Abstract
Legume-rhizobium symbiosis is achieved by two major events evolutionarily acquired: root hair infection and organogenesis. Infection thread (IT) development is a distinct element for rhizobial infection. Through ITs, rhizobia are efficiently transported from infection foci on root hairs to dividing meristematic cortical cells. To unveil this process, we performed genetic screening using Lotus japonicus MG-20 and isolated symbiotic mutant lines affecting nodulation, root hair morphology, and IT development. Map-based cloning identified an AP2/ERF transcription factor gene orthologous to Medicago truncatula ERN1. LjERN1 was activated in response to rhizobial infection and depended on CYCLOPS and NSP2. Legumes conserve an ERN1 homolog, ERN2, that functions redundantly with ERN1 in M. truncatula. Phylogenetic analysis showed that the lineages of ERN1 and ERN2 genes originated from a gene duplication event in the common ancestor of legume plants. However, genomic analysis suggested the lack of ERN2 gene in the L. japonicus genome, consistent with Ljern1 mutants exhibited a root hair phenotype that is observed in ern1/ern2 double mutants in M. truncatula. Molecular evolutionary analysis suggested that the nonsynonymous/synonymous rate ratios of legume ERN1 genes was almost identical to that of non-legume plants, whereas the ERN2 genes experienced a relaxed selective constraint.
Keywords: infection thread development, ERN1, Lotus japonicus, molecular evolutionary analysis
1. Introduction
Legumes produce root nodules as a consequence of a successful interaction with nitrogen-fixing Gram negative bacteria, collectively termed rhizobia. The interaction with bacterial symbionts enables the host plant to utilize atmospheric nitrogen. Therefore, the nodule symbiosis affects agricultural ecosystems. For establishing a symbiotic relationship, host cells guide rhizobia from the infection foci on root hair surfaces into cortical nodule primordia through membranous and tubular paths known as infection threads (ITs), which basipetally develop into infected root hairs and further penetrate into dividing cortical cells. The tip-growth of ITs is correlated with bacterial cell division and file movement.1 Rhizobia endocytically released from ITs into cortical cells and differentiate into bacteroids and can reduce dinitrogen into ammonium ion. Thus, IT progression is important for efficiently establishing the endosymbiosis. IT development has been observed not only in the root nodule symbiosis of legumes but also in the non-legume nodule symbiosis of actinorhizal plants, which are a group of a nitrogen-fixing lineage of angiosperms and interact with nitrogen fixing actinobacteria Frankia.2 The acquisition of nodule symbioses of legumes and actinorhizal plants is suggested to have been independent evolutionary events, further implying advantages of this infection system.
In the legume nodule symbiosis, Nod factors, lipochitoligosaccharides secreted by rhizobia, act as initial signals of nodulation.3 The Nod factor perception by host receptors (LjNFR1, LjNFR5, MtNPF, and MtLYS3) triggers the oscillation of perinuclear Ca2+ concentration and root hair deformation to entrap rhizobia.4–10 This Ca2+ spiking requires host factors that are also involved in mycorrhizal symbioses, the so-called common SYMs,11 which are essential for the IT development. The calcium signal is believed to be decoded by LjCCaMK (calcium and calmodulin-dependent protein kinase)/MtDMI3 because gain-of-function versions of this protein kinase suppress symbiosis-defective phenotypes observed in the loss-of-function of common SYMs that are required for the Ca2+ spiking.12–15 Transcriptome analyses of actinorhizal plants have revealed that Casuarina glauca, Datisca glomera, and Alnus glutinosa possess common SYM factors including SymRK and CCaMK,16–18 which are involved in the nodule symbiosis of actinorhizal plants.19–21 These findings indicate that common SYM factors also act as signaling components of actinorhizal nodule symbiosis. In addition, a recent study further showed that the C.glauca, homolog of a nodulation-specific legume transcription factor, NODULE INCEPTION (NIN), is involved in the IT development,22–24 implying some nodulation-specific factors commonly required for both types of the nodule symbiosis. Homologous genes among legumes and actinorhizal plants may play a common important role in the development of nodules. Several homologs in a genome (paralogs) are also crucial for the nodulation. The above-mentioned receptors of Nod factors are such kind of paralogous genes and are believed to have originated from gene duplications.25,26 Duplication events in the gene- and genome-levels were suggested to associate with the evolution of symbiosis between legumes and rhizobia.27–29 Possible outcomes of functional divergence of the duplicated genes such as subfunctionalization and neofuntionalization might have adopted a key role in the legume evolution.
The above-mentioned L. japonicus and M. truncatula mutants exhibit deficiencies in IT progression and cortical cell division because these factors are responsible for early nodulation signaling pathways. Nodulation includes coordinately regulated processes of IT progression and cortical cell division producing nodule primordia. However, cortical cell division does not always associate with IT progression into the cortex. L. japonicus mutants, cerberus and cyclops, display infection defective phenotypes, but produce nodule primordia. CERBERUS and CYCLOPS encode a U-box protein containing WD-40 repeats and a transcription factor, respectively.30,31 The later protein is targeted by LjCCaMK, and its activity is regulated by phosphorylation at critical amino acid residues.32 Similar to L. japonicus mutants, loss-of-functions of M. truncatula counterparts also exhibit infection-deficient phenotype.33,34 These data indicate factors specifically regulating IT progression are conserved in legumes.
Many factors regulated with nodulation have been identified to date and have been used to create a basic model of nodulation pathways. Furthermore, common genetic features related with nodulation have been clarified between legumes and actinorhizal plants. However, our knowledge regarding regulatory pathways of IT progression remains fragmented. For elucidating molecular mechanisms underlying rhizobial infection processes, identifying novel factors involved in IT progression is indispensable. For this purpose, we screened L. japonicus mutants exhibiting infection-deficient phenotypes but producing nodule primordia. Here, we report a novel L. japonicus mutant for which infection processes are remarkably affected. Map-based cloning of the mutation locus revealed that the causative gene encodes homolog of M. truncatula ERN1.35–37 Our genomic analysis of ERN in legumes showed a unique feature of ERN genes of L. japonicus. Phylogenetic analysis indicated the gene duplication of the ERN gene in the common ancestor of legumes. Molecular evolutionary analysis suggested the difference of functional constraint between the genes of ERN1 and ERN2 lineages after the duplication event.
2. Materials and Methods
2.1. Plant materials
The F29 mutant was obtained by ethyl methanesulfonate (EMS) mutagenesis. Approximately 5,000 seeds were immersed in 0.4% EMS overnight and allowed to germinate. M1 plants were grown in a greenhouse, and M2 seeds were collected. A screen for mutants that form nodules but exhibit retarded plant growth was performed in approximately 25,000 M2 plants. A single individual of the M3 self progeny from the M2 candidate mutant was backcrossed with the parent MG-20. The 1699-1 mutant line was produced by carbon-ion beams. Ion-beam irradiation was conducted as previously described.38 The energy of carbon ions was 320 MeV, and the mean linear energy transfer within the seed was estimated to be 86 keV/μm. Approximately 5,000 dry seeds of wild-type (WT) L. japonicus MG-20 were irradiated with carbon ions (C6+) at a dose of 80 Gy. Approximately 84,000 M2 plants derived from approximately 2,900 M1 plants were screened for the nodulation phenotype. The heredity of the mutant candidates was evaluated by segregation in the next generation. The mutants nin-2, cyclops-3, cerberus-5, nsp2-1, and alb1-3 were all derived from L. japonicus Gifu and described previously.22,30,31,39
2.2. Observation of the rhizobial infection
Seeds were scarified with sandpaper and sterilized with sodium hypochlorite. Sterilized seeds were germinated on 1.0% agar plates under dark condition at 24°C in a growth cabinet (LH-220S, NK System). After 2 days, seedlings were transferred to inoculation pods containing vermiculite supplied with Broughton & Dilworth (B&D) medium,40 10 μM potassium nitrate and Mesorhizobium loti MAFF 303099 constitutively expressing red fluorescent protein (DsRed) gene.41 Plants were grown under 16 h light/8 h dark cycles at 24°C in the growth cabinet. The morphology of each infection events on roots infected with M. loti MAFF 303099 expressing DsRed was observed by a SZX16 microscope (Olympus).
2.3. Map-based cloning
The F29 mutant line was crossed with L. japonicus Gifu B-129 for obtaining the F2 population. A total of 1,015 F2 plants were analysed by DNA markers flanking the F29 locus. The DNA markers are detailed below: CM0371I1 is a polymerase chain reaction (PCR) marker distinguished by an insertion/deletion. CM0371SN2 is a marker detecting single-nucleotide polymorphism by a sequence analysis. TM0371 is a simple sequence repeat marker. CM0371C1 is a cleaved amplified polymorphic sequence digested by MseI. Candidate genes on TAC clone LjT46E19 were predicted by GENSCAN (http://genes.mit.edu/GENSCAN.html).
F2 seeds were scarified with sandpaper and sterilized with 10% hypochlorite. The seeds were maintained in sterile water for water absorption. The seeds were then germinated on 1.0% agar plates for 2 days. Seedlings were transferred to nitrogen-deficient soil and inoculated with M. loti MAFF303099. After 2 weeks, nodules were observed.
2.4. Plasmid construction and transformation
pCAMBIA1300GFP was constructed detailed below. The HPTII gene was replaced with the green fluorescent protein (GFP) gene, and an AscI site was introduced into the SmaI site in pCAMBIA1300 (Cambia). ERN1 promoter region (3,047 bp) and terminator region (1,109 bp) were amplified by PCR using specific primer sets (ERN1 promoter-F, AACTGCAGCGTGCATT GC AC GG ATATAC; ERN1 promoter-R, AAGGCGCGCCCACAAATTG TTCAAATTT AGTAA T T GAGTG; ERN1 terminator-F, AAGGC GCGC CACTTGATCTT GAAGGTCTTAAGTTAATG; ERN1 ter mi nator-R, TTGAGCTCT GCCTTTGAACTGTGAGCAG). The promoter fragment was digested with PstI and AscI and ligated into pCAMBIA1300GFP. The terminator fragment was ligated into the vector after digesting with AscI and SacI. To utilize Gateway technology, a destination vector named pCGERNGW was then constructed by inserting reading frame cassette A of the Gateway vector conversion system (Thermo Fisher Scientific) into the AscI site treated by the Klenow Fragment (Takara) of the vector. For complementation tests, ERN1 gene amplified twice by PCR (ERN1 gene 1st-F, AAAAAGCAGG CTCCGTGATG GAG AT TCAAT TCCAGCA ; ERN1 gene 1st-R, AGAAAGCTGGGT TT A A CA GAACAATGA GCACAAGG; ERN1 gene 2nd-F, GGGGACAA G TT T G TACAAAAAAGCAGGCT; ERN1 gene 2nd-R, GGGG AC C ACTTTGTACAAGAAAGCTGGGT) was cloned into pDONR/ZEO (Thermo Fisher Scientific) via Gateway BP reaction (Thermo Fisher Scientific). A binary vector for complementation tests was constructed by introducing the ERN1 gene into pCGERNGW using the Gateway LR reaction (Thermo Fisher Scientific). For promoter analysis, a β-glucuronidase (GUS)-Plus was amplified from pCAMBIA1305.1 (Cambia) by two times by PCR (GUS Plus gene 1st-F, AA AAAGCAGGCTCCACCATGGTAGATCTGAGGGTAA; GUS Plus gene 1st-R, AGAAAGCTGG GTTCA CACGT GATG GTG AT GG T; GUS Plus gene 2nd-F, GGGGACAA GTTTGTACA AAAA AG C AG GCT; GUS Plus gene 2nd-R, GGGGACCACTTT GTACAAG AAA GCTGGGT). This fragment was then cloned into pDONR/ZEO (Thermo Fisher Scientific) via the Gateway BP reaction (Thermo Fisher Scientific). A binary vector for the promoter-GUS analysis was constructed by introducing GUS Plus gene into pCGERNGW using the Gateway LR reaction (Thermo Fisher Scientific).
Plasmid constructs were introduced into Agrobacterium rhizogenes AR1193 and transformed into WT, F29, or 1699-1 plants by the hairy root transformation method with minor modifications.42
2.5. Histochemical GUS staining and quantified reverse transcription-PCR
For histochemical GUS staining, transformed plants with the promoter-GUS construct were inoculated with M. loti MAFF303099. The plants were grown for 7 days after inoculation. Roots were stained with 0.5 mg/ml 5-bromo-4-chloro-3-indolyl β-d-glucuronic acid cyclohexylammonium salt, 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, and 10 mM ethylenediaminetetraacetic acid (EDTA) in 100 mM sodium phosphate (pH 7.0). For observation of cortical cell division, roots were cleared by a chloral hydrate/glycerol/water (8w/1v/2v) solution.
For quantified reverse transcription (qRT)-PCR analysis, plants were grown for 0, 0.5, 1, and 3 days after inoculation with M. loti MAFF303099, and their roots were sampled from them. Total RNA was extracted from roots using the RNeasy Plant Mini Kit (Qiagen). Reverse transcription was conducted with the QuantiTect Reverse Transcription Kit (Qiagen). qRT-PCR was performed using the ABI Prism 7000 (Applied Biosystems) with a QuantiTect SYBR Green RT-PCR Kit (Qiagen). The following primer sets were used for detection of ERN1 and ATP synthase expressions (ERN1-F,TCCA AT GAAACTCTGTGGGATC; ERN1-R,GGGAAGAAACA TTTGC AG GA; ATP synthase-F, GGTGATAAGCAGAGTGAAAGCA; ATP synthase-R,AAGACCAGTGAGACCAACACG).
2.6. Ortholog and synteny analysis
The most recent versions of genome, protein, and complementary DNA (cDNA) sequences of each species were downloaded from the respective genome sequence sites as follows: L. japonicus (version 3.0) from the L. japonicus Genome Sequencing Project (http://www.kazusa.or.jp/lotus/, November 2016, date last accessed), M. truncatula (Mt4.0v1), and Glycine (Wm82.a2.v1) from Phytozome 11 (https://phytozome.jgi.doe.gov/pz/portal.html#, November 2016, date last accessed).
The analysis of microsynteny across species was based on comparisons of the specific regions containing ERN genes. Based on the LjERN1 and ERN genes in M. truncatula, the ERN genes of L. japonicus and G. max were set as the anchor points according to their physical locations. The protein-coding sequences assigned to the flanking regions of each ERN gene in one species were compared with those in the other species by the Basic Local Alignment Search Tool for protein databases (BLASTP) program. The reciprocal best hit method was used to define orthologous relationship among the genes in L. japonicus, M. truncatula, and G. max in the same ERN gene region. For the gene conservation among the three ERN gene regions, a BLASTP E-value <1e−20 was considered to be significant.
2.7. Phylogenetic analysis
Sequences similar to LjERN1 were collected with similarity searches against all the 68 plant genomes in the refseq database of the National Center for Biotechnology Information (NCBI) using BLASTP on 9 April 2016. The amino acid sequences similar to MtERN1, MtERN2, MtERN3, Sesamum indicum XP_011075059 and XP_011089231, Oryza sativa XP_015645889, and Amborella trichopoda XP_011628828 were selected with the option-max_target_seqs 100. The resulting 213 sequences and three other sequences (LjERN1, Cajanus cajan KYP69442.1, and KYP62096.1) were used for the preliminary analysis. Those containing large indels or those that were highly diverged were omitted from the dataset. Finally, 36 sequences of fabids (eurosid I) were used for the phylogenetic analysis. The amino acid sequences were aligned with the MAFFT program43 using the strategy E-INS-i, followed by manual adjustment. The maximum likelihood (ML) analysis was conducted with the LG model implemented in the RAxML program.44 The best model of evolution was determined using the option –auto-prot = aic in the program. The Bayes method was performed with MrBayes 3.2.6 using a WAG + I+G model.45 Neighbor-Joining (NJ) analysis was conducted with the JTT model of MEGA5.2.2 using the option of partial deletion of site coverage cutoff 80%.46
2.8. Molecular evolutionary analysis
To examine the pattern of ω (dn/ds: nonsynonymous/synonymous substitution rate ratio) acting on the ERN1 and ERN2 homologs at lineage-specific levels, ML tests implemented in Phylogenetic Analysis by Maximum Likelihood (PAML) were applied to the 36 sequences.47 The branch-specific model of PAML includes the one-, two-, and three-ratio models to represent the different presumed hypotheses. The one-ratio model assumed a constant ratio ω across all branches. The two-ratio models assigned the ratio ωERN1, ωERN2, or ωERN1 + ERN2 to the branches of the ERN1, ERN2, or ERN1 and ERN2 clades, respectively, and the ratio ωoutgroup to the remaining branches. The three-ratio model assigned the ratios ωERN1, ωERN2, and ωoutgroup to the corresponding branches. For any pair of models that were nested together, the goodness of fit was tested using the likelihood ratio test (LRT) with the CODEML program in PAML. The branch-site and clade models were also used for the analysis of amino acid sites under positive selection.
3. Results
3.1. Mutant lines defective in IT progression
During the early process of nodulation, rhizobia are entrapped by curled or deformed root hairs to form an infection pocket. From the site as the starting point, they enter into the plant cells through the development of ITs. In L. japonicus, mutants such as alb1, cerberus, crinkle, cyclops, epr3, and nin are shown to be defective in the infection process after the curling.22,30,31,48–50 To identify a new component involved in the infection process, we have performed an extensive screening of symbiotic mutants using L. japonicus. F29, a mutant line of infection processes that does not form effective nodules, was originally isolated by the treatment of EMS to L. japonicus Miyakojima MG-20. On the other hand, the 1699-1 mutant line was isolated through the screening of progeny of ion-beam-mutagenized L. japonicus MG-20 seeds. Both lines did not display a completely non-nodulating phenotype or form an increased number of bump-like structures on the root such as alb1 and epr3 (Fig. 1A).
Figure 1.
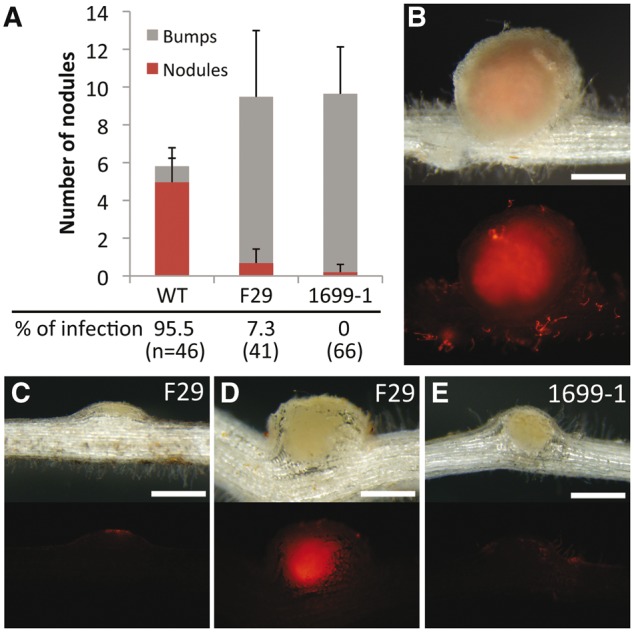
Nodule phenotypes of F29 and 1699-1 mutant lines. (A) Means and SDs of nodule and bump numbers 14 DAI (n = 25), and percentages of nodules/bumps that DsRed-labeled M. loti invaded into (10 DAI) are shown. (B–E) Nodules and bumps formed on WT (B), F29 (C and D), and 1699-1 (E) roots at 10 DAI with DsRed-labeled M. loti. Upper and bottom panels are bright field images and corresponding fluorescent images. Bars = 0.5 mm.
To visualize infection events, we inoculated F29 and 1699-1 mutant lines with Ds-Red-labeled M. loti MAFF303099. The nodulation phenotype of 1699-1 mutants was relatively severe compared with that of F29 mutants. The difference was clearly detected at 10 days after inoculation (DAI), when most of bumps or nodules formed on WT MG-20 roots have already displayed IT progression and release of rhizobia into cortical cells (Fig. 1A and B). Several bumps with IT progression into cortical cells were found in F29 roots although nodule development was remarkably delayed compared with WT nodules (Fig. 1A, C, and D). On the other hand, IT progression into the cortex was not observed in bumps formed on 1699-1 roots at this time point (Fig. 1A and E). Thus, both mutant lines were defective in IT progression into the cortex and nodule development.
The mutant lines were mapped near 53 cM chromosome 1, where no known symbiotic gene is located. We, therefore, conducted the allelism test. F1 plants did not form mature nodules, but instead developed many bumps or ineffective nodules in wider range of roots. This result indicates that the two mutant loci are allelic.
3.2. Root hair morphology and IT development in two mutant lines
Mutants that can form bumps but do not produce mature nodules have been reported many times. They have usually shown defects in the infection processes including IT development. Therefore, we evaluated the infection events visualized by DsRed-labeled M. loti. In the WT (MG-20), long ITs were observed in root hairs at 10 DAI (Fig. 2A). The WT mean number was 44.4, and shorter types were only 1.3% of total ITs (Fig. 2C). Frequencies of IT formation in F29 and 1699-1 roots were remarkably lower than WT roots (Fig. 2C). Statistical analysis showed that the numbers of ITs on F29 and 1699-1 roots were 8% and 0.8% of WT roots, respectively. Over 10% of ITs formed in F29 roots were of the shorter type (Fig. 2B and C), indicating that IT development in root hairs was inhibited in mutant lines. These results suggested that the mutated gene is essential for IT development in root hairs and in cortical cells.
Figure 2.
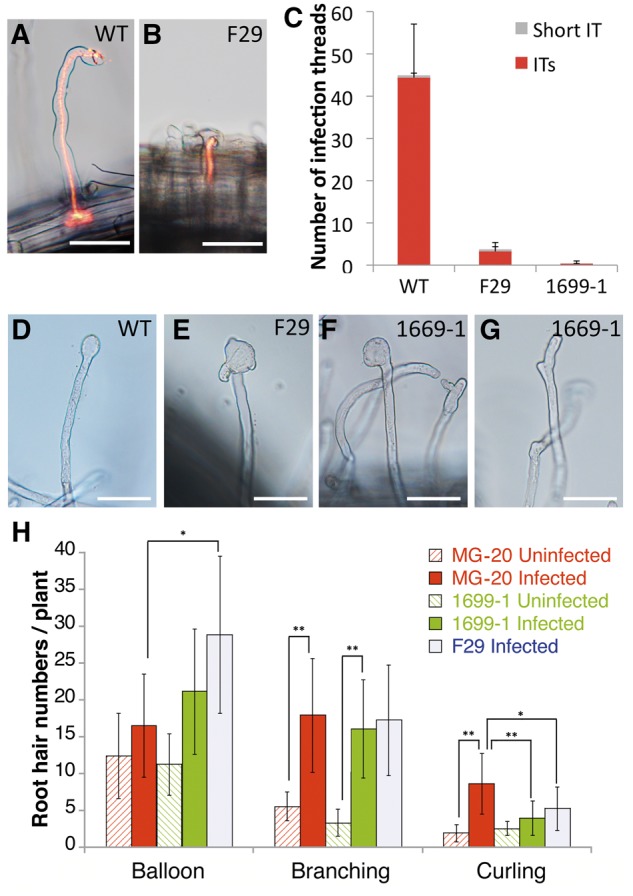
Root hair phenotypes of F29 and 1699-1 mutant lines. (A and B) WT (A) and F29 (B) root hairs infected by DsRed-labeled M. loti (10 DAI). ITs were visualized by DsRed fluorescence. Bars = 50 µm. (C) Means and SDs of IT numbers at 7 DAI (n = 16) are shown. ITs and short ITs represented in (A) and (B), respectively, were counted. (D–G) Root hair morphologies of WT (D), F29 (E), and 1699-1 (F and G) roots that were inoculated with M. loti for 4 days. Typical balloon-shaped root hairs found in WT (D) and mutant lines (E and F) and typical branched root hairs (G) are shown. Bars = 50 µm. (H) Statistic analysis of root hair structures. Means and SDs of root hair numbers were shown. Over 8 plants were observed (4 DAI). One-way ANOVA followed by a Tukey Honestly Significant Difference (HSD) test of the values was performed (*P < 0.05; **P < 0.01).
Furthermore, we investigated root hair responses at earlier stages after inoculation because fewer ITs in F29 and 1699-1 roots might be due to abnormal symbiotic root hair responses. In WT roots, curled, branched, and balloon-shaped root hairs comprised major morphological alterations at 4 DAI. Although the mean numbers of branched root hairs were not significantly different between WT and mutant lines, curled root hairs were decreased in mutant roots compared with those of WT roots (Fig. 2A, G, and H). In contrast, the mean numbers of balloon-shaped root hairs were increased in mutant lines (Fig. 2H). These misshapen root hairs expanded to an extreme compared with balloon-shaped root hairs observed in WT roots (Fig. 2D–F), and often generated new tips from lateral sides of the expanded tip (Fig. 2E). Depolarization and repolarization of root hair tips after rhizobial infection may be disorganized in mutant lines. These results indicated that the causative gene of F29 and 1699-1 lines is involved in correct symbiotic root hair responses in addition to IT development.
3.3. Complementation by an ERF/AP2 transcription factor
To generate a mapping population, the F29 mutants (Miyakojima MG-20 background) were crossed with L. japonicus Gifu B-129. Fine mapping using the F2 plants allowed us to encompass the causal gene between two makers, TM0371 and CM0371C1, on LjT46E19 of chromosome 1 (http://www.kazusa.or.jp; Fig. 3A). Eleven open reading frames were predicted in this region, and sequence comparison of WT and the F29 mutant revealed a missense mutation from Gly to Glu in the AP2 domain of an AP2/ERF transcription factor with a sequence similar to that of M. truncatula ERN1 (Fig. 3B). The sequence of the AP2/ERF transcription factor of L. japonicus showed high similarity to the M. trancatula ERN1 and other homologs of legumes (Supplementary Table S1). On the other hand, approximately 10 kb of genomic region including this AP2/ERF gene was deleted in the 1699-1 mutant. We then performed a complementation test by hairy roots via A. rhizogenes. For six of eight F29 plants, an ineffective nodule phenotype was rescued by introducing the AP2/ERF gene carrying its 3,047 bp promoter and 1,109 bp terminator (Fig. 3C) but not by introducing a GUS gene (Fig. 3D). Based on the results, we concluded that L. japonicus ERN1 gene is responsible for F29 and 1699-1 mutants. Hereafter, we describe F29 and 1699-1 as Ljern1-5 and Ljern1-6, respectively.
Figure 3.
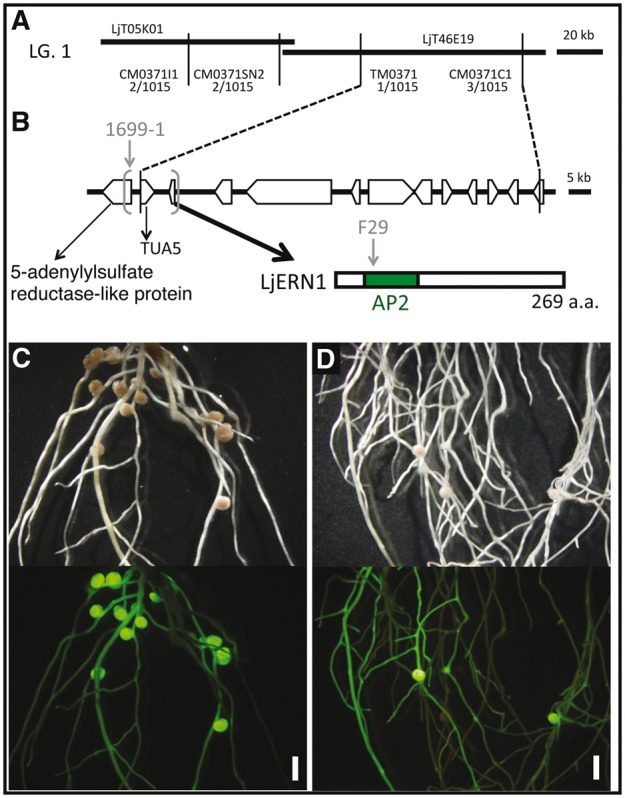
Positional cloning of F29 and 1699-1. (A) Gentic linkage map of the F29 causative locus on the linkage group 1. LjT05K01 and LjT46E19 are TAC clones. DNA markers and their positions are indicated with the numbers of recombinants (in parentheses). (B) ORFs in the F29 locus delimited by recombination events and LjERN1 protein structure predicted by the nucleotide sequence are shown. The mutation site on F29 and the large deletion found in 1699-1 was indicated by a grey arrowhead and grey parentheses. (C and D) Complementation test. F29 hairy roots transformed with an ERN1 construct (C) and with an GUS vector as a negative control (D) were inoculated with M. loti for 32 days. Upper and bottom panels are bright field images and fluorescent images from a GFP transformation marker, respectively. Bars = 2 mm.
3.4. CYCLOPS and NSP2-dependent gene activation of ERN1
We then investigated the ERN1 expression pattern in WT in response to M. loti inoculation. The ERN1 gene expressed constitutively in roots, even in the absence of M. loti, and was clearly induced at 12 h after inoculation (Fig. 4). The induction was approximately 3-fold, the transcription levels were maintained at early stages, without being elevated such as the NIN and CLE-RS genes.51 To further explore the spatial expression pattern of ERN1, we constructed a GUS reporter carrying the same promoter and terminator used for the complementation, and transformed to the hairy roots via A. rhizogenes. GUS expression from LjERN1 promoter was detectable for whole uninoculated roots (Fig. 5A). Interestingly, ProERN1::GUS::TerERN1 showed a spatial gradient that the signals were especially high in a developmentally younger region of the root, becoming gradually lower towards the base (Fig. 5A). The root region near the root tip is known to be a susceptible zone for rhizobial infection.52,53 In the root tip region shown in Figure 5C, strong GUS signals were detected in the developing root, root hairs, and a root cap, whereas no signal was detected in the root meristem that undergoes cell division. We then evaluated the expression of ProERN1::GUS::TerERN1 in inoculated roots at 7 DAI. The expression pattern near root tips at 7 DAI was not significantly changed when compared with uninoculated roots (Fig. 5B), whereas the GUS signal was observed on the curled root hair and was more distinct on the dividing cells in the outer cortex (Fig. 5D). These expression patterns are in good agreement with those of M. truncatula ERN1 and ERN2 genes.37LjERN1 expression in nodule primordia resembled that of MtERN1 rather than the other because MtERN2 expression is closely associated with IT progression. On the other hand, regions expressing LjERN1 were broadly expanded within primordia similar to MtERN1. In developing nodules, the GUS signal was restricted to the inside where the infection zone is formed, whereas it was not detected significantly in the peripheral regions of the inoculated root (Fig. 5E).
Figure 4.
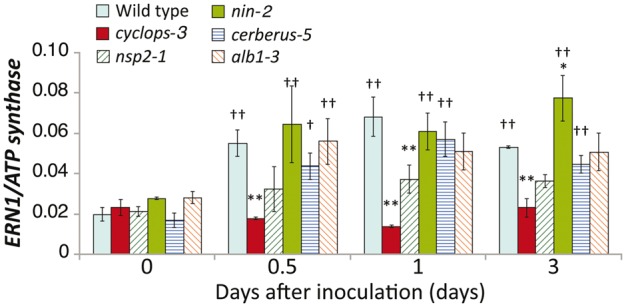
qRT-PCR analysis of ERN1 expression in WT and symbiotic mutants. Total RNA was extracted from roots at 0, 0.5, 1, and 3 days after inoculation with M. loti MAFF303099. ATP synthase was used as an internal standard. Means and SDs of three independent experiments are shown. One-way ANOVA followed by a Tukey HSD test of the values was performed. Significant differences from WT plants at each time points (*P < 0.05; **P < 0.01), and from mock controls (†P < 0.05; ††P < 0.01) are indicated.
Figure 5.
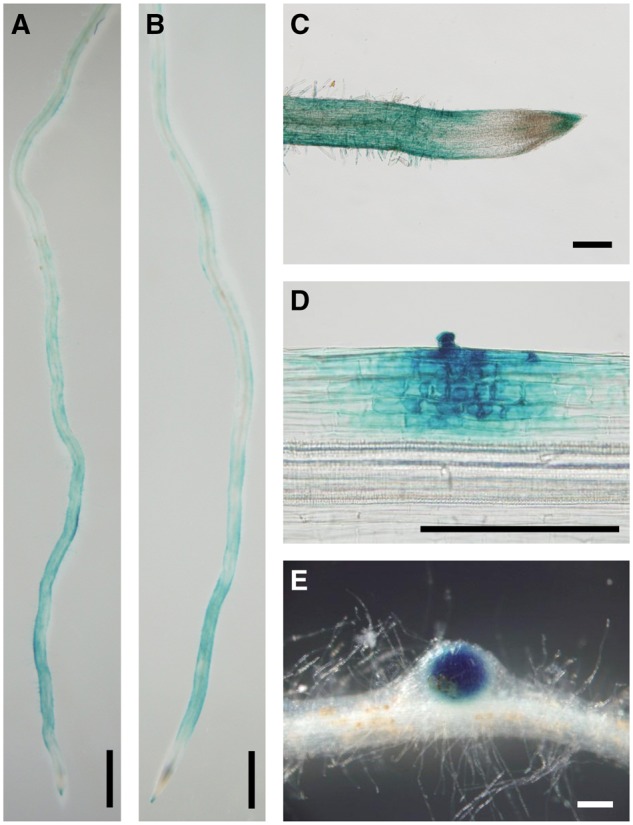
GUS expression from ERN1 promoter after inoculation with M. loti. hairy roots carrying ProERN1::GUS::TerERN1 were inoculated without (A and C) or with M. loti MAFF303099 (B, D, and E) for 7 days. Bars: 1.0 mm in (A) and (B), 0.2 mm in C–E.
To identify what components of the Nod factor signaling pathway activate ERN1 induction in response to rhizobia, we performed an expression analysis of ERN1 by qRT-PCR using five mutants defective in IT development positioned downstream of CCaMK. ERN1 was induced at early stages in nin-2, cerberus-5, and alb-3 mutants, up to the similar levels as the WT. The induction levels of ERN1 in nsp2-1 mutant were almost one-third or one-half that in the WT (Fig. 4). Notably, no induction was observed in the cyclops-3 mutant (Fig. 4). These results indicate a CYCLOPS and NSP2-dependent induction of ERN1 expression.
3.5. Syntenic relationship of the genome regions around ERN genes of L. japonicus, M. truncatula and G. max
ERN2, an ERF/AP2 transcription factor closely related to ERN1, exists wildly in legumes.54 In M. truncatula, it has been shown to be able to bind to the NF box, and activate ENOD11 transcription36 and to possess similar biological activities with ERN1.54 The double-mutant analyses indicated that ERN1/ERN2 play essential roles for initiating root hair infection and root nodule development.55 Thus, we investigated the ERN2 ortholog in L. japonicus. By searching the latest genome sequences of L. japonicus (Lj_ver3.0) using MtERN2 (Medtr6g029180.1) as query, a top BLASTP hit was obtained against LjERN1 (Lj1g3v3975310.1). The same result was obtained in the tBLASTn search against the transcriptome assembly of L. japonicus.
To confirm an orthologus relationship between LjERN1 (Lj1g3v3975310.1) and MtERN1 (Medtr7g085820.1), the syntenic relationships between the genome regions flanking to these genes were analysed. Using a stepwise gene-by-gene reciprocal comparison of the regions, we observed strong conserved microsynteny between these regions (Table 1). As these genome regions correspond to the large synteny block between chromosome 1 of L. japonicus and chromosome 7 of M. truncatula reported by Cannon et al.,56 the synteic relation extended to chromosomal level. Therefore, the orthologous relationship between LjERN1 and MtERN1 was confirmed from the viewpoint of genome synteny. The syntenic relationship surrounding the ERN1 gene was also observed in the corresponding genome regions in G. max (flanking regions of Glyma.16G040000.1 and Glyma.19G113200.1).
Table 1.
Syntenic relation in the genome regions of ERN genes of L. japonicus, M. truncatula, and G. max
| Annotation | ERN1 |
ERN2 |
||||||
|---|---|---|---|---|---|---|---|---|
| Lj_chr3 | Mt_chr7 | Gm_chr16 | Gm_chr19 | Lj | Mt_chr6 | Gm_chr2 | Gm_chr16 | |
| Transmembrane protein, putative | Lj1g3v3975270.2 | Medtr7g085790.1 | – | Glyma.19G112800.1 | – | Medtr6g029250.1 | Glyma.02G072500.1 | Glyma.16G153700.1 |
| 5-Adenylylsulphate reductase-like protein | Lj1g3v3975280.1 | – | Glyma.16G040200.1 | Glyma.19G112900.1 | – | Medtr6g029240.1 | Glyma.02G072600.1 | |
| Tubulin/FtsZ family protein | Lj1g3v3975290.1 | Medtr7g085800.1 | Glyma.16G040100.1 | Glyma.19G113000.1 | – | Medtr6g029190.1 | – | Glyma.16G154000.1 |
| AP2/ERF domain transcription factor | Lj1g3v3975310.1 (LjERN1) | Medtr7g085810.1 (MtERN1) | Glyma.16G040000.1 | Glyma.19G113100.1 | – | Medtr6g029180.1 (MtERN2) | Glyma.02G072800.1 | Glyma.16G154100.1 |
| Importin-like protein | Lj1g3v3975340.2 | Medtr7g085820.1 | Glyma.16G039900.1 | Glyma.19G113200.1 | – | – | – | – |
| glycosyltransferase family protein | Lj1g3v3975450.1 | Medtr7g085840.1 | Glyma.16G039800.1 | Glyma.19G113300.1 | – | – | – | – |
| Hypothetical protein | Lj1g3v3975460.1 | Medtr7g085850.1 | – | Glyma.19G113400.1 | – | – | – | – |
Regarding the genome region of the ERN2 gene, genes orthologous to MtERN2 were identified in the G. max genome (Glyma.02G072800.1 and Glyma.16G154100.1), and a syntenic relationship between ERN1 and ERN2 genome regions was observed (Table 1). Since the candidate gene orthologous to any of these flanking genes was not identified in L. japonicus genome sequences, the genome region corresponding to the ERN2 gene would probably be deleted in the genome of L. japonicus.
3.6. ERN1 and ERN2 clades resulting from gene duplication in legume evolution
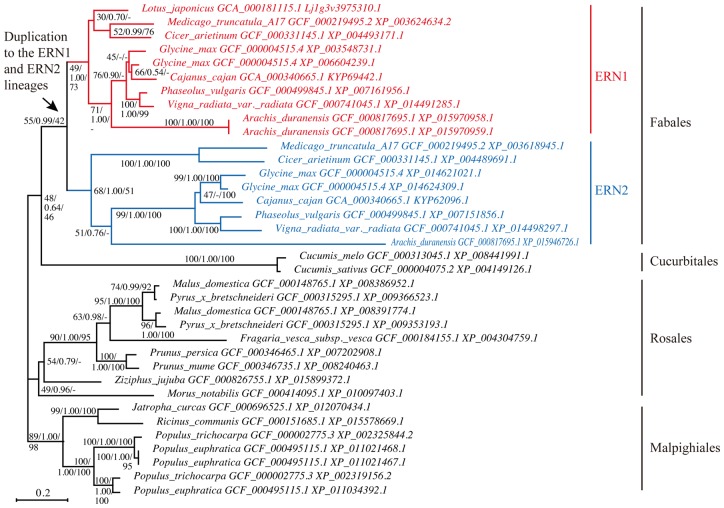
We estimated the phylogenetic tree of ERN genes using all genomic datasets in NCBI RefSeq (Fig. 6). ML, Bayesian inference, and NJ methods estimated essentially similar tree topologies (Fig. 6 and Supplementary Figures S1 and S2). The estimated trees strongly suggested that a gene duplication event produced the ERN1 and ERN2 clades after the divergence of legume plants in Fabales from the others (rosid I)57. It should be noted that LjERN1 of L. japonicus is included in the ERN1 clade. All the genome-analysed legume species harbored both the ERN1 and ERN2 genes except for L. japonicus.
Figure 6.
Phylogenetic tree from the homologs of ERN1 and ERN2 genes inferred by ML method with LG model using RAxML program. The arrow indicates the duplication to the ERN1 and ERN2 lineages. ERN1 and ERN2 genes of Fabaceae are indicated with red and blue letters, respectively. The first and third numbers given at the relevant nodes correspond to the bootstrap support from the ML and NJ methods, respectively, and the second to the posterior probability from the Bayesian method. Phylogenetic trees inferred by the Bayesian and NJ methods are available in supplementary figures. The assembly accession number and accession version of the genes is following to the species name. The scale bar represents substitutions per site.
3.7. Difference of functional constraint between ERN1 and ERN2 genes
The phylogenetic tree shape of the ERN genes suggests divergence of evolutionary rates between the ERN1 and ERN2 genes (Fig. 6); the branches in the ERN2 clade are longer (i.e. more substitutions per site) than those in the ERN1 clade. In general, variations in substitution rates can be due to different nonsynonymous/synonymous rate ratios (ω = dn/ds).27,58–60 The branch model of PAML was adopted to consider different ω in the tree and five following models were compared: (i) one ratio across the tree; (ii) two different ratios between the ERN1 clade and the other parts; (iii) two different ratios between the ERN2 clade and the other parts; (iv) two different ratios between the ERN1/ERN2 clade and the other parts; and (v) three different ratios between the ERN1, ERN2 clade, and the other parts (Table 2).47,61 The LRT was conducted to compare nested models and showed no significant likelihood differences between the models 1 and 2 and between 3 and 5. On the other hand, the tests rejected the null hypotheses for all other combinations (Table 2) and Akaike's and Bayesian information criteria favored the model 3. These results suggested that the ω of the genes in the ERN2 clade is greater than the background ratio and that ω of the genes in the ERN1 clade is almost identical to that of the genes of nonlegumes. The clade model of PAML also showed similar results (Supplementary Table S2). No positively selected site was not detected in the ERN2 genes with branch-site and clade models (Supplementary Table S2).
Table 2.
Model comparison of ERN genes estimate by PAML using branch model
| Model | (i) One-ratio model (M0 of Yang et al.47) | Two ratio model |
(v) Three ratio model | ||
|---|---|---|---|---|---|
| (ii) ERN1 | (iii) ERN2 | (iv) ERN1 + ERN2 | ERN1, ERN2 | ||
| np | 72 | 73 | 73 | 73 | 74 |
| ln L | −15088.32041 | −15087.59693 | −15063.74053 | −15076.07402 | −15063.08349 |
| kappa (ts/tv) | 1.58345 | 1.5825 | 1.58201 | 1.58408 | 1.58288 |
| omega (dN/dS) | 0.28104 | ω0 = 0.28866, ω1 = 0.25656 | ω0 = 0.23609, ω1 = 0.45111 | ω0 = 0.22744, ω1 = 0.34265 | ω0 = 0.22749, ω1 = 0.25621, ω2 = 0.45081 |
| LRT with 2ΔL and P value | |||||
| Two-ratio model (ERN1) | −1.447 (P = 0.229) | ||||
| Two-ratio model (ERN2) | −49.16 (P < 10−11) | ||||
| Two ratio model (ERN1+ERN2) | −24.493 (P < 10−6) | ||||
| Three ratio model (ERN1, ERN2) | −50.474 (P < 10−10) | −49.027 (P < 10−10) | −1.314 (P = 0.518) | −25.981 (P < 10−5) | |
| AIC | 30320.641 | 30321.194 | 30273.481 | 30298.148 | 30274.167 |
| BIC | 30618.089 | 30622.773 | 30575.060 | 30599.728 | 30579.878 |
NOTE.—np is the number of parameters. ln L is the log-likelihood. 2ΔL is twice the log-likelihood difference between the two models. Kappa indicates the transitions/transversions ratio. Omega shows the nonsynonymous/synonymous rate ratio. The Akaike’s information criterion (AIC) and Bayesian information criterion (BIC) were estimated according to Aoki et al.27. The best model is underlined. LRT allows one to compare nested models only and is defined as minus two times the logarithm of the ratio of the likelihoods.
4. Discussion
Phenotypic analyses of L. japonicus ern1 mutants showed that this gene is essential for symbiotic root hair responses and IT development. Although Ljern1 mutants produced nodule primordia equivalently to the WT, its development was restricted (Fig. 1). Nodule primordia formed in Ljern1 mutants did not develop ITs into cortical cells (Fig. 1). Thus, the defect in IT development is a major Ljern1 phenotype. This symbiotic phenotype of Ljern1 mutants appeared to be intermediate between the M. truncatula ern1 single and ern1/erm2 double mutants. Mtern1 produces a limited number of ITs from infection foci on root hairs. These ITs are arrested within root hairs and do not progress into nodule primordia.35 Similar to the M. truncatula counterpart, IT formation was remarkably downregulated in Ljern1 mutants (Fig. 2). Regarding the defect in IT development and production of nodule primordia, the Ljern1 phenotype resembles that of the Mtern1 single mutant rather than the ern1/ern2 double mutant, which exhibits a more severe phenotype than respective single mutants. The double mutant does not produce ITs and nodule primordia,55 suggesting the implication of ERN transcription factors in nodule organogenesis. Alternatively, cortical cells may not be stimulated due to loss of infection events (infection pockets and ITs) in the root epidermis of the double mutant. Host loss-of-function symbiotic mutants able to produce nodule primordia without any infection events in the epidermis have not been identified so far.
The loss-of-infection events in the double mutant are partially explained by aberrant symbiotic root hair responses. Unlike Mtern1, root hairs of the double mutants are unable to entrap rhizobia because of a defect in tip curling.55 Root hair tips of the double mutants are abnormally expanded, suggesting that its polarity is deregulated. Similar abnormality was observed in Ljern1 single mutants after inoculation with rhizobia (Fig. 2). The number of curled root hairs was decreased, and balloon-shaped root hairs were increased in the mutant, indicating that LjERN1 is involved in symbiotic root hair responses besides IT development. Thus, the root hair phenotype of Ljern1 includes the feature characteristic to ern1/ern2 double mutants in M. truncatula, which is compatible with the absence of ERN2 in L. japonicus. However, the Ljern1 phenotype is weaker than that of the M. truncatula double mutants. Other factor may partially compensate the loss of ERN function during early symbiotic root hair responses in L. japonicus.
GUS reporter analysis reveals gradients of LjERN1 expression across uninfected primary roots, namely high expression levels within root tip regions of the roots, whereas levels are lower towards the base. As the root tip region is susceptible to rhizobial infection,52 this expression gradient is believed to represent an infection potential of rhizobia; in other words, the distribution of infectable root cells. When M. loti was inoculated, the expression of ProERN1::GUS::TerERN1 was observed only in root hair cells undergoing rhizobial infection (via IT development) and in cortical cells initiating cell division (Fig. 5). On the other hand, ERN1 expression was suppressed in the peripheral epidermal regions where few infection events occurred. These findings suggest that ERN1 expression patterns are in good agreement with rhizobial infection potential and infection events. Strong expression of ERN1 is also distinct in dividing cortical cells. Within this interpretation, two possibilities are conceivable: (i) ERN1 is directly involved in cortical cell division; and (ii) ERN1 expression confers the ability to accept ITs to dividing cortical cells. In the former case, ERN1 is unlikely to be involved in universal cell division because no expression was found in root meristem where cell division is always active (Fig. 5). In the latter case, ERN1 expression may be involved in the navigation of developing ITs to cortical cells. Further analysis will be required to clarify what kinds of functions ERN1 has in dividing cortical cells.
Quantitative expression analysis using symbiotic mutants shows that the early induction of ERN1 by M. loti inoculation is required for CYCLOPS and partially for NSP2. Recently, Cerri et al.55 clearly showed that NSP2 in the presence of NSP1 can act as a strong activator of ERN1 transcription in transient expression assays in Nicotiana benthamiana, and our data are consistent with their results regarding NSP2. On the other hand, ERN1 induction was clearly canceled in the cyclops mutant background. Although CYCLOPS was originally identified as a component of the common symbiosis pathway that directly interacts with CCaMK, it has been demonstrated that it acts as a transcription activator of the NIN gene via direct binding of CYCLOPS-box in its promoter region. Since NIN is required for early infection events via IT development and its target NF-YA1 directly binds with MtERN1 promoter,62,63 it is possible that ERN1 acts downstream of NIN to regulate IT progression. However, our results show that ERN1 is normally induced in response to the inoculation of M. loti in the nin mutant background similar to the WT, indicating that ERN1 activation is not controlled by NIN. NIN is likely to act parallel with ERN1 under the control of CYLOPS or may act downstream of ERN1. The balloon-shaped root hair phenotype characteristic to Ljern1 mutants has not been reported in nin mutants, which display excess curled and branched root hairs in response to rhizobial infection. The frequency of branched root hairs in Ljern1 mutants was equivalent to that of the WT. The different root hair morphology may suggest that ERN1 and NIN play distinct roles to positively regulate root hair responses and IT progression. In any case, the elucidation of transcription networks involving CYCLOPS, ERN1, and NIN is a future challenge.
The phylogenetic analyses showed that ERN1 and ERN2 genes were originated from the gene duplication after the divergence of Fabales. The duplication event and subsequent molecular evolution of the genes might associate with the evolution of nodulation symbiosis of legume plants although we do not know the full details of the evolutionary history of ERN genes because of the lack of information for plants other than symbiotic legumes of Faboideae (Fabaceae) in Fabales (e.g. Caesalpinioideae, Mimosoideae, Quillajaceae, Polygalaceae, and Surianaceae). The comparative genomic analysis of the synteny of ERN genes showed that ERN1 of legumes had a similar gene order to the ERN genes of nonlegume plants, rather than that of ERN2 did. In contrast to the previous suggestion that ERN1 was derived from ERN2 via duplication,54,55,64 the synteny analysis proposed that ERN1 might have an orthologous role to ERN genes of plants of the outgroup. Molecular evolutionary analysis of ω ratio supported this result. The LRT failed to reject the alternative hypothesis of different ω ratio between ERN1 and all other genes in the phylogeny of Figure 6 against the null hypothesis of the same ω ratios of all genes (Table 2). The test also showed that the ω ratio of ERN2 was significantly greater than the background ratio of ERN1 and other ERN genes. It has been believed that variable nonsynonymous/synonymous rate ratios among lineages may associate with the difference of functional constraint along certain lineages.65,66 According to this proposition, the functional constraint appears to be conserved between legume ERN1 and nonlegume ERN genes, whereas the genes in the ERN2 lineage may have suffered relaxed selective constraints. It is possible that ERN1 took over the main molecular function of original ERN genes and that ERN2 has subfunctionalized and/or neofunctionalized the ERN function, and is a helpful adjunct to the ERN1 gene during nodulation. A recent article appears to support this proposition.55 Although the ern1 mutants of M. truncatula were unable to nodulate, ern2 mutants can nodulate but with signs of premature senescence. Although the ern1 mutant still exhibited early symbiotic responses including rhizobial infection, ern1/ern2 double mutants were completely unable to initiate infection and nodule development. ERN2 showed partially overlapping expression patterns with ERN1.55
Our genomic analysis suggested the possibility of a lack of the ERN2 gene in the genome of L.japonicus. LjERN1 might have a sufficient role in IT development of L. japonicus. Alternatively, the function and expression of the LjERN1 may be complement in those of ERN2 genes of other legumes. This notion is consistent with the fact that Ljern1 single mutants display balloon-shaped root hairs that are observed in only ern1/ern2 double mutants in M. truncatula.
Supplementary Material
Acknowledgements
This work was supported by a Research Fellow of the Japan Society for the Promotion of Science (JSPS) (to K.Y.; 22·3459), Grant-in-Aid for Challenging Exploratory Research from JSPS (Grant Number 26660057), the Canon foundation (to W.I.), The Ministry of Education, Culture, Sports, Science and Technology KAKENHI Grant Number 16H06279 (to W.I.), and Grant for Basic Science Research Projects 150574 from the Sumitomo Foundation (to T.S.).
Conflict of interest
None declared.
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
References
- 1. Fournier J., Timmers A. C., Sieberer B. J., Jauneau A., Chabaud M., Barker D. G.. 2008, Mechanism of infection thread elongation in root hairs of Medicago truncatula and dynamic interplay with associated rhizobial colonization, Plant Physiol., 148, 1985–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Svistoonoff S., Hocher V., Gherbi H.. 2014, Actinorhizal root nodule symbioses: what is signalling telling on the origins of nodulation?, Curr. Opin. Plant Biol., 20, 11–18. [DOI] [PubMed] [Google Scholar]
- 3. Downie J. A., Walker S. A.. 1999, Plant responses to nodulation factors, Curr. Opin. Plant Biol., 2, 483–9. [DOI] [PubMed] [Google Scholar]
- 4. Ehrhardt D. W., Wais R., Long S. R.. 1996, Calcium spiking in plant root hairs responding to Rhizobium nodulation signals, Cell, 85, 673–81. [DOI] [PubMed] [Google Scholar]
- 5. Amor B. B., Shaw S. L., Oldroyd G. E., et al. 2003, The NFP locus of Medicago truncatula controls an early step of Nod factor signal transduction upstream of a rapid calcium flux and root hair deformation, Plant J., 34, 495–506. [DOI] [PubMed] [Google Scholar]
- 6. Esseling J. J., Lhuissier F. G., Emons A. M.. 2003, Nod factor-induced root hair curling: Continuous polar growth towards the point of nod factor application, Plant Physiol., 132, 1982–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Madsen E. B., Madsen L. H., Radutoiu S., et al. 2003, A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals, Nature, 425, 637–40. [DOI] [PubMed] [Google Scholar]
- 8. Radutoiu S., Madsen L. H., Madsen E. B., et al. (2003). Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases, Nature, 425, 585–92. [DOI] [PubMed] [Google Scholar]
- 9. Smit P., Limpens E., Geurts R., et al. 2007, Medicago LYK3, an entry receptor in rhizobial nodulation factor signaling, Plant Physiol., 145, 183–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Broghammer A., Krusell L., Blaise M., et al. 2012, Legume receptors perceive the rhizobial lipochitin oligosaccharide signal molecules by direct binding, Proc. Natl. Acad. Sci. USA, 109, 13859–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kistner C., Parniske M.. 2002, Evolution of signal transduction in intracellular symbiosis, Trends Plant Sci., 7, 511–8. [DOI] [PubMed] [Google Scholar]
- 12. Gleason C., Chaudhuri S., Yang T., Muñoz A., Poovaiah B. W., Oldroyd G. E.. 2006, Nodulation independent of rhizobia induced by a calcium-activated kinase lacking autoinhibition, Nature, 441, 1149–52. [DOI] [PubMed] [Google Scholar]
- 13. Tirichine L., Imaizumi-Anraku H., Yoshida S., et al. 2006, Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development, Nature, 441, 1153–6. [DOI] [PubMed] [Google Scholar]
- 14. Hayashi T., Banba M., Shimoda Y., Kouchi H., Hayashi M., Imaizumi-Anraku H.. 2010, A dominant function of CCaMK in intracellular accommodation of bacterial and fungal endosymbionts, Plant J., 63, 141–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Madsen L.H., Tirichine L., Jurkiewicz A., et al. 2010, The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus, Nat. Commun., 1, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hocher V., Alloisio N., Auguy F., et al. 2011, Transcriptomics of actinorhizal symbioses reveals homologs of the whole common symbiotic signaling cascade, Plant Physiol., 156, 700–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Demina I. V., Persson T., Santos P., Plaszczyca M., Pawlowski K.. 2013, Comparison of the nodule vs. root transcriptome of the actinorhizal plant Datisca glomerata: actinorhizal nodules contain a specific class of defensins, PLoS One, 8, e72442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Diédhiou I., Tromas A., Cissoko M.. et al. 2014, Identification of potential transcriptional regulators of actinorhizal symbioses in Casuarina glauca and Alnus glutinosa, BMC Plant Biol., 14, 342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gherbi H., Markmann K., Svistoonoff S., et al. 2008, SymRK defines a common genetic basis for plant root endosymbioses with arbuscular mycorrhiza fungi, rhizobia, and Frankia bacteria, Proc. Natl. Acad. Sci. USA, 105, 4928–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Markmann K., Giczey G., Parniske M.. 2008, Functional adaptation of a plant receptor-kinase paved the way for the evolution of intracellular root symbioses with bacteria, PLoS Biol., 6, e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Svistoonoff S., Benabdoun F. M., Nambiar-Veetil M., et al. 2013, The independent acquisition of plant root nitrogen-fixing symbiosis in Fabids recruited the same genetic pathway for nodule organogenesis, PLoS One, 8, e64515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schauser L., Roussis A., Stiller J., Stougaard J.. 1999, A plant regulator controlling development of symbiotic root nodules, Nature, 402, 191–5. [DOI] [PubMed] [Google Scholar]
- 23. Marsh J. F., Rakocevic A., Mitra R. M., et al. 2007, Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase, Plant Physiol., 144, 324–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Clavijo F., Diedhiou I., Vaissayre V., et al. 2015, The Casuarina NIN gene is transcriptionally activated throughout Frankia root infection as well as in response to bacterial diffusible signals, New Phytol., 208, 887–903. [DOI] [PubMed] [Google Scholar]
- 25. Limpens E., van Zeijl A., Geurts R.. 2015, Lipochitoologosaccharides modulate plant host immunity to enable endosymbiosis, Annu. Rev. Phytopathol., 53, 311–4. [DOI] [PubMed] [Google Scholar]
- 26. De Mita S., Streng A., Bisseling T., Geurts R.. 2014, Evolution of a symbiotic receptor through gene duplications in the legume-rhizobium mutualism, New Phytol., 201, 961–72. [DOI] [PubMed] [Google Scholar]
- 27. Aoki S., Ito M., Iwasaki W.. 2013, From β- to α-Proteobacteria: The origin and evolution of rhizobial nodulation genes nodIJ, Mol. Biol. Evol., 30, 2494–508. [DOI] [PubMed] [Google Scholar]
- 28. Cannon S. B., McKain M. R., Harkess, et al. 2015, Multiple polyploidy events in the early radiation of nodulating and nonnodulating legumes, Mol. Biol. Evol., 32, 193–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li Q.-G., Zhang L., Li C., Dunwell J. M., Zhang Y.-M.. 2013, Comparative genomics suggests that an ancestral polyploidy event leads to enhanced root nodule symbiosis in the Papilionoideae, Mol. Biol. Evol., 30, 2602–11. [DOI] [PubMed] [Google Scholar]
- 30. Yano K., Yoshida S., Müeller J., et al. 2008, CYCLOPS, a mediator of symbiotic intracellular accommodation, Proc. Natl. Acad. Sci. USA, 105, 20540–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yano K., Shibata S., Chen W. L., et al. 2009, CERBERUS, a novel U-box protein containing WD-40 repeats, is required for formation of the infection thread and nodule development in the legume-Rhizobium symbiosis, Plant J., 60, 168–80. [DOI] [PubMed] [Google Scholar]
- 32. Singh S., Katzer K., Lambert J., Cerri M., Parniske M.. 2014, CYCLOPS, a DNA-binding transcriptional activator, orchestrates symbiotic root nodule development, Cell Host Microbe, 15, 139–52. [DOI] [PubMed] [Google Scholar]
- 33. Messinese E., Mun J. H., Yeun L. H., et al. 2007, A novel nuclear protein interacts with the symbiotic DMI3 calcium- and calmodulin-dependent protein kinase of Medicago truncatula, Mol. Plant Microbe. Interact., 20, 912–21. [DOI] [PubMed] [Google Scholar]
- 34. Kiss E., Oláh B., Kaló P., et al. 2009, LIN, a novel type of U-box/WD40 protein, controls early infection by rhizobia in legumes, Plant Physiol., 151, 1239–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Middleton P. H., Jakab J., Penmetsa R. V., et al. 2007, An ERF transcription factor in Medicago truncatula that is essential for nod factor signal transduction, Plant Cell, 19, 1221–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Andriankaja A., Boisson-Dernier A., Frances L., et al. 2007, AP2-ERF transcription factors mediate Nod factor dependent MtENOD11 activation in root hairs via a novel cis-regulatory motif, Plant Cell, 19, 2866–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cerri M. R., Frances L., Laloum T., et al. 2012, Medicago truncatula ERN transcription factors: Regulatory interplay with NSP1/NSP2 GRAS factors and expression dynamics throughout rhizobial infection, Plant Physiol., 160, 2155–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Magori S., Oka-Kira E., Shibata S., et al. 2009, TOO MUCH LOVE, a root regulator associated with the long-distance control of nodulation in Lotus japonicus, Mol. Plant Microbe. Interact., 22, 259–68. [DOI] [PubMed] [Google Scholar]
- 39. Kawaguchi M., Imaizumi-Anraku H., Koiwa H., et al. 2002, Root, root hair, and symbiotic mutants of the model legume Lotus japonicus, Mol. Plant-Microbe Interact., 15, 17–26. [DOI] [PubMed] [Google Scholar]
- 40. Broughton W. J., Dilworth M. J.. 1971, Control of leghaemoglobin synthesis in snake beans, Biochem J., 125, 1075–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Maekawa T., Maekawa-Yoshikawa M., Takeda N., Imaizumi-Anraku H., Murooka Y., Hayashi M.. 2009, Gibberellin controls the nodulation signaling pathway in Lotus japonicus, Plant J., 58, 183–94. [DOI] [PubMed] [Google Scholar]
- 42. Diaz C., Gronlund M., Schlaman H. R. M., Spaink H.P.. 2005, Induction of hairy roots for symbiotic gene expression studies In: Marquez A.J. (ed), Lotus japonicus Handbook. Springer: Dordrecht, pp. 261–77. [Google Scholar]
- 43. Katoh K., Kuma K., Toh H., Miyata T.. 2005, MAFFT version 5: improvement in accuracy of multiple sequence alignment, Nucleic Acids Res., 33, 511–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stamatakis A., Ludwig T., Meier H.. 2005, RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees, Bioinformatics, 21, 456–63. [DOI] [PubMed] [Google Scholar]
- 45. Huelsenbeck J. P., Ronquist F.. 2001, MRBAYES: Bayesian inference of phylogeny, Bioinformatics, 17, 754–755. [DOI] [PubMed] [Google Scholar]
- 46. Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S.. 2011, MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods, Mol. Biol. Evol., 28, 2731–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yang Z. 2007, PAML 4: Phylogenetic analysis by maximum likelihood, Mol. Biol. Evol ., 23, 7–9. [DOI] [PubMed] [Google Scholar]
- 48. Imaizumi-Anraku H., Kouchi H., Syono K., Akao S., Kawaguchi M.. 2000, Analysis of ENOD40 expression in alb1, a symbiotic mutant of Lotus japonicus that forms empty nodules with incompletely developed nodule vascular bundles, Mol. Gen. Genet., 264, 402–10. [DOI] [PubMed] [Google Scholar]
- 49. Tansengco M. L., Hayashi M., Kawaguchi M., Imaizumi-Anraku H., Murooka Y.. 2003, crinkle, a novel symbiotic mutant that affects the infection thread growth and alters the root hair, trichome, and seed development in Lotus japonicus, Plant Physiol., 131, 1054–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kawaharada Y., Kelly S., Nielsen M. W., et al. 2015, Receptor-mediated exopolysaccharide perception controls bacterial infection, Nature, 523, 308–12. [DOI] [PubMed] [Google Scholar]
- 51. Okamoto S., Ohnishi E., Sato S., et al. 2009, Nod factor/nitrate-induced CLE genes that drive HAR1-mediated systemic regulation of nodulation, Plant Cell Physiol., 50, 67–77. [DOI] [PubMed] [Google Scholar]
- 52. Bhuvaneswari T. V., Turgeon B. G., Bauer W. D.. 1980, Early Events in the Infection of Soybean (Glycine max L. Merr) by Rhizobium japonicum: I. LOCALIZATION OF INFECTIBLE ROOT CELLS, Plant Physiol., 66, 1027–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Murakami Y., Yokoyama H., Fukui R., Kawaguchi M.. 2013, Down-regulation of NSP2 expression in developmentally young regions of Lotus japonicus roots in response to rhizobial inoculation, Plant Cell Physiol., 54, 518–27. [DOI] [PubMed] [Google Scholar]
- 54. Cerri M. R., Gamas P., de Carvalho-Niebel F.. 2015, AP2/ERF transcription factors and root nodulation In: de Bruijn F.J. (ed), Biological Nitrogen Fixation, vol. 2, Wiley-Blackwell: Hoboken, pp. 609–22. [Google Scholar]
- 55. Cerri M. R., Frances L., Kelner A., et al. 2016, The symbiosis-related ERN transcription factors act in concert to coordinate rhizobial host root infection, Plant Physiol., 171, 1037–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cannon S. B., Sterck L., Rombauts S., et al. 2006, Legume genome evolution viewed through the Medicago truncatula and Lotus japonicus genomes, Proc. Natl. Acad. Sci. USA, 103, 14959–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. The Angiosperm Phylogeny Group 2016. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV, Bot. J. Linn. Soc. 181, 1095–8339. [Google Scholar]
- 58. Arbiza L., Dopazo J., Dopazo H.. 2006, Positive selection, relaxation, and acceleration in the evolution of the human and chimp genome, PLoS Comput. Biol., 2, e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhong B., Yonezawa T., Zhong Y., Hasegawa M.. 2009, Episodic evolution and adaptation of chloroplast genomes in ancestral grasses, PLoS One, 4, e529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Neiman M., Hehman G., Miller J. T., Logsdon J. M., Taylor D. R.. 2010, Accelerated mutation accumulation in asexual lineages of a fresh-water snail, Mol. Biol. Evol., 27, 954–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yang Z. 1998, Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution, Mol. Biol. Evol., 15, 568–73. [DOI] [PubMed] [Google Scholar]
- 62. Soyano T., Kouchi H., Hirota A., Hayashi M.. 2013, NODULE INCEPTION directly targets NF-Y subunit genes to regulate essential processes of root nodule development in Lotus japonicus, PLoS Genet., 9, e1003352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Laloum T., Baudin M., Frances L., et al. 2014, Two CCAAT-box-binding transcription factors redundantly regulate early steps of the legume-rhizobia endosymbiosis, Plant J., 79, 757–68. [DOI] [PubMed] [Google Scholar]
- 64. Young N. D., Debellé F., Oldroyd G. E., et al. 2011, The Medicago genome provides insight into the evolution of rhizobial symbioses, Nature, 480, 520–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Crandall K. A., Hillis D. M.. 1997, Rhodopsin evolution in the dark, Nature, 387, 667–8. [DOI] [PubMed] [Google Scholar]
- 66. Yang Z., Nielsen R.. 2000, Estimating synonymous and non-synonymous substitution rates under realistic evolutionary models, Mol. Biol. Evol., 17, 32–43. [DOI] [PubMed] [Google Scholar]
- 67. Nakano T., Suzuki K., Fujimura T., Shinshi H.. 2006, Genome-wide analysis of the ERF gene family in Arabidopsis and rice, Plant Physiol., 140, 411–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.