KP116299 |
| S. Typhimurium | SGI1c | − | A | G | AF261825 |
| S. Cerro | SGI1-Fd | − | A | T | KU847976 |
| S. Emek | SGI2e | − | A | G | AY963803 |
SNP positions in the ORF of the TA system sgiAT are given according to ENA accession no. LT630458.
The SGI1-PmMAT variant harbored the deletion created by ISVch4 and extending from within ORF S005 to within ORF S009 but without the presence of ISVch4 (18).
The original SGI1 sequence (AF261825) showed 6 other specific SNP positions.
The SGI1-F variant showed 5 other specific SNP positions.
The whole-genome sequence analysis of M. morganii LIM90 revealed the presence of other antibiotic resistance determinants in the chromosome: (i) chromosomal AmpC β-lactamase gene blaDHA-17, conferring resistance to amoxicillin ± clavulanic acid, cephalotin, and cefuroxim (further confirmed by the phenotypic cloxacillin disk diffusion test; data not shown), and (ii) a class 2 integron carrying the sat2 and aadA1 gene cassettes, conferring resistance to streptothricin and streptomycin/spectinomycin, respectively. No plasmid of Enterobacteriaceae was detected by the PlasmidFinder tool, indicating that M. morganii LIM90 did not carry a conjugative IncA/C plasmid known to specifically mobilize SGI1 (2, 3). This observation is in accordance with the recently described incompatibility between SGI1 and members of the IncA/C plasmid family (30).
The identification of SGI1 in a M. morganii clinical isolate is of great interest, as the spread of this multidrug-resistant genomic island, especially in a naturally β-lactam-resistant species such as M. morganii, is a nonnegligible threat to public health. The important role of the horizontal transfer of SGI1 is crucial in the dissemination of multidrug resistance and may increase through pathogenic or nonpathogenic bacterial genera as well.
Accession number(s).
The sequence of SGI1 was deposited in the European Nucleotide Archive (ENA) under accession number LT630458.
ACKNOWLEDGMENTS
We thank the LABGeM and the National Infrastructure France Genomique for technical support and Karine Praud for technical assistance. We thank the members of the staff of the Limoges University Genolim platform for technical and bioinformatic assistance.
This work was supported by public funds from the French National Institute of Agricultural Research, from Ministère de l’Enseignement Supérieur et de la Recherche, and from Institut National de la Santé et de la Recherche Médicale (Inserm). E.S. is supported by a PhD fellowship from Anses and INRA.
REFERENCES
- 1.Boyd D, Peters GA, Cloeckaert A, Boumedine KS, Chaslus-Dancla E, Imberechts H, Mulvey MR. 2001. Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J Bacteriol 183:5725–5732. doi: 10.1128/JB.183.19.5725-5732.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doublet B, Boyd DA, Mulvey MR, Cloeckaert A. 2005. The Salmonella genomic island 1 is an integrative mobilizable element. Mol Microbiol 55:1911–1924. doi: 10.1111/j.1365-2958.2005.04520.x. [DOI] [PubMed] [Google Scholar]
- 3.Douard G, Praud K, Cloeckaert A, Doublet B. 2010. The Salmonella genomic island 1 is specifically mobilized in trans by the IncA/C multidrug resistance plasmid family. PLoS One 5:e15302. doi: 10.1371/journal.pone.0015302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carraro N, Matteau D, Luo P, Rodrigue S, Burrus V. 2014. The master activator of IncA/C conjugative plasmids stimulates genomic islands and multidrug resistance dissemination. PLoS Genet 10:e1004714. doi: 10.1371/journal.pgen.1004714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mulvey MR, Boyd DA, Olson AB, Doublet B, Cloeckaert A. 2006. The genetics of Salmonella genomic island 1. Microbes Infect 8:1915–1922. doi: 10.1016/j.micinf.2005.12.028. [DOI] [PubMed] [Google Scholar]
- 6.Hall RM. 2010. Salmonella genomic islands and antibiotic resistance in Salmonella enterica. Future Microbiol 5:1525–1538. doi: 10.2217/fmb.10.122. [DOI] [PubMed] [Google Scholar]
- 7.Levings RS, Djordjevic SP, Hall RM. 2008. SGI2, a relative of Salmonella genomic island SGI1 with an independent origin. Antimicrob Agents Chemother 52:2529–2537. doi: 10.1128/AAC.00189-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doublet B, Chu C, Chiu CH, Fan YC, Cloeckaert A. 2009. Truncated tni module adjacent to the complex integron of Salmonella genomic island 1 in Salmonella enterica serovar Virchow. Antimicrob Agents Chemother 53:824–827. doi: 10.1128/AAC.01015-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doublet B, Praud K, Bertrand S, Collard JM, Weill FX, Cloeckaert A. 2008. Novel insertion sequence and transposon-mediated genetic rearrangements in genomic island SGI1 of Salmonella enterica serovar Kentucky. Antimicrob Agents Chemother 53:3745–3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doublet B, Praud K, Weill FX, Cloeckaert A. 2009. Association of IS26-composite transposons and complex In4-type integrons generates novel multidrug resistance loci in Salmonella genomic island 1. J Antimicrob Chemother 63:282–289. doi: 10.1093/jac/dkn500. [DOI] [PubMed] [Google Scholar]
- 11.Wilson NL, Hall RM. 2010. Unusual class 1 integron configuration found in Salmonella genomic island 2 from Salmonella enterica serovar Emek. Antimicrob Agents Chemother 54:513–516. doi: 10.1128/AAC.00895-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Le Hello S, Weill FX, Guibert V, Praud K, Cloeckaert A, Doublet B. 2012. Early strains of multidrug-resistant Salmonella enterica serovar Kentucky sequence type 198 from Southeast Asia harbor Salmonella genomic island 1-J variants with a novel insertion sequence. Antimicrob Agents Chemother 56:5096–5102. doi: 10.1128/AAC.00732-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamidian M, Holt KE, Hall RM. 2015. The complete sequence of Salmonella genomic island SGI1-K. J Antimicrob Chemother 70:305–306. doi: 10.1093/jac/dku331. [DOI] [PubMed] [Google Scholar]
- 14.Hamidian M, Holt KE, Hall RM. 2015. The complete sequence of Salmonella genomic island SGI2. J Antimicrob Chemother 70:617–619. doi: 10.1093/jac/dku407. [DOI] [PubMed] [Google Scholar]
- 15.Ahmed AM, Hussein AI, Shimamoto T. 2007. Proteus mirabilis clinical isolate harbouring a new variant of Salmonella genomic island 1 containing the multiple antibiotic resistance region. J Antimicrob Chemother 59:184–190. doi: 10.1093/jac/dkl471. [DOI] [PubMed] [Google Scholar]
- 16.Doublet B, Poirel L, Praud K, Nordmann P, Cloeckaert A. 2010. European clinical isolate of Proteus mirabilis harbouring the Salmonella genomic island 1 variant SGI1-O. J Antimicrob Chemother 65:2260–2262. doi: 10.1093/jac/dkq283. [DOI] [PubMed] [Google Scholar]
- 17.Siebor E, Neuwirth C. 2011. The new variant of Salmonella genomic island 1 (SGI1-V) from a Proteus mirabilis French clinical isolate harbours blaVEB-6 and qnrA1 in the multiple antibiotic resistance region. J Antimicrob Chemother 66:2513–2520. doi: 10.1093/jac/dkr335. [DOI] [PubMed] [Google Scholar]
- 18.Siebor E, Neuwirth C. 2013. Emergence of Salmonella genomic island 1 (SGI1) among Proteus mirabilis clinical isolates in Dijon, France. J Antimicrob Chemother 68:1750–1756. doi: 10.1093/jac/dkt100. [DOI] [PubMed] [Google Scholar]
- 19.Lei CW, Zhang AY, Liu BH, Wang HN, Guan ZB, Xu CW, Xia QQ, Cheng H, Zhang DD. 2014. Molecular characteristics of Salmonella genomic island 1 in Proteus mirabilis isolates from poultry farms in China. Antimicrob Agents Chemother 58:7570–7572. doi: 10.1128/AAC.03992-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schultz E, Haenni M, Mereghetti L, Siebor E, Neuwirth C, Madec JY, Cloeckaert A, Doublet B. 2015. Survey of multidrug resistance integrative mobilizable elements SGI1 and PGI1 in Proteus mirabilis in humans and dogs in France, 2010–13. J Antimicrob Chemother 70:2543–2546. doi: 10.1093/jac/dkv154. [DOI] [PubMed] [Google Scholar]
- 21.Schultz E, Cloeckaert A, Doublet B, Madec JY, Haenni M. 2017. Detection of SGI1/PGI1 elements and resistance to extended-spectrum cephalosporins in Proteae of animal origin in France. Front Microbiol 8:32. doi: 10.3389/fmicb.2017.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lei CW, Zhang AY, Liu BH, Wang HN, Yang LQ, Guan ZB, Xu CW, Zhang DD, Yang YQ. 2015. Two novel Salmonella genomic island 1 variants in Proteus mirabilis isolates from swine farms in China. Antimicrob Agents Chemother 59:4336–4338. doi: 10.1128/AAC.00120-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qin S, Qi H, Zhang Q, Zhao D, Liu ZZ, Tian H, Xu L, Xu H, Zhou M, Feng X, Liu HM. 2015. Emergence of extensively drug-resistant Proteus mirabilis harboring a conjugative NDM-1 plasmid and a novel Salmonella genomic island 1 variant, SGI1-Z. Antimicrob Agents Chemother 59:6601–6604. doi: 10.1128/AAC.00292-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siebor E, Neuwirth C. 2014. Proteus genomic island 1 (PGI1), a new resistance genomic island from two Proteus mirabilis French clinical isolates. J Antimicrob Chemother 69:3216–3220. doi: 10.1093/jac/dku314. [DOI] [PubMed] [Google Scholar]
- 25.Girlich D, Dortet L, Poirel L, Nordmann P. 2015. Integration of the blaNDM-1 carbapenemase gene into Proteus genomic island 1 (PGI1-PmPEL) in a Proteus mirabilis clinical isolate. J Antimicrob Chemother 70:98–102. doi: 10.1093/jac/dku371. [DOI] [PubMed] [Google Scholar]
- 26.The European Committee on Antimicrobial Susceptibility Testing 2016. Breakpoint tables for interpretation of MICs and zone diameters, Version 6.0 http://www.eucast.org.
- 27.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vallenet D, Engelen S, Mornico D, Cruveiller S, Fleury L, Lajus A, Rouy Z, Roche D, Salvignol G, Scarpelli C, Médigue C. 2009. MicroScope: a platform for microbial genome annotation and comparative genomics. Database (Oxford) 2009:bap021. doi: 10.1093/database/bap021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harmer CJ, Hamidian M, Ambrose SJ, Hall RM. 2016. Destabilization of IncA and IncC plasmids by SGI1 and SGI2 type Salmonella genomic islands. Plasmid 87–88:51–57. doi: 10.1016/j.plasmid.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 30.Huguet KT, Gonnet M, Doublet B, Cloeckaert A. 2016. A toxin antitoxin system promotes the maintenance of the IncA/C-mobilizable Salmonella genomic island 1. Sci Rep 6:32285. doi: 10.1038/srep32285. [DOI] [PMC free article] [PubMed] [Google Scholar]