Abstract
Fibroblasts in 2D cultures differ dramatically in behavior from those in the 3D environment of a multicellular organism. However, the basis of this disparity is unknown. A key difference is the spatial arrangement of anchored extracellular matrix (ECM) receptors to the ventral surface in 2D cultures and throughout the entire surface in 3D cultures. Therefore, we asked whether changing the topography of ECM receptor anchorage alone could invoke a morphological response. By using polyacrylamide-based substrates to present anchored fibronectin or collagen on dorsal cell surfaces, we found that well spread fibroblasts in 2D cultures quickly changed into a bipolar or stellate morphology similar to fibroblasts in vivo. Cells in this environment lacked lamellipodia and large actin bundles and formed small focal adhesions only near focused sites of protrusion. These responses depend on substrate rigidity, calcium ion, and, likely, the calcium-dependent protease calpain. We suggest that fibroblasts respond to both spatial distribution and mechanical input of anchored ECM receptors. Changes in cell shape may in turn affect diverse cellular activities, including gene expression, growth, and differentiation, as shown in numerous previous studies.
Keywords: adhesion, cell migration, integrin, morphology
Cells in the tissues of multicellular organisms show a wide spectrum of sizes and shapes, reflective of their diverse functions. For example, a neuron is highly polarized and elongated, as required for the transmission of information over long distances, whereas epithelial cells, also highly polarized, are generally columnar or cubical in shape, reflective of their barrier functions. It has long been recognized that such variability in cell shape and behavior is coupled to differences in cell growth, differentiation, and other important functions (1, 2).
A long-standing question is how the diverse cell shapes are derived. It is generally recognized that chemical signals play an important role in defining the cell shape and migration, as demonstrated by chemotaxis. However, it is also becoming clear that physical and topographical parameters may have equally significant effects (3–5). A dramatic example is the shape change that occurs when a cell is removed from the tissue and is grown on glass or plastic surfaces (6, 7). These 2D cultures, as conveniently used for the maintenance of cells and for biological studies, impose highly unnatural geometric and mechanical constraints to many types of cells (8, 9). Fibroblasts, which are normally bipolar or stellate in shape when embedded in flexible, fibrous networks of the extracellular matrix (ECM), adhere to these stiff, nonpermeable surfaces and adopt a dramatic spread morphology (10).
A key aspect that distinguishes fibroblasts in 2D and 3D cultures is the degree and spatial distribution of receptors anchored to the ECM. Integrins, the primary receptors interacting with the ECM, are known to induce the formation of focal adhesions (11, 12) and to modulate multiple signaling pathways that regulate such important processes as cell migration, cytoskeletal organizations, cell growth, gene expression, and apoptosis (13, 14). These effects led us to suspect that the lack of anchorage of dorsal ECM receptors and the artificial dorsal-ventral asymmetry might be responsible for the morphological differences between fibroblasts in vivo and in 2D cultures.
Decades of studies with fibroblasts embedded in collagen gels (15–17) and tissue-derived matrices (18) have indeed shown an elongated bipolar or stellate morphology similar to what was found in vivo. Although important insight has been gained from these studies, they involve changes in multiple physical and chemical parameters from conventional surface cultures, making it difficult to separate the effects of dorsal integrin anchorage from potential effects of other parameters. The effects of dorsal receptor anchorage and mechanical stressing have also been probed by using ECM-coated microbeads (19, 20); however, the observed effects were localized, and the relationship to global cell behavior in a 3D environment was difficult to assess (21).
To specifically address the effects of dorsal receptor anchorage, we have compared fibroblasts cultured on the surface of a flexible, ECM-coated polyacrylamide sheet (22) with those cultured between two layers of polyacrylamide substrates under otherwise identical conditions. Our results demonstrate that the difference in the extent and topography of ECM receptor anchorage is sufficient to induce a striking shape change and cytoskeletal organization similar to what were found of fibroblasts in vivo. In addition, we show that these effects require signaling through calcium and, possibly, the calcium-dependent protease calpain.
Materials and Methods
Preparation of Polyacrylamide Substrates. Flexible polyacrylamide substrates were prepared and coated with bovine plasma fibronectin (Sigma, catalog no. 4759), calf skin collagen (United States Biochemical), or BSA (Sigma A-7638, at 10 mg/ml) as described (22). All substrates used in this study were 5% acrylamid/0.1% N,N-methylene-bis-acrylamide unless otherwise specified. The Young's modulus of the substrate was estimated to be 2.8 × 104 N/m2, which was determined as described (5). To prepare the double substrate, extraneous media on the polyacrylamide sheets were removed by aspiration with a Pasteur pipette. The top polyacrylamide sheet, attached to a coverslip, was gently laid over the bottom substrate. A square piece of glass (2 × 2 × 5 mm, 4.4 g) was then placed on the coverslip attached to the top substrate to stabilize the sandwich. To reduce the distance between the top and bottom substrates, a weight of 30 g was applied for 30 sec with a minimal amount of medium. The medium was then replenished, and cells were observed on an Axiovert 100TV microscope (Zeiss) equipped with a custom stage incubator. Distance between the substrates was determined by using calibrated microscope focusing mechanism, moving from the top surface of the ventral substrate to the bottom surface of the dorsal substrate, as indicated by the embedded fluorescent microbeads. For time-lapse recordings, phase images were collected every 5 min as described (23).
Cell Culture, Transfection, and Immunofluorescence. NIH 3T3 mouse embryonic fibroblasts were obtained from the American Type Culture Collection and maintained as described (23). The GFP-paxillin plasmid was described (24). Cells were plated to 90% confluency on 60-mm dishes and transiently transfected with Superfect (Qiagen, Valencia, CA) and 8–10 μg of DNA for GFP-paxillin, following the protocol provided. After transfection, cells were allowed to recover for 4 h in fresh serum-containing medium before replating onto polyacrylamide substrates.
EGFP-calpastatin was kindly provided by A. Huttenlocher (University of Wisconsin, Madison) (25). Transient transfection of NIH 3T3 fibroblasts was performed by nucleofection using the Amaxa nucleofector and kit R (Amaxa, Gaithersburg, MD), following the protocol recommended by the manufacturer. After transfection, cells were plated directly onto fibronectin-coated substrates and given 4 h to adhere and spread before sandwiching in the double substrate. Images were recorded after 12–16 h of incubation.
To fix cells within the double-substrate culture, the medium around the sandwich was first removed, and the fixative was added with the top substrate still in place. The top coverslip with substrate attached was then gently removed with a pair of tweezers. Subsequent fixation and immunostaining were performed as for 2D cultures (26) by using anti-Arp3 polyclonal at 1:100 (Cytoskeleton, Denver), anti-vinculin at 1:100 (Sigma), and Alexa Fluor 546-labeled secondary antibodies.
All images were acquired with a Zeiss Axiovert S100 inverted microscope, equipped with a 10×/0.25 numerical aperture CP-Achromat lens and a 40×/0.75 numerical aperture Plan-Neofluar lens for phase-contrast images, and a Nikon 60×/1.20 numerical aperture PlanApo water-immersion lens with a Zeiss adaptor lens for fluorescence images and differential interference contrast microscopy. Live cells were imaged at 37°C, whereas fixed cells were imaged at room temperature. All images were acquired with a 512BFT cooled charge-coupled device camera (Roper Scientific, Trenton, NJ) and an ST133 controller driven by custom software. Dark counts were subtracted from the images before images were stored as TIFF files. The contrast and brightness of some images were adjusted with coreldraw (Corel, Ottawa) or photoshop (Adobe Systems, San Jose, CA).
To characterize the topography of initial cell–substrate interactions, fibroblasts were plated on fibronectin-coated polyacrylamide substrates embedded with 0.1-μm green fluorescent beads. The cells were incubated with fibronectin-coated 0.5-μm polychromatic red fluorescent beads (Polysciences) for 1 h at 37°C, then fixed with 4% formaldehyde and sandwiched underneath a second sheet of identical substrate. Optical sections were acquired with a ×63 water-immersion lens (1.20 numerical aperture, Zeiss) and a spinning-disk confocal microscope with a 488-nm argon laser (Solamere Technologies, Salt Lake City), which allowed simultaneous detection of both beads distinguishable by their different sizes. Conventional epifluorescence with a narrow bandwidth filter set, consisting of a 540DF23 excitation filter, 565DRLP dichroic mirror, and a 590DF35 emission filter (Omega Optical, Brattleboro, VT), was used to visualize the polychromatic beads alone.
Cell shape was measured by calculating the aspect ratio of equivalent rectangles. The spread area of the cell and the cumulative length of all of the projections were measured by using a custom program. The equivalent width of the cell was then calculated by dividing the area with the cumulative length, and the aspect ratio was obtained as the ratio between the cumulative length and equivalent width.
Measurements of Traction Stress. Data for analysis of traction stress were collected after 12–16 h of incubation in the double-substrate culture as described (23). Microbeads of different colors were embedded in the top and bottom substrates to track the deformation. Images of null force were obtained after soaking the double-substrate culture in 1% Triton X-100 overnight. Traction stresses were calculated and rendered as described (23, 27).
Calcium Depletion. NIH 3T3 mouse embryonic fibroblasts were plated on fibronectin-coated substrates the night before the experiments. Cells were rinsed with 37°C Hepes-buffered saline (HBS) and incubated for 25 min at 37°C in HBS containing 30 μM 5,5′-dimethyl 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetate acetoxymethyl ester (Molecular Probes) and 3 μM thapsigargin (Sigma). The cells were then rinsed with 37°C HBS and sandwiched under a second fibronectin-coated substrate as described above. Complete DME containing 2 mM EGTA, 30 μM 5,5′-dimethyl 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetate acetoxymethyl ester, and 3 μM thapsigargin was then added to the dish after the sandwiching. Cells in the double substrate were then placed on a 37°C microscope stage under CO2. Regions in the sandwich with a gap of 3–6 μm, as determined by focusing on the microbeads embedded in the substrates, were imaged as described above.
Results
Anchorage of Dorsal ECM Receptors Changes Fibroblast Morphology. To address how the degree and spatial distribution of ECM receptor anchorage affect fibroblast behavior, we compared NIH 3T3 cells cultured on polyacrylamide surfaces coated with ECM proteins and cells sandwiched between two similar layers of polyacrylamide substrates (Fig. 1A). Attention was focused on regions where the top and bottom substrates were separated by 3–6 μm, which allowed top substrates to interact with a portion of the dorsal surface of most cells.
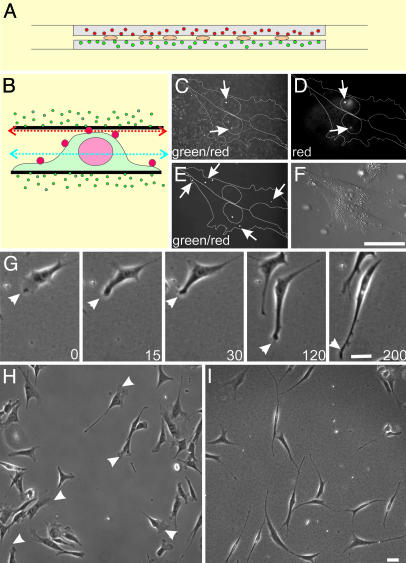
Fig. 1.
The double-substrate culturing system and its effects on fibroblast morphology. (A) NIH 3T3 cells are cultured between two sheets of protein-coated polyacrylamide, each ≈75 μm in thickness and embedded with 0.1-μm red (top substrate) or green (bottom substrate) fluorescent microbeads. (B) Cells were surface-labeled with polychromatic beads of 0.5 μm, fixed, and placed in the double substrate with a gap distance of 3.6 μm, to document the initial cell-substrate contact. (C) Confocal image recorded in the green channel and focused slightly underneath the top substrate (red arrow in B) shows both polychromatic beads on the dorsal surface above the nucleus and green beads embedded within the top substrate. (D) Image taken in the red channel on the same focal plane identifies the 0.5-μm polychromatic beads. Beads that bind to more peripheral regions of the dorsal surface are out of focus in this image but are visible on a lower plane of focus (E, taken in the green channel; blue arrow in B), where green beads within the substrates are out of focus. Over a period of 3 h (time in minutes as indicated), long extensions form out of existing lamellipodia (G, arrowhead), such that, at steady state, fibroblasts in the double-substrate culture become highly elongated and lack lamellipodia (I). In contrast, fibroblasts on fibronectin-coated single substrate maintain typical lamellipodia (H, arrowheads). (Bar, 30 μm.)
In most experiments, NIH 3T3 cells were first allowed to adhere and spread overnight on a fibronectin-coated polyacrylamide sheet. An identical sheet was then placed over their top surfaces. The tallest region of an NIH 3T3 cell that came close to the top substrate during initial engagement was above the nucleus and was typically 3–6 μm in height (K.A.B., unpublished observation). Confocal optical sectioning of cells surface-labeled with 0.5-μm polychromatic beads and embedded in substrates with 0.1-μm green beads verified that the top substrate interacted primarily with the nuclear region upon initial application, without engulfing the cell or contacting the lateral borders (Fig. 1 B–F). However, within 2 h, thin extensions grew out of existing broad lamellipodia, which then collapsed laterally and merged into the cell body (Fig. 1G). At steady state, the cells became highly elongated, showing one or several long, thin extensions but no lamellipodium (Fig. 1 H and I). Active protrusions were limited to small regions at the tip or along the side of the extensions. Analysis of the aspect ratio (length divided by width in an equivalent rectangular shape) indicated a 10-fold increase from 7.0 ± 3.8 for cells on 2D surfaces to 68 ± 35 for cells in the double-substrate culture. This steady state was maintained for >24 h with no detectable deterioration of the cells. Similar observations were made with NIH 3T3 cells on collagen-coated substrates and with primary cultures of chick embryonic fibroblasts (data not shown). In collagen-coated double substrates, NIH 3T3 cells were more highly branched compared with those in fibronectin-coated double-substrate cultures, suggesting that different ECM receptors invoked at least quantitatively different responses (see Supporting Text and Fig. 6, which are published as supporting information on the PNAS web site).
Whereas initial contact between the dorsal surface of the cell and the top substrate was limited to the region above the nucleus (Fig. 1 B–F), at steady state contacts with the top substrate were found to include some peripheral branches. Mechanical interactions at such branches were detected by measuring traction forces with double substrates embedded with fluorescent beads of different colors on the top and bottom (Fig. 2 A and B). Strong, inward traction forces were concentrated primarily near the tips of active protrusions (see Fig. 7, which is published as supporting information on the PNAS web site), with an average stress similar to what was observed on 2D cultures (≈2 × 104 dyne/cm2; 1 dyne = 10 μN). Moreover, a given extension rarely exerts forces on both surfaces (Fig. 2 A and B), which is consistent with the small size of cellular extensions relative to the gap between substrates. This configuration was most likely reached by the extension of thin processes in an upward or downward manner after the initial engagement of the top substrate with the dorsal membrane.
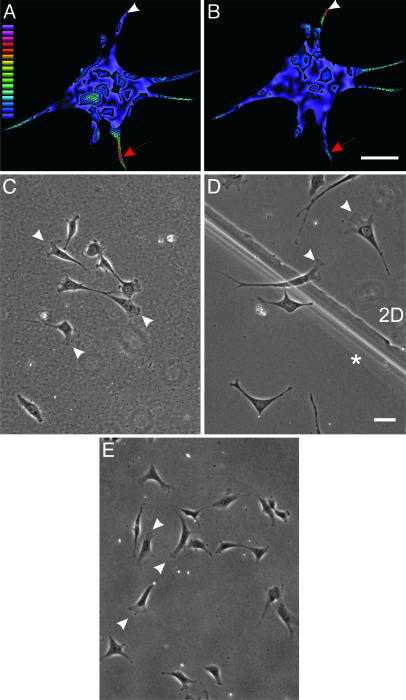
Fig. 2.
Involvement of anchorage of dorsal ECM receptors in morphological changes observed at steady state. NIH 3T3 cells were cultured in the double-substrate system, coated with fibronectin, and embedded with red and green beads in bottom and top substrates, respectively. After overnight incubation, the magnitude of traction stress exerted by the cell on the bottom (A) and top (B) substrate was measured based on displacements of the embedded beads and rendered as color images. Warm (red) colors designate strong traction forces, and cool (blue) colors indicate weaker forces. Red arrows indicate an extension where the cell exerts stronger traction forces on the top substrate than on the bottom substrate. Arrowheads indicate the converse situation for another extension. Color-coded bar represents the magnitude of stress from 1 × 102 to 2.7 × 105 dyne/cm2. (C) When only the bottom sheet is coated with fibronectin, after overnight incubation in the double-substrate culture, cells maintain a morphology similar to those in 2D cultures. (D) In addition, at the boundary between fibronectin-coated single and double substrates, lamellipodia are found where cells make contact with only the bottom substrate, whereas elongated morphology is seen on the double-substrate side (indicated with an asterisk). (E) Cells also maintain a morphology similar to those in 2D cultures when the bottom sheet has been coated with fibronectin and the top sheet has been coated with BSA. (Bar, 30 μm.)
To verify that simple physical contact with the top substrate was not responsible for the morphological response, we placed fibroblasts in a double substrate where the top substrate was either uncoated or coated with BSA. Fibroblasts in this environment maintained a 2D morphology, confirming that anchorage of dorsal ECM receptors was essential for the response (Fig. 2 C and E). Furthermore, clear differences in morphology were observed between cells in regions of different distances between the top and bottom substrates and across the boundary of single and double substrates (Fig. 2D). These controls demonstrate that neither global chemical factors nor the weight of the substrate was responsible for the morphological changes and that anchorage of dorsal receptors was the key factor.
Anchorage of Dorsal ECM Receptors Affects Cell Migration and Cytoskeletal Organization. Fibroblasts in double-substrate cultures migrated by lengthening their extensions and smoothly pulling the cell body forward, with no apparent cycles of elongation and retraction typical of cells on rigid 2D surfaces (see Movies 1 and 2, which are published as supporting information on the PNAS web site). The speed in the double-substrate culture was reduced by 54% from that on single substrates (0.28 ± 0.3 μm/min versus 0.61 ± 0.3 μm/min as determined by tracking the center of the nucleus), likely because of the anchorage on the dorsal surface. However, other experimental parameters, including fibronectin density and substrate rigidity, may also have affected the rate of migration (28).
The actin cytoskeleton and focal adhesions are known to respond to the anchorage of integrins and effect the changes in cell shape and migration (1, 12, 29). Phalloidin staining showed fewer prominent stress fibers for cells in double-substrate cultures (average number of prominent fibers in double-substrate cultures was 4 ± 1.4, n = 20) compared with cells on 2D cultures (average number of prominent fibers in single-substrate cultures was 24.3 ± 5.2, n = 20; Fig. 3A). Although there were short bundles in the cell body, most actin bundles were found along the lateral borders of the extensions (Fig. 3 B and C), similar to what was observed with fibroblasts in collagen matrices (16, 17).
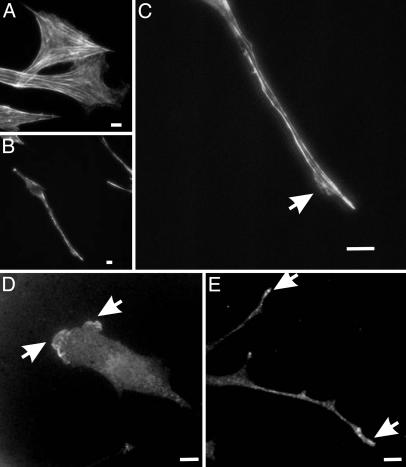
Fig. 3.
Organization of actin filaments and Arp3 in fibroblasts cultured in fibronectin-coated 2D and double-substrate cultures. (A) Rhodamine phalloidin staining of actin filaments show many large actin bundles in fibroblasts cultured on 2D polyacrylamide sheets. In contrast, in the double-substrate culture, most actin bundles are located along lateral borders of the extensions (B and C), although there are a few short actin bundles near the nucleus. Arrow indicates a small protrusive region where actin organization is more diffuse. Arp3 staining shows a band of concentration along the leading edge on a fibronectin-coated 2D substrate (D) and discrete foci at the tips of extensions in a double-substrate culture (E). (Bar, 10 μm.)
The disappearance of lamellipodium in double-substrate culture prompted us to examine also the distribution of Arp2/3, which is believed to nucleate actin filaments in the lamellipodium (30). Arp2/3 was found as small foci scattered along long projections of cells in double substrates and was more concentrated at tips of advancing extensions. The lack of a broad band of Arp2/3 was consistent with the inhibition of lamellipodia activities (Fig. 3 D and E).
Unlike focal adhesions in 2D cultures, the organization of focal adhesions in previous 3D culture systems was highly variable (17, 18), whereas their existence in vivo was unclear (11, 8, 21). We found that cells in double-substrate cultures formed fewer prominent focal adhesions (average number of detectable adhesions was 26.5 ± 8.2, n = 20) than those on 2D substrates (average number of detectable adhesions was 174.8 ± 26.5; n = 20), despite the presence of adhesive substrates for both dorsal and ventral cell surfaces (Fig. 4 A and B). Moreover, most focal adhesions appeared as small dots near protrusive regions at the tip and side of the extensions (Fig. 4 B and C). Imaging of living cells expressing GFP-paxillin showed striking appearance, movement, and maturation/stabilization of focal adhesions over a broad lamellipodial region for cells in 2D cultures (Movie 3, which is published as supporting information on the PNAS web site). In contrast, cells in the double-substrate culture formed focal adhesions in a highly localized manner at protrusive regions, with most nascent focal adhesions disappearing within 30 min near the ruffling edge (Movie 4, which is published as supporting information on the PNAS web site). Only a small fraction of focal adhesions persisted beyond a 30-min period and became separated from protrusive regions as the cell migrated forward.
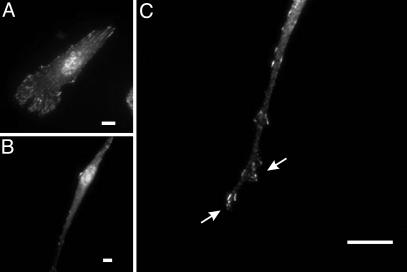
Fig. 4.
Organization of focal adhesions in fibroblasts cultured in fibronectin-coated 2D and double-substrate cultures. Immunofluorescence of paxillin at a low magnification shows many prominent focal adhesions in cells on 2D substrates (A) but few focal adhesions in double-substrate cultures (B). At a higher magnification, small dot-like focal adhesions are observed in an extension, many of which are located near protrusive edges (C, arrows). (Bar, 10 μm.)
Responses to Double Substrate Require Mechanical Input and Calcium Signaling. Because mechanical input from the substrate has substantial effects on cellular behavior in 2D cultures (31), we asked how the flexibility of the top substrate affects morphological responses. Cells were cultured in double substrates of mixed rigidity, which was controlled by changing the concentration of bis-acrylamide crosslinker. Cells in this hybrid environment adopted a morphology intermediate between those in uniformly rigid double-substrate culture and on single polyacrylamide sheets (Fig. 5A), indicating that mechanical signals are essential for the responses.
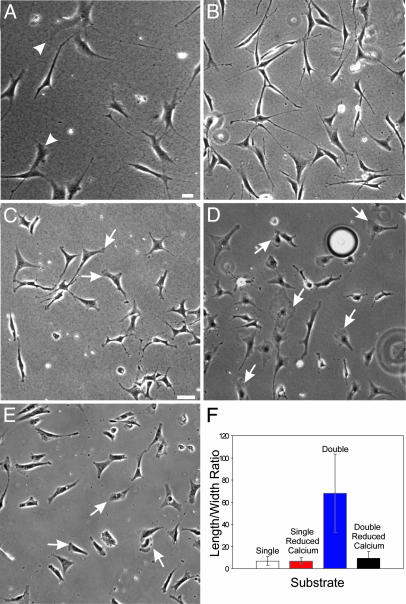
Fig. 5.
Mechanical input and calcium are required for the response to double-substrate culture. Fibroblasts cultured in a hybrid double substrate of soft (5% acrylamide, 0.06 bis-acrylamide; Young's modulus 1.4 × 104 N/m2) and stiff (5% acrylamide, 0.1% bis-acrylamide; Young's modulus 2.8 × 104 N/m2) polyacrylamide show both elongated morphology and lamellipodia (A, arrowheads). (B) Vehicle-treated control cells in fibronectin-coated double-substrate cultures elongate normally after 2 h in culture. (C and D) In contrast, cells cultured in the presence of 30 μM 5,5′-dimethyl 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetate acetoxymethyl ester, 3 μM thapsigargin, and 2 mM EGTA maintained a more spread morphology and lamellipodia activities, as indicated by arrows, after 2 h in the double-substrate cultures. (E) Fibroblasts cultured on fibronectin-coated 2D substrates and depleted of calcium maintain lamellipodia (arrows) and show a normal phase morphology. (F) The difference in cell shape is further quantified by measuring the aspect ratio in single- and double-substrate cultures with and without calcium depletion (n = 25 cells, from three to five experiments for each condition). (Bar, 30 μm.)
Previous studies have implicated calcium in the transduction and propagation of mechanical signals from the site of receptor anchorage (26, 32, 33). To address the involvement of calcium, we used a mixture of EGTA, thapsigargin and 5,5′-dimethyl 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetate acetoxymethyl ester in the double-substrate culture, a treatment used previously to deplete both intra- and extracellular calcium stores of fibroblasts (34). Cells in this environment maintained an apparently normal morphology in 2D culture; 31% of cells showed lamellipodia (n = 241; arrows in Fig. 5 C and D) as compared with 23% of vehicle treated cells (n = 325). However, these cells were unable to respond to the application of top substrate by collapsing their lamellipodia and developing the elongated morphology over a period of 2 h (Fig. 5 B–F). Measurement of cell shape indicated a low aspect ratio (8.8 ± 4), as with cells in calcium-depleted single-substrate cultures (7.2 ± 3, t test, P = 0.09; Fig. 5 E and F). In addition, preliminary results with gene knockout cells (data not shown) and with cells expressing calpastatin (an endogenous inhibitor of calpain) indicated that calpain, a potential protease effector of calcium, is essential for the response to dorsal substrate (Fig. 8, which is published as supporting information on the PNAS web site).
Discussion
The goal of this study was to address the striking range of cell shape and behavior, as observed when fibroblasts were grown in vivo and in different culture environments in vitro. We have designed an approach where cells are sandwiched between two thin sheets of porous, flexible polyacrylamide. This double-substrate culture not only allowed us to manipulate the anchorage of dorsal ECM receptors and to control important parameters such as substrate rigidity and ligand coating but also provided excellent image quality. The approach should also greatly facilitate future studies of other events affected by integrin anchorage, such as cell differentiation and gene expression (13, 14).
By using this approach, we demonstrated that both the topography of ECM receptor anchorage and the mechanical characteristics of the substrate play an important role in defining fibroblast morphology. We suggest that the lack of dorsal receptor anchorage in 2D cultures tips the balance between spreading and retraction and creates an overall stimulatory environment for the organization of lamellipodia, stress fibers, and focal adhesions. This imbalance would then cause cells to spread out, possibly in search of an environment that allows the engagement of ECM receptors on the dorsal surface. Conversely, anchorage of dorsal receptors in 3D environment may enhance a global retraction signal, causing fibroblasts to adopt a highly elongated morphology without lamellipodia (10, 15, 18). Interestingly, the response appears to be different in epithelial cells that lose their apical/basal polarity and become fibroblast-like when placed in collagen matrices (35). These observations underscore the potential importance of differentiation states and organ environment in cellular responses to physical and topographical parameters (9).
Calcium represents a logical candidate for mediating the mechanical signals triggered by ECM receptor anchorage because of its ability to propagate throughout the cell and trigger a number of downstream signaling pathways. Consistent with this possibility, our data showed that cells cultured in a calcium-depletion medium maintained their ability to form lamellipodia and failed to respond to dorsal receptor anchorage within a 2-h period. The effects of calcium is certain to involve both an upstream mechanism that causes its release and downstream effectors that mediate the effects on actin cytoskeleton and focal adhesions. A potential mechanism for calcium release is through stretch-activated channels that were suggested to regulate cellular traction forces and focal adhesions in response to mechanical forces exerted on integrins (26, 36). The initial entry of calcium may subsequently trigger calcium-initiated calcium release from intracellular stores over long distances (33). Potential effectors of the calcium signal include calmodulin, myosin light-chain kinase, and actin-binding proteins such as gelsolin (33). It is particularly interesting that cells transfected with calpastatin failed to respond to the anchorage of dorsal ECM receptors. Calpain is known to associate with focal adhesions (37) and to cleave several focal adhesion proteins, including talin, vinculin, and paxillin upon calcium activation (38). It may play a role in the release of substrate adhesions for the cell to assume an elongated shape (25, 39). Thus, upon dorsal receptor anchorage, increased calpain-mediated degradation of focal adhesions may lead to reduced adhesions and dramatic changes in cell shape.
It is important to emphasize that calcium likely works in conjunction with other adhesion-mediated signaling molecules such as small GTPases (40) and tyrosine kinases (41–43). For example, calcium has been reported to regulate the translocation of Rac (43), whereas calpain is able to degrade FAK, pp60Src, and RhoA (38, 44). Consistent with this idea, we found that manipulation of the activities of small GTPases had dramatic effects on cellular response to the double substrate (K.A.B., personal observation). Finally, changes in cell shape and migration likely represent only one of many profound effects of dorsal receptor anchorage in fibroblasts (13, 14). In conjunction with effects on cell growth and differentiation, these responses are likely to play a central role in both physiological processes such as embryonic development and pathological processes such as cancer metastasis.
Supplementary Material
Acknowledgments
We thank Sandy Cho for technical assistance, J. Wehland (Gesellschaft für Biotechnologishe Forschung, Braunschweig, Germany) and Anna Huttenlocher for providing plasmids. This work was supported by National Aeronautics and Space Administration Grant NAG2-1495 and National Institutes of Health Grants GM-32476 (to Y.-L.W.) and GM-61806 (to M.D.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: ECM, extracellular matrix.
References
- 1.Folkman, J. & Moscona, A. (1978) Nature 273, 345–349. [DOI] [PubMed] [Google Scholar]
- 2.Watt, F. M., Jordan, P. W. & O'Neill, C. H. (1988) Proc. Natl. Acad. Sci. USA 85, 5576–5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dunn, G. A. & Brown, A. F. (1986) J. Cell Sci. 83, 313–340. [DOI] [PubMed] [Google Scholar]
- 4.Curtis, A. S. G. & Wilkinson, C. (1997) Biomaterials 18, 1573–1583. [DOI] [PubMed] [Google Scholar]
- 5.Lo, C.-M., Wang, H.-B., Dembo, M. & Wang, Y.-L. (2000) Biophys. J. 79, 144–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lewis, W. H. & Lewis, M. R. (1924) in General Cytology, ed. Cowdry, E.V. (Univ. of Chicago Press, Chicago), pp. 384–447.
- 7.Weiss, P. (1959) Rev. Mod. Phys. 31, 11–20. [Google Scholar]
- 8.Walpita, D. & Hay, E. D. (2002) Nat. Rev. Mol. Cell Biol. 3, 137–141. [DOI] [PubMed] [Google Scholar]
- 9.Schmeichel, K. L. & Bissell, M. J. (2003) J. Cell Sci. 116, 2377–2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bard, J. B. & Hay, E. D. (1975) J. Cell Biol. 67, 400–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jockusch, B. M., Bubeck, P., Giehl, K., Kroemker, M., Moschner, J., Rothkegel, M., Rudiger, M., Schluter, K., Stanke, G. & Winkler, J. (1995) Annu. Rev. Cell Dev. Biol. 11, 379–416. [DOI] [PubMed] [Google Scholar]
- 12.Hynes, R. O. (2002) Cell 110, 673–687. [DOI] [PubMed] [Google Scholar]
- 13.Burridge, K. & Chrzanowska-Wodnicka, M. (1996) Annu. Rev. Cell Dev. Biol. 12, 463–519. [DOI] [PubMed] [Google Scholar]
- 14.Schwartz, M. A. & Ginsberg, M. H. (2002) Nat. Cell Biol. 4, E65–E68. [DOI] [PubMed] [Google Scholar]
- 15.Elsdale, T. & Bard, J. (1972) J. Cell Biol. 54, 626–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomasek, J. J. & Hay, E. D. (1984) J. Cell Biol. 99, 536–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grinnell, F. (2003) Trends Cell Biol. 13, 264–268. [DOI] [PubMed] [Google Scholar]
- 18.Cukierman, E., Pankov, R., Stevens, D. R. & Yamada, K. M. (2001) Science 294, 1708–1712. [DOI] [PubMed] [Google Scholar]
- 19.Wang, N., Butler, J. P. & Ingber, D. E. (1993) Science 260, 1124–1127. [DOI] [PubMed] [Google Scholar]
- 20.Choquet, D., Felsenfeld, D. P. & Sheetz, M. P. (1997) Cell 88, 39–48. [DOI] [PubMed] [Google Scholar]
- 21.Friedl, P. & Brocker, E. B. (2000) Cell. Mol. Life Sci. 57, 41–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beningo, K. A., Lo, C.-M. & Wang, Y.-L. (2002) Methods Cell Biol. 69, 325–339. [DOI] [PubMed] [Google Scholar]
- 23.Munevar, S., Wang, Y.-L. & Dembo, M. (2001) Biophys. J. 80, 1744–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rottner, K., Krause, M., Gimona, M., Small, J. V. & Wehland, J. (2001) Mol. Biol. Cell 12, 3103–3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhatt, A., Kaverina, I., Otey, C. & Huttenlocher, A. (2002) J. Cell Sci. 115, 3415–3425. [DOI] [PubMed] [Google Scholar]
- 26.Munevar, S., Wang, Y.-L. & Dembo, M. (2004) J. Cell Sci. 117, 85–92. [DOI] [PubMed] [Google Scholar]
- 27.Dembo, M. & Wang, Y.-L. (1999) Biophys. J. 76, 2307–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palecek, S. P., Loftus, J. C., Ginsberg, M. H., Lauffenburger, D. A. & Horwitz, A. F. (1997) Nature 385, 537–540. [DOI] [PubMed] [Google Scholar]
- 29.Chen, C. S., Alonso, J. L., Ostuni, E., Whitesides, G. M. & Ingber, D. E. (2003) Biochem. Biophys. Res. Commun. 307, 355–361. [DOI] [PubMed] [Google Scholar]
- 30.Welch, M. D. (1999) Trends Cell Biol. 9, 423–427. [DOI] [PubMed] [Google Scholar]
- 31.Bershadsky, A. D., Balaban, N. Q. & Geiger, B. (2003) Annu. Rev. Cell Dev. Biol. 19, 677–695. [DOI] [PubMed] [Google Scholar]
- 32.Schwartz, M. A. (1993) J. Cell Biol. 120, 1003–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berridge M. J., Lipp, P. & Bootman, M. D. (2000) Nat. Rev. 1, 11–21. [DOI] [PubMed] [Google Scholar]
- 34.Emmert, D. A., Fee, J. A., Goeckeler, Z. M., Grojean, J. M., Wakatsuki, T., Elson, E. L., Herring, B. P., Gallagher, P. J. & Wysolmerski, R. B. (2004) Am. J. Physiol. 286, C8–C21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Greenberg, G. & Hay, E. D. (1982) J. Cell Biol. 95, 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee, J., Ishihara, A., Oxford, G., Johnson, B. & Jacobson, K. (1999) Nature 400, 382–386. [DOI] [PubMed] [Google Scholar]
- 37.Beckerle, M. C., Burridge, K., DeMartino, G. H. & Croal, D. E. (1987) Cell 51, 569–577. [DOI] [PubMed] [Google Scholar]
- 38.Glading, A., Lauffenburger, D. A. & Wells, A. (2002) Trends Cell Biol. 12, 46–54. [DOI] [PubMed] [Google Scholar]
- 39.Huttenlocher, A., Palecek, S. P., Lu, Q., Zhang, W., Mellgren, R. L., Lauffenburger, D. A., Ginsberg, M. H. & Horwitz, A. F. (1997) J. Biol. Chem. 272, 32719–32722. [DOI] [PubMed] [Google Scholar]
- 40.Etienne-Manneville, S. & Hall, A. (2002) Nature 420, 629–635. [DOI] [PubMed] [Google Scholar]
- 41.Glogaur, M., Arora, P., Yao, G., Sokholov, I., Ferrier, J. & McCulloch, C. A. (1997) J. Cell Sci. 110, 11–21. [DOI] [PubMed] [Google Scholar]
- 42.Schlaepfer, D. D. & Hunter, T. (1998) Trends Cell Biol. 8, 151–157. [DOI] [PubMed] [Google Scholar]
- 43.Price, L. S., Langeslag, M., ten Klooster, J. P., Hordjik, P. L., Jalink, K. & Collard, J. G. (2003) J. Biol. Chem. 278, 39413–39421. [DOI] [PubMed] [Google Scholar]
- 44.Kulkarni S., Goll, D. E. & Fox, J. E. (2002) J. Biol. Chem. 277, 24435–24441. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.