Abstract
Synthesis of progeny DNA genomes in cells infected by human subgroup C adenoviruses leads to several changes in viral gene expression. These changes include transcription from previously silent, late promoters, such as the IVa2 promoter, and a large increase in the efficiency of major-late (ML) transcription. Some of these changes appear to take place sequentially, because the product of the IVa2 gene has been implicated in stimulation of ML transcription. Our previous biochemical studies suggested that IVa2 transcription is regulated by viral DNA synthesis-dependent relief of transcriptional repression by a cellular protein that we termed IVa2-RF. To test the relevance of such a repressor-titration mechanism during the viral infectious cycle, we introduced into the endogenous IVa2 promoter two mutations that impair in vitro-binding of IVa2-RF, but introduce no change (Rep7) or one conservative amino acid substitution (Rep6) into the overlapping coding sequence for the viral DNA polymerase. The results of run-on transcription assays indicated that both mutations induced earlier-than-normal and more efficient IVa2 transcription. Both mutations were also observed to result in modest increases in the efficiency of viral DNA synthesis. However, measurement of the concentration of IVa2 transcripts as a function of IVa2 template concentration demonstrated that the Rep mutations increased by up to 60-fold the efficiency with which IVa2 templates were used during the initial period of the late phase of infection, as predicted by the repressor titration hypothesis. These mutations also increased the efficiency of ML transcription in infected cells.
Keywords: late transcription, repressor titration
The infectious cycles of animal viruses with DNA genomes are characterized by transcription of viral genes in a stereotyped temporal sequence, regardless of whether transcription is carried out by cellular or viral DNA-dependent RNA polymerases. The complexity of such temporal regulation of transcription increases with viral genome size. Nevertheless, transcription of viral late genes encoding structural proteins invariably depends on synthesis of progeny viral DNA genomes in the infected cell (reviewed in ref. 1). In some cases, the activities of a single viral protein control both viral DNA synthesis and late gene transcription. For example, the simian virus 40 early protein large T antigen is not only the origin-recognition protein required for initiation of viral DNA synthesis but also an activator of late transcription (see refs. 2-5). The transition from the early to the late transcriptional program is considerably more intricate in cells infected by DNA viruses with larger genomes, such as human adenoviruses.
Synthesis of viral DNA in cells infected by human subgroup C adenoviruses, such as adenovirus type 2 or 5 (Ad2 or Ad5), results in transcription by cellular RNA polymerase II from three previously inactive viral promoters, those of the IX and IVa2 genes and the E2 late promoter (reviewed in ref. 6) Such off-to-on switches in transcription are analogous to viral DNA synthesis-dependent activation of simian virus 40 late transcription and may be mediated by a similar mechanism (see Results and Discussion). Entry into the late phase of adenoviral infection is also accompanied by a dramatic change in termination of major-late (ML) transcription. This promoter is active before the onset of viral DNA synthesis (7), when ML transcription terminates within a broad region spanning the middle of the transcription unit. In contrast, ML transcription terminates close to the right-hand end of the genome late in infection (8-11). In conjunction with regulation of alternative polyadenylation and alternative splicing, this difference results in production of a single ML mRNA early in infection, that encoding the L1 52/55-kDa protein, but synthesis of at least a dozen during the late phase (see refs. 6 and 12). The mechanism(s) responsible for this alteration in termination of ML transcription are not understood. However, the observation that coding sequences for viral structural proteins present in the ML transcription unit can be expressed only from replicated viral DNA molecules (13) indicates that viral DNA synthesis is required.
Another transcriptional change characteristic of the late phase of adenovirus infection is an increase of some 20- to 30-fold in the efficiency of ML transcription (7). Such stimulation of ML transcription requires two intragenic sequences, termed DE1 (+85 to +98) and DE2 (+100 to +120) both in vitro and in infected cells (14-18). These sequences function cooperatively (19) with the upstream binding site for upstream stimulatory factor (20-22). Binding to the intragenic sequences of two infected cell-specific proteins, designated DEF-A and DEF-B, stimulates ML transcription in vitro (16, 19). Purification of DEF-B established that it is a dimer of the viral IVa2 protein (22), whereas DEF-A was found to comprise both this same viral protein and one or more additional proteins, as yet unidentified (23). Overexpression of the IVa2-coding sequence in cells infected with Ad5-stimulated expression of a ML-luciferase reporter gene and such stimulation required the intragenic sequences described above (22). In toto, these observations strongly indicate that a IVa2 protein dimer is one infected cell-specific activator of ML transcription. The results of more recent genetic experiments are consistent with this conclusion: mutations in the Ad5 genome that prevent production of the IVa2 protein or eliminate the upstream binding site for the cellular transcriptional activator upstream stimulatory factor in the ML promoter singly did not prevent the recovery of viable virus, but were lethal when combined (24).
The observations summarized above indicate that the late phase of adenovirus infection comprises two transcriptional switches, such that viral DNA synthesis and expression of coding sequences for the viral structural proteins, which are contained within the ML transcription unit, are not directly coupled. Viral DNA replication results in activation of IVa2 transcription and production of the IVa2 protein, and subsequently, the IVa2 protein participates in stimulation of ML transcription. Activation of IVa2 transcription is therefore the crucial trigger for completion of the adenoviral infectious cycle.
IVa2 transcription is initiated at one major and one minor site, which lie some 200 bp upstream of, and in the opposite strand to, the ML promoter (see refs. 6 and 12). Previous in vitro studies have established that the organization of the IVa2 promoter is unusual: it lacks a TATA sequence (25-27), or indeed, any upstream sequence that strongly influences the efficiency of transcription in vitro (27-29), or in infected cells (C.S.H. Young, A. Timko, and S.J.F., unpublished observations). An initiator element spanning positions -4 to +11 specifies recognition of the major initiation site (29-31), but efficient initiation from this site in vitro requires the intragenic sequence shown in Fig. 1 (29, 30). Superimposed on these IVa2 promoter sequences is a binding site for a cellular repressor of transcription, termed IVa2-RF (31, 32). Because adenovirus infection does not lead to inactivation of this repressor, we proposed that activation of IVa2 transcription is the direct consequence of the increase in concentration of IVa2 promoters as progeny viral genomes are synthesized (31). We have used transient expression assays to demonstrate that this repressor titration mechanism can regulate IVa2 transcription in mammalian cells (33). To investigate its relevance during the viral infectious cycle, we have now examined the effects of mutations that specifically impair binding of the cellular repressor to the endogenous IVa2 promoter on IVa2 and ML transcription in infected human cells.
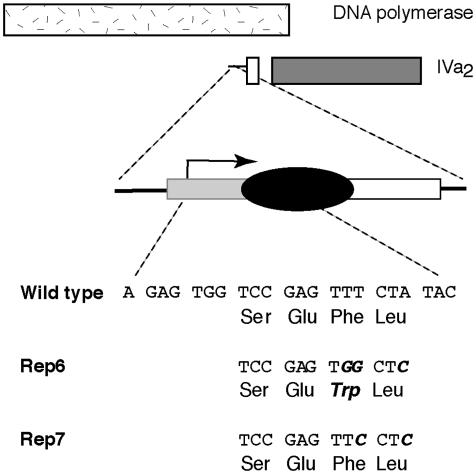
Fig. 1.
Location of the Rep mutations in the IVa2 promoter. (Top) The relationship of the coding sequence for the DNA polymerase and the IVa2 gene in the l-strand of the viral genome. The IVa2 promoter (Middle) comprises an initiator element (gray box) and an intragenic sequence (downstream boundary +29) necessary for efficient initiation of transcription (white box). The major site of initiation (+1) is indicated by the jointed arrow drawn in the direction of transcription. The binding site for the cellular transcriptional repressor IVa2-RF is indicated by the black oval. The Ad2 IVa2 sequence from positions +1 to +22 is shown below, written as codons of the DNA polymerase coding sequence. The substitutions introduced in the Rep6 and Rep7 mutants and their effects (if any) on the amino acid sequence of the DNA polymerase are shown in bold italics below the WT sequence.
Materials and Methods
Construction of Recombinant Adenoviruses. Precise substitution mutations, termed Rep6 and Rep7 (31), previously introduced into the binding site for IVa2-RF, were recovered into the viral genome by a double-homologous recombination strategy in Escherichia coli BJ5183, based on the procedure of Chartier et al. (34). The pTG3602 plasmid (34) containing the full-length Ad5 genome was cut with BstZ17I, which cleaves the viral genome at position 5,766 adjacent to the IVa2 promoter (positions 5,818-5,829) where mutations were to be introduced. The cut at this BstZ17I site was rescued by recombination with fragments comprising positions 4,122-6,585 of the Ad2 genome and carrying WT or mutated Ad2 IVa2 promoter sequences (positions 5,776-5,991 in Ad2). Cleavage at the second BstZ17I site within the Ad5 genome (position 29,012) was rescued by recombination with an Ad5 fragment comprising positions 25,352-32,127. The plasmids generated by this double-recombination method were designated pTG RepWT, pTGRep6, and pTGRep7. They were examined for correct introduction of mutations by sequencing of PCR products spanning the IVa2 promoter region. Ad5/Ad2 recombinant viruses carrying the WT or mutant IVa2 promoters were isolated after transfection of PacI-restricted pTGRep plasmids into 293 cells (35) and purified by two cycles of plaque purification in HeLa cells. The presence of the desired mutations was again confirmed by sequence analysis of the recombinant adenoviral genomes.
Cells and Viruses. HeLa and 293 cells were maintained in mono-layer cultures in DMEM supplemented with 5% FBS, 5% calf serum, and 1% (vol/vol) glutamine. The typical multiplicity of infection was 10 plaque-forming units (pfu)/ml. To analyze the growth characteristics of recombinant versus WT viruses, infections were performed at both 0.5 and 10 pfu/ml, and plaque assays were performed as described (36).
Isolation and Analysis of DNA and RNA. Nuclear and cytoplasmic fractions from infected HeLa cells were separated essentially as described by Greenberg and Bender (37). Cells were washed with ice-cold PBS and then extracted twice with 50 mM Tris·HCl, pH 7.5, containing 100 mM KCl, 2 mM EDTA, and 0.5% (vol/vol) Nonidet P-40. After centrifugation, cytoplasmic RNA was isolated from the pooled supernatants as described (27), whereas DNA was isolated from nuclear pellets by a modification of the procedure described by Strauss (38). Infected HeLa cell nuclei were suspended in 10 mM Tris·HCl, pH 8.0, containing 100 mM NaCl, 25 mM EDTA, 0.5% (wt/vol) SDS, and 0.1 mg/ml proteinase K, and incubated at 37°C for 30 min. The high salt-precipitated DNA was then treated with 20 μg/ml RNase A and purified by ethanol precipitation. To quantify the viral DNA accumulated in cells infected with WT or mutant adenoviruses, slot-blot analysis was performed as described (7, 13) by using 20 μg of total DNA. The membranes were hybridized at 65°C overnight with 10 ng/ml of a linearized plasmid containing a full-length copy of E1A 13S cDNA labeled with [α-32P]dCTP (3,000 Ci/mmol; 1 Ci = 37 GBq, New England Nuclear) by the random priming method (39) at 65°C. Washed and air-dried membranes were analyzed by using a Molecular Dynamics PhosphorImager.
Primer Extension Assays. Primer extension assays were performed as described (27) by using 10 μg of cytoplasmic RNA and 20 fmol of primers complementary to viral E2E (positions +60 to +37), IVa2 (positions +65 to +40), and ML (positions +40 to +15) mRNAs, and to cellular β-actin (positions +89 to +113) mRNA. Primer extension products were separated by electrophoresis in 8% polyacrylamide sequencing gels, visualized by autoradiography, and quantified by using a Molecular Dynamics PhosphorImager.
Run-On Transcription Assays. Nuclei from HeLa cells were isolated as described (40) and resuspended at a concentration of 108 nuclei per ml. The RNA chains initiated in vivo were elongated at 30°C for 30 min in the presence of 100 μCi of [α-32P]CTP (800 Ci/mmol, New England Nuclear). The labeled RNA was then purified and hybridized (37) for 40 h at 65°C to membrane-bound plasmid DNAs and antisense RNAs by using Church buffer (41). In all cases, the quantity of labeled RNA per pmol of probe was equivalent to that produced by 8 × 105 nuclei. After hybridization, the membranes were washed sequentially four times for 1 h each, with 2× SSC (1× SSC is 0.015 M sodium citrate, pH 7.0, containing 0.15 M NaCl) at 65°C, 2× SSC containing 0.1% (wt/vol) SDS at 65°C, 2× SSC containing 10 μg/ml RNase A at 37°C, and 2× SSC containing 5 μg/ml tRNA at 37°C. Hybridization signals were analyzed and quantified as described above. The DNA probes were the pETS RS(B) plasmid, which contains a fragment (positions -500 to + 700) of the gene for human ribosomal RNA, linearized with EcoRI, and the pSE420 plasmid, which contains the entire Ad2 IVa2 coding sequence, linearized with HindIII. The antisense RNA probes were complementary to positions +12 to +104 of the E2E transcription unit and positions +1 to +1,057 of the ML L3 region and were synthesized by using SP6 or T7 RNA polymerases (New England Biolabs), respectively, according to the manufacturer's instructions.
Results and Discussion
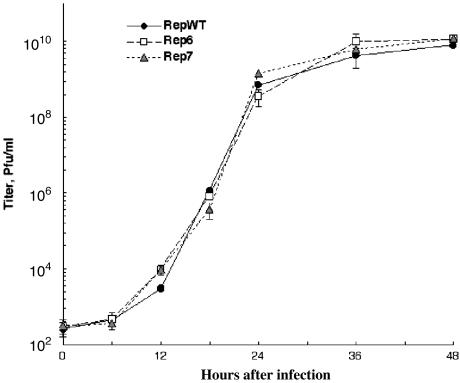
Isolation and Characterization of the Replication Kinetics of Mutant Viruses. We previously identified a number of substitution mutations that impair binding of the cellular transcriptional repressor to the Ad2 adenoviral IVa2 promoter and increase the activity of this promoter in vitro (31). Two of these mutations, Rep6 and Rep7, which introduce one conservative amino acid substitution, and no change, respectively, into the overlapping coding sequence of the viral DNA polymerase (Fig. 1), were recovered into the Ad5 genome by homologous recombination in E. coli, as described in Materials and Methods. Viruses carrying these mutations were recovered as readily as a recombinant containing the WTAd2 IVa2 promoter, after transfection of viral genomes into human 293 cells. Indeed, both mutants, designated Rep6 and Rep7, replicated as efficiently as the recombinant WT (RepWT), in HeLa cells infected at 0.5 or 10 pfu per cell (Fig. 2 and data not shown, respectively): the Rep mutations induced no significant differences in either virus yield at 48 h after infection or the kinetics of replication (Fig. 2).
Fig. 2.
Kinetics of replication of the RepWT and mutant viruses. HeLa cells were infected with 0.5 pfu per cell of the viruses indicated. Infected cells were harvested at 6-h intervals, and virus titers were determined by plaque assay as described in Materials and Methods.
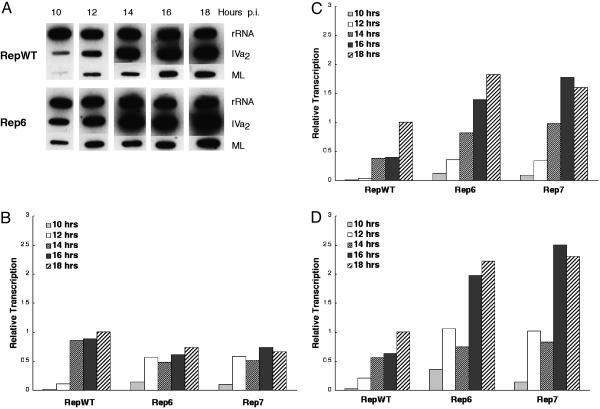
The Rep Mutations Accelerate IVa2 and ML Transcription. The effects of the Rep mutations on IVa2 transcription were assessed by using run-on transcription in nuclei isolated at 2-h intervals during the early-to-late transition from HeLa cells infected in parallel with the WT and mutant recombinant viruses. The RNA labeled during incubation of the nuclei in vitro under the conditions described in Materials and Methods was hybridized to membranes carrying DNA or RNA probes complementary to the viral transcription units of interest, and to the internal transcribed spacer of the human rRNA transcription unit, which served as the internal control. Representative results of these analyses are shown in Fig. 3. Quantification of hybridization signals like those shown in Fig. 3A indicated that E2E transcription was more efficient in cells infected by Rep6 or Rep7 than in RepWT-infected cells at 10 and 12 h after infection but somewhat less efficient from 14 h after infection (Fig. 3B). In contrast, IVa2 transcription was more efficient throughout the period examined, and was detected earlier in cells infected by the mutants than in WT-infected cells (Fig. 3C). At 10 and 12 h after infection, the rate of IVa2 transcription was some 10-fold higher in cells infected by Rep6 or Rep7 than in RepWT-infected cells. Furthermore, the rate of IVa2 transcription seen at 18 h after infection in cells infected by RepWT was attained by 14-15 h after infection in mutant virus-infected cells (Fig. 3C). These data establish that the mutations that impair binding of IVa2-RF to the IVa2 promoter lead to both earlier-than-normal and more efficient transcription from this viral promoter. As shown in Fig. 3D, the Rep mutations also increased the efficiency of transcription from the viral ML promoter.
Fig. 3.
The Rep mutations increase the efficiency of IVa2 and ML transcription. Transcription of the IVa2 and ML genes was examined at the times indicated in cells infected by RepWT, Rep6, or Rep7 using run-on transcription in isolated nuclei, as described in Materials and Methods. (A) An example of hybridization of RNA labeled in isolated nuclei to DNA or RNA probes for genes listed at the right. Such signals were quantified and corrected by using rRNA as the internal control as described in Materials and Methods. They are expressed relative to the value observed at 18 h after infection with RepWT. (B) E2E. (C) IVa2. (D) ML.
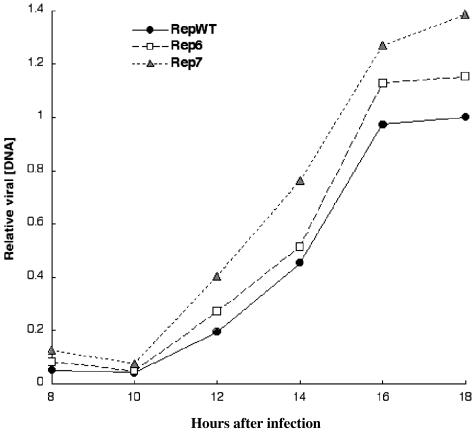
The Rep Mutations Increase the Efficiency of IVa2 Template Use. The results described in the previous section appear consistent with the repressor titration hypothesis summarized in the Introduction: impaired binding of IVa2-RF to the Rep6 and Rep7 mutant promoters resulted in more efficient IVa2 transcription, particularly during the initial period of the late phase of infection. However, in view of the higher rates of E2E transcription observed at 10-12 h after infection, it remained possible that these mutations altered the timing or efficiency of viral DNA synthesis to increase the concentration of IVa2 templates above some threshold required for recognition by the cellular transcriptional machinery. To assess the contribution, if any, of such indirect effects of the Rep mutations on IVa2 transcription, we first compared viral DNA synthesis in cells infected by the recombinant WT and mutant viruses. Representative results of determination of the intranuclear concentrations of viral DNA, as described in Materials and Methods, are shown in Fig. 4. Modest increases in viral DNA concentration were observed reproducibly from 12 h after infection in cells infected by each of the Rep mutants. Because the Rep7 mutation does not alter the sequence encoding the viral DNA polymerase (Fig. 1), these differences are unlikely to stem from increased activity of this viral enzyme. They are, therefore, likely to be a trivial result of minor inaccuracies in the measurement of viral titers, and hence, differences in input viral concentrations (Fig. 4). It is also possible that the more efficient expression of the E2E transcription unit, which encodes the viral replication proteins (see refs. 6 and 12), initially observed in Rep mutant virus-infected cells (Fig. 3A), accounts for the modest increases in viral DNA synthesis shown in Fig. 4. When the concentration of the IVa2 protein incorporated into virions is reduced, entry of the viral genome into the infected cell nucleus and expression of all early genes is delayed (42). This property suggests that our Rep mutants that accelerate expression of the IVa2 gene (Fig. 3B) might exhibit the opposite phenotype, that is, accelerated expression of early genes because the mutant virus particles contain higher-than-normal quantities of the IVa2 protein.
Fig. 4.
Kinetics of viral DNA synthesis in cells infected by the RepWT and mutant viruses. The concentrations of intranuclear viral DNA at the times indicated were determined as described in Materials and Methods. All values are expressed relative to that observed at 18 h after infection in RepWT-infected cells.
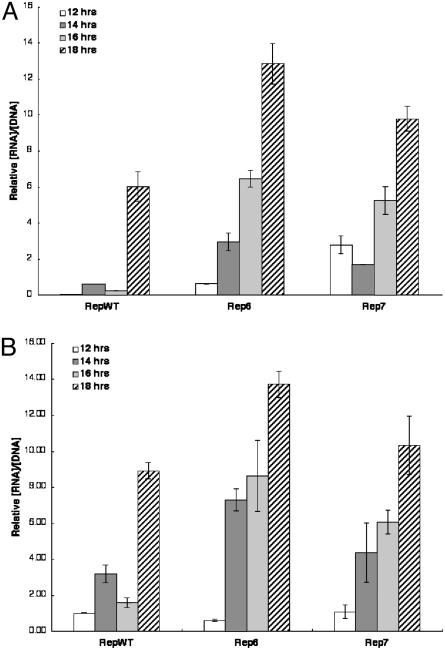
To account for the differences in the kinetics of viral DNA synthesis shown in Fig. 4, we compared the efficiencies with which the WT and mutant viral DNA templates for IVa2 transcription were used. In these experiments, the concentration of IVa2 mRNA in the cytoplasm was used as a surrogate measure of transcription, so that the quantities of template (nuclear viral DNA) and RNA product (cytoplasmic mRNA) could be measured in the same infected cell samples. Concentrations of IVa2 mRNA were determined by primer extension as described in Materials and Methods, and the raw values were corrected for any differences in cell number or RNA recovery by using β-actin mRNA as the internal control. The corrected values were then expressed as a function of the concentration of viral DNA present in the nuclei of the same infected cells samples, determined as described. This approach established unambiguously that the Rep6 and Rep7 IVa2 templates were used more efficiently than the WT (Fig. 5A). For example, at 12 h after infection, the two mutant templates were transcribed some 60-fold more efficiently than the WT, whereas the differences were no more than a factor of 2 by 18 h after infection. This temporal pattern is exactly as predicted by the repressor titration hypothesis. When the intranuclear concentration of viral DNA is low, the mutations that reduce the affinity with which IVa2-RF binds to the IVa2 promoter relieve transcriptional repression to allow IVa2 transcription under conditions in which the WT promoter remains fully repressed. However, with continuing viral DNA synthesis, the number of mutant and WT template molecules that cannot be bound by IVa2-RF becomes more similar, and, therefore, so does the efficiency of IVa2 transcription. The data shown in Fig. 5A therefore establish unequivocally that viral DNA synthesis-dependent titration of a cellular transcriptional repressor is responsible for activation of IVa2 transcription during the adenoviral infectious cycle, as predicted on the basis of biochemical analyses (31, 32). Such a mechanism of temporal control of viral transcription is extremely efficient: no dedicated viral gene products are required to activate transcription of late genes, but rather the essential production of progeny viral genomes is coupled to the activity of specific viral promoters.
Fig. 5.
The Rep mutations increase the efficiency with which IVa2, and ML templates are transcribed. The concentrations of IVa2 (A) and ML (B) cytoplasmic mRNAs in HeLa cells infected in parallel with RepWT, Rep6, or Rep7 at the times indicated were quantified as described in Materials and Methods. The concentration of viral DNA in the nuclei of each infected sample was determined as described in the legend to Fig. 4 and was used to calculate the quantities of the viral mRNAs produced per unit concentration of viral DNA. The values shown (in arbitrary units) for such a measure of the efficiency with which viral transcriptional templates were used represent the mean of two independent experiments.
Although activation of transcription from a viral late promoter by viral DNA synthesis-dependent titration of a cellular transcriptional repressor has not been directly demonstrated in previous studies, this mechanism of temporal control of viral gene expression is probably not a unique property of the adenoviral IVa2 gene. Substitutions within a binding site for the cellular RBP-2 protein within the adenoviral p(IX) promoter have been reported to increase the concentration of the corresponding mRNA detected at 12 h after infection (43). Furthermore, by using in vitro transcription and transient expression assays, Mertz and colleagues (44) demonstrated that binding of orphan members of the steroid/thyroid hormone receptor superfamily represses transcription from the major initiation site of the simian virus 40 late gene during the early, but not the late, phase of infection. In the context of the simian virus 40 infectious cycle, a mutation that impairs binding of the cellular proteins to this viral late promoter leads to overproduction of late mRNA during the first few hours of the late phase of infection (45), consistent with activation of late transcription as a result of repressor titration.
The Efficiency of ML Template Use Is also Increased in Rep Mutant Virus-Infected Cells. Because ML transcription was increased by the Rep mutations (Fig. 3), we also compared the efficiencies with which ML templates are transcribed in cells infected by the Rep WT and mutant viruses by the method described in the previous section. The results of this analysis (Fig. 5B) indicate that the ML promoter was also used more efficiently in cells infected by both Rep6 and Rep7 than in RepWT-infected cells. This effect on ML transcription was specific because neither of the Rep mutations resulted in more efficient use of E2E templates (data not shown). However, the increase in the efficiency of transcription of ML templates was delayed relative to the more efficient use of Rep mutant IVa2 templates, and not detected until 14 h after infection (compare Fig. 5 A and B). Such a temporal difference is consistent with stimulation of ML transcription in Rep mutant-infected cells as an indirect result of earlier-than-normal IVa2 gene expression (Fig. 5A) (see the Introduction). On the other hand, the Rep mutant ML templates were transcribed no more than 2.5-fold more efficiently than the WT (Fig. 5B). These modest effects of the Rep mutations could therefore be a direct result of increased activity of the ML promoter, analogous to those previously observed in vitro (31). Experiments are in progress to distinguish such cis and trans mechanisms of stimulation of ML transcription by the Rep mutations.
Acknowledgments
We thank Barbara Sollner-Webb (Johns Hopkins University School of Medicine, Baltimore) for the pETS RS(B) plasmid, and Dara Whalen and Wenying Huang for excellent technical assistance. This work was supported by a grant from the National Institute of General Medical Science, National Institutes of Health.
Author contributions: C.I. and S.J.F. designed research; C.I. performed research; C.I. and S.J.F. analyzed data; and S.J.F. wrote the paper.
Abbreviations: ML, major-late; Ad2, adenovirus type 2; Ad5, adenovirus type 5; pfu, plaque-forming unit.
References
- 1.Flint, J., Enquist, L., Racaniello, V. & Skalka, A. (2003) Principles of Virology: Molecular Biology, Pathogenesis, and Control (Am. Soc. Microbiol., Washington, DC), 2nd Ed.
- 2.Kelly, T. J. (1988) J. Biol. Chem. 263, 17889-17892. [PubMed] [Google Scholar]
- 3.Diffley, J. F. X. (1992) Trends Cell Biol. 2, 298-303. [DOI] [PubMed] [Google Scholar]
- 4.Keller, J. M. & Alwine, J. C. (1984) Cell 36, 381-389. [DOI] [PubMed] [Google Scholar]
- 5.Brady, J. & Khoury, G. (1985) Mol. Cell. Biol. 5, 1391-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flint, S. J. (1986) Adv. Virus Res. 31, 169-228. [DOI] [PubMed] [Google Scholar]
- 7.Shaw, A. R. & Ziff, E. B. (1980) Cell 22, 905-916. [DOI] [PubMed] [Google Scholar]
- 8.Fraser, N. & Ziff, E. B. (1978) J. Mol. Biol. 124, 27-51. [DOI] [PubMed] [Google Scholar]
- 9.Fraser, N. W., Nevins, J. R., Ziff, E. & Darnell, J. E. J. (1979) J. Mol. Biol. 129, 643-656. [DOI] [PubMed] [Google Scholar]
- 10.Nevins, J. R. & Wilson, M. C. (1981) Nature 280, 113-118. [DOI] [PubMed] [Google Scholar]
- 11.Iwamoto, S., Eggerding, F., Falck-Pederson, E. & Darnell, J. E., Jr. (1986) J. Virol. 59, 112-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shenk, T. (2001) in Virology, ed. Fields B., Howley, P. & Knipe, D. (Raven, New York), 4 th Ed., pp. 2265-2300.
- 13.Thomas, G. B. & Mathews, M. B. (1980) Cell 22, 523-533. [DOI] [PubMed] [Google Scholar]
- 14.Mansour, S. L., Grodzicker, T. & Tjian, R. (1986) Mol. Cell. Biol. 6, 2684-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jansen-Durr, P., Boeuf, H. & Kedinger, C. (1988) Nucleic Acids Res. 16, 3771-3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jansen-Durr, P., Mondésert, G. & Kedinger, C. (1989) J. Virol. 63, 5124-5132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leong, K., Lee, W. & Berk, A. J. (1990) J. Virol. 64, 51-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mason, B. B., Davis, A. R., Bhat, B. M., Chengalvala, M., Lubeck, M. D., Zandle, G., Kostek, B., Cholodofsky, S., Dheer, S., Molnar-Kimber, K., et al. (1990) Virology 177, 452-461. [DOI] [PubMed] [Google Scholar]
- 19.Mondesert, G., Tribouley, C. & Kedinger, C. (1992) Nucleic Acids Res. 20, 3881-3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carthew, R. W., Chodosh, L. A. & Sharp, P. A. (1985) Cell 43, 439-448. [DOI] [PubMed] [Google Scholar]
- 21.Sawadogo, M. & Roeder, R. G. (1985) Cell 43, 165-175. [DOI] [PubMed] [Google Scholar]
- 22.Tribouley, C., Lutz, P., Staub, A. & Kedinger, C. (1994) J. Virol. 68, 4450-4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lutz, P. & Kedinger, C. (1996) J. Virol. 70, 1396-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pardo-Mateos, A. & Young, C. S. (2004) Virology 324, 151-164. [DOI] [PubMed] [Google Scholar]
- 25.Natarajan, V., Madden, M. J. & Salzman, N. P. (1984) Proc. Natl. Acad. Sci. USA 81, 6290-6294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carcamo, J., Maldonado, E., Cortes, P., Ahn, M.-H., Ha, I., Kasai, Y., Flint, S. J. & Reinberg, D. D. (1990) Genes Dev. 4, 1611-1622. [DOI] [PubMed] [Google Scholar]
- 27.Kasai, Y., Chen, H. & Flint, S. J. (1992) Mol. Cell. Biol. 12, 2884-2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Natarajan, V. & Salzman, N. P. (1985) Nucleic Acids Res. 13, 4067-4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen, H. & Flint, S. J. (1992) J. Biol. Chem. 267, 25457-25465. [PubMed] [Google Scholar]
- 30.Carcamo, J., Buckbinder, L. & Reinberg, D. (1991) Proc. Natl. Acad. Sci. USA 88, 8052-8056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin, H. J. & Flint, S. J. (2000) Virology 277, 397-410. [DOI] [PubMed] [Google Scholar]
- 32.Chen, H., Vinnakota, R. & Flint, S. J. (1994) Mol. Cell. Biol. 14, 676-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang, W. & Flint, S. J. (2003) J. Virol. 77, 4015-4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chartier, C., Degryse, E., Gantzer, M., Dieterle, A., Pavirani, A. & Mehtali, M. (1996) J. Virol. 70, 4805-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Graham, F. L. & van der Eb, A. J. (1973) Virology 54, 536-539. [DOI] [PubMed] [Google Scholar]
- 36.Williams, J. F. (1973) Nature 243, 162-163. [DOI] [PubMed] [Google Scholar]
- 37.Greenberg, M. & Bender, T. (1997) in Current Protocols in Molecular Biology, eds. Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A. & Struhl, K. (Wiley, New York), pp. 4.10.1-4.10.11.
- 38.Strauss, M. (1998) in Current Protocols in Molecular Biology, eds. Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A. & Struhl, K. (Wiley, New York), pp. 2.2.1-2.2.3.
- 39.Feinberg, A. P. & Vogelstein, B. (1983) Anal. Biochem. 137, 266-267. [DOI] [PubMed] [Google Scholar]
- 40.Huang, W., Pruzan, R. & Flint, S. J. (1994) Proc. Natl. Acad. Sci. USA 91, 1265-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Church, G. M. & Gilbert, W. (1984) Proc. Natl. Acad. Sci. USA 81, 1991-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang, W. & Imperiale, M. J. (2003) J. Virol. 77, 3586-3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dou, S., Zeng, X., Cortes, P., Erdjument-Bromage, H., Tempst, P., Honjo, T. & Vales, L. D. (1994) Mol. Cell. Biol. 14, 3310-3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wiley, S. R., Kraus, R. J., Zuo, F., Murray, E. E., Lovitz, K. & Mertz, J. E. (1993) Genes Dev. 7, 2206-2219. [DOI] [PubMed] [Google Scholar]
- 45.Farrell, M. L. & Mertz, J. E. (2002) Virology 297, 307-318. [DOI] [PubMed] [Google Scholar]