Abstract
Streptococcus pyogenes secretes many toxins that facilitate human colonization, invasion, and dissemination. NADase (SPN) and streptolysin O (SLO) are two toxins that play important roles in pathogenesis. We previously showed that increased production of SPN and SLO in epidemic serotype M1 and M89 S. pyogenes strains is associated with rapid intercontinental spread and enhanced virulence. The biological functions of SPN and SLO have been extensively studied using eukaryotic cell lines, but the relative contribution of each of these two toxins to pathogenesis of epidemic M1 or M89 strains remains unexplored. Herein, using a genetically representative epidemic M1 strain and a panel of isogenic mutant derivative strains, we evaluated the relative contributions of SPN and SLO toxins to virulence in mouse models of necrotizing myositis, bacteremia, and skin and soft tissue infection. We found that isogenic mutants lacking SPN, SLO, and both toxins are equally impaired in ability to cause necrotizing myositis. In addition, mutants lacking either SPN or SLO are significantly attenuated in the bacteremia and soft tissue infection models, and the mutant strain lacking production of both toxins is further attenuated. The mutant strain lacking both SPN and SLO production is severely attenuated in ability to resist killing by human polymorphonuclear leukocytes. We conclude that both SPN and SLO contribute significantly to S. pyogenes pathogenesis in these virulence assays.
Streptococcus pyogenes (alias group A Streptococcus) is a major bacterial pathogen causing human morbidity and mortality globally. Streptococcus pyogenes produces many virulence factors that facilitate transmission, colonization, invasion, and dissemination.1 NADase (SPN) and streptolysin O (SLO) are two potent cytotoxins secreted by S. pyogenes, and multiple roles in virulence have been attributed to each protein. SPN is actively translocated into the host cells, resulting in cell injury and death via depletion of host cell NAD+.2, 3, 4 SLO is a cholesterol-dependent cytolysin that forms pores in host cell membranes. SPN and SLO inhibit phagocytosis,3 inhibit maturation of autophagosomes,5, 6 impair neutrophil oxidative burst,7 and prevent the killing of S. pyogenes by a variety of host immune cells.3, 5, 6, 7, 8
SPN and SLO are interconnected in many ways. Several lines of evidence suggest that the SPN and SLO proteins interact physically.4, 9, 10, 11 Moreover, SPN and SLO toxins are functionally related. For example, translocation of SPN into host cells requires SLO.2 In addition, binding of SPN to the host cell membrane promotes SLO-mediated pore formation.12 Furthermore, the NADase activity of SPN augments the cytotoxic and hemolytic activity of SLO.4
The chromosomal region encoding SPN and SLO contains three contiguous genes organized as an operon: nga, ifs, and slo. nga and slo encode SPN and SLO, respectively.13 ifs encodes an endogenous inhibitor of SPN, which inhibits the NADase activity before SPN secretion.14 nga, ifs, and slo are cotranscribed from a promoter located upstream of nga.13 We recently showed that single-nucleotide polymorphisms in the nga promoter region confer increased nga promoter activity and thereby increased production of SPN and SLO in contemporary epidemic M1 and M89 strains.8, 15 We also demonstrated that increased toxin production is associated with enhanced bacterial fitness during upper respiratory tract infections, increased resistance to innate immunity, and increased tissue destructions.8, 15 These features contribute to rapid clonal expansion and epidemics of severe streptococcal infections caused by the epidemic M1 and M89 clones.8, 15, 16, 17 Investigating the relative contributions of SPN and SLO to streptococcal pathogenesis may provide useful information to develop effective vaccines and therapeutic strategies to blunt the virulence of S. pyogenes. In this study, by using a genetically representative epidemic M1 strain MGAS2221, and a panel of isogenic mutant strains deficient in SPN, SLO, or both, we compared the abilities of the mutants and the wild-type strain to cause invasive muscle infections, bacteremia, and soft tissue infections in mice, and to resist human neutrophil-mediated killing.
Materials and Methods
Bacterial Strains and Growth Conditions
The S. pyogenes strains used in this study are listed in Table 1. Strain MGAS2221 is genetically representative of contemporary epidemic M1 strains.17, 18 Four isogenic mutant strains were derived from MGAS2221. SPN mutant strains Δnga and Δnga-ifs and SLO mutant strain Δslo were generated for the current study. SPN, SLO mutant strain Δnga-ifs-slo was described previously.8 All S. pyogenes strains were grown in Todd-Hewitt broth supplemented with 0.2% yeast extract (THY broth) at 37°C with 5% CO2.
Table 1.
Streptococcus pyogenes Strains Used in this Study
| Strains | Description | Reference |
|---|---|---|
| MGAS2221 | Representative epidemic serotype M1 isolate | 8, 17 |
| Δnga | MGAS2221 with a 1314-bp deletion in nga | This article |
| Δnga-ifs | MGAS2221 with a 1777-bp deletion that includes nga and ifs | This article |
| Δslo | MGAS2221 with a 473-bp deletion in slo | This article |
| Δnga-ifs-slo | MGAS2221 with a deletion that includes nga, ifs, and slo | 8 |
Construction of Isogenic Mutant Strains
MGAS2221 was the wild-type parental strain used to generate all isogenic deletion-mutant strains. MGAS2221 is genetically representative of contemporary epidemic M1 strains and has wild-type virulence-related transcriptional regulators, including CovR/CovS, Mga, RopB, and RocA.8, 17 SPN mutant strain Δnga was constructed by in-frame deletion of a 1314-bp internal region of nga. In this mutant, the nga promoter and nga ribosome binding site were preserved to avoid potential polar effects on the downstream genes ifs and slo. Briefly, using MGAS2221 genomic DNA as template, primer sets NGA-1/2 and NGA-3/4 (Table 2) were used to amplify two 1184-bp fragments upstream and downstream of the region of deletion, respectively. The two PCR fragments were merged by combinatory PCR and ligated into the BamHI site of suicide vector pBBL740. The recombinant plasmid containing a truncated nga gene (with a 1314-bp deletion) was transformed into strain MGAS2221 to replace the native nga gene via allelic exchange, as described previously.8 The nga-ifs double-deletion mutant Δnga-ifs was constructed by deleting a 1777-bp DNA sequence that includes the nga and ifs genes. In this mutant, the nga promoter and the intergenic region between ifs and slo were retained to avoid polar effects on slo. Briefly, primer sets NGA-IFS-1/2 and NGA-IFS-3/4 were used for combinatory PCR to generate a 2094-bp fragment with a deletion that includes both nga and ifs. The resulting PCR product was ligated into the BamHI site of plasmid pBBL740 and transformed into strain MGAS2221 to replace the native nga and ifs genes via allelic exchange using the previously mentioned method.8 The SLO-negative mutant Δslo was constructed by deleting a 473-bp 5′ portion of the slo coding sequence using an analogous strategy. Primer sets SLO-1/2 and SLO-3/4 were used for recombinant PCR to generate a 2380-bp fragment with an internal deletion in slo. Allelic exchange was performed to replace the native slo gene. Whole genome sequence analysis was performed on the four isogenic mutant strains to confirm the absence of spurious mutations.
Table 2.
Primers Used for Constructing Isogenic Mutant Strains
| Primers | Sequences | Remarks |
|---|---|---|
| NGA-1 | 5′-GCGTGGATCCGCTCTGACACAAGACACTAAAAAAG-3′ | Underlined: BamHI site |
| NGA-2 | 5′-TTACTTCCTATCTTGCATTCTATCTACTTTTTTGTTTCTCATGTAAAC-3′ | |
| NGA-3 | 5′-GTTTACATGAGAAACAAAAAAGTAGATAGAATGCAAGATAGGAAGTAA-3′ | |
| NGA-4 | 5′-GCGTGGATCCTCAACCGTTGCTTTGTCTCCCATAC-3′ | Underlined: BamHI site |
| NGA–IFS-1 | 5′-GCGTGGATCCGATACGTTGACACCAGCCCTGTCTT-3′ | Underlined: BamHI site |
| NGA–IFS-2 | 5′-TGTTTCTATTGTTCTTTCGACCATTGCGACAATATGAGCTAATGTTAC-3′ | |
| NGA–IFS-3 | 5′-GTAACATTAGCTCATATTGTCGCAATGGTCGAAAGAACAATAGAAACA-3′ | |
| NGA–IFS-4 | 5′-GCGTGGATCCTGCTGACACCTTTTCGTTGCAACTC-3′ | Underlined: BamHI site |
| SLO-1 | 5′-GCGTGGATCCCGGATGTATGTTAGCCGCAAATACC-3′ | Underlined: BamHI site |
| SLO-2 | 5′-CCTTTATCAGCCAGCTGAAGGGCTGGGTTAGTAGCCCAGCGACGCGACTGTA-3′ | |
| SLO-3 | 5′-TACAGTCGCGTCGCTGGGCTACTAACCCAGCCCTTCAGCTGGCTGATAAAGG-3′ | |
| SLO-4 | 5′-GCGTGGATCCCTACTTATAAGTAATCGAACCATAT-3′ | Underlined: BamHI site |
Western Immunoblot Analysis of SPN, SLO, and IFS in the Midexponential Cultures
Western immunoblot analysis was done as described previously.8 Briefly, to detect SPN and SLO, cell-free supernatants of S. pyogenes cultures grown to an OD600 of 0.5 were collected by centrifugation at 4000 × g for 10 minutes and filtered through a 0.2-μm filter. Culture supernatant proteins were separated by SDS-PAGE, transferred to a nitrocellulose membrane, and probed with anti-SPN (Abcam, Cambridge, MA) and anti-SLO antibodies (American Research Products, Waltham, MA). To detect IFS, bacterial cell pellets were collected from S. pyogenes cultures grown to OD600 of 0.5, suspended in 1× Laemmli sample buffer, and boiled for 10 minutes. The resulting samples were used for Western immunoblot analysis with anti-IFS antibody (B-Bridge International, Inc., Santa Clara, CA).
NADase Activity Assay and SLO Activity Assay
NADase activity and SLO activity present in the culture supernatant were measured as described previously.18 NADase activity was determined by measuring the highest twofold dilution of the culture supernatant capable of hydrolyzing 100 nmol of exogenous NAD+. SLO activity was determined by measuring the hemolytic activity of culture supernatants reduced by dithiothreitol.
Quantitative RT-PCR Analysis of slo Expression in S. pyogenes Strains
Streptococcus pyogenes strains were grown in THY to OD600 = 0.5. RNA from S. pyogenes cultures was extracted with an RNAeasy Mini Kit (Qiagen, Hilden, Germany) and converted into cDNA using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA). Quantitative PCR was performed with the TaqMan Fast Universal PCR Master Mix (Applied Biosystems) and an ABI 7500 Fast System (Life Technologies, Carlsbad, CA) instrument. The sequences of the TaqMan primers and probes for slo and rpsL are listed in Table 3. Each experiment was performed in triplicate, with data expressed as means ± SD. Statistical differences between strains were determined using one-way analysis of variance and Dunnett's post test.
Table 3.
Primers and Probes Used for TaqMan Quantitative RT-PCR
| Primers and probes | Sequences |
|---|---|
| slo | |
| Forward | 5′-TGGATGATATGCTTAACTCTAACGA-3′ |
| Reverse | 5′-TCAGTGTGATCTTCTTCGCTCT-3′ |
| Probe | 5′FAM-AGCTTGCTCCCAAAGAAATGCCA-3′TAMRA |
| rpsL | |
| Forward | 5′-CGTGTTGGAACAATGACACCTAA-3′ |
| Reverse | 5′-CTTCGATAAGGTTGCTCAAACGT-3′ |
| Probe | 5′FAM-CCTAACTCAGCCCTTCGTAAATTCGCTCGT-3′TAMRA |
Virulence Studies Using Mouse Models of Necrotizing Myositis, Bacteremia, and Skin and Soft Tissue Infection
Mouse necrotizing myositis challenge studies were performed as described previously.19 Briefly, immunocompetent 6-week-old female CD1 mice (Envigo Laboratories, Houston, TX) were randomly assigned to treatment groups and inoculated in the right hind limb with 1 × 107 colony-forming units (CFUs) of each S. pyogenes strain. For mouse survival experiments (n = 20 mice per bacterial strain), infected mice were monitored for near mortality at least once daily for 7 days. Near mortality was determined using internationally recognized criteria.20 Results were expressed as Kaplan-Meier curves, and differences between strain treatment groups were determined with the log-rank test (GraphPad Prism software version 6; GraphPad Software, Inc., La Jolla, CA). For histopathology evaluation (n = 3 mice per strain), lesions were excised, visually inspected, and photographed. The tissue was fixed in 10% phosphate-buffered formalin, decalcified, and embedded in paraffin using standard automated instruments.
For the bacteremia model, immunocompetent 6-week-old female CD1 mice (n = 20 mice per strain) were inoculated i.p. with 1 × 107 CFUs of each strain. Mice were monitored at least once daily for near mortality for 7 days. The near-mortality results were expressed as Kaplan-Meier curves, and differences between treatment groups were determined with the log-rank test.
The mouse model of skin and soft tissue infection was performed as described previously.21 Briefly, immunocompetent 6-week-old female SKH1-hrBR hairless mice (Charles River BRF, Houston, TX) were injected s.c. on the neck with 1 × 107 CFUs of each S. pyogenes strain. Lesion areas (n = 20 mice per strain) were measured daily for 7 days with a digital caliper. Lesion areas caused by each strain were compared using two-way analysis of variance (Prism6). For histopathology evaluation (n = 3 mice per strain), lesions were excised on day 3 after inoculation, fixed in 10% phosphate-buffered formalin, and embedded in paraffin using standard automated instruments. Animal study protocols AUP-0416-0019, AUP-0615-0041, and AUP-0716-0038 were approved by the Institutional Animal Care and Use Committee, Houston Methodist Research Institute (Houston, TX).
Streptococcus pyogenes–Human PMN Assays
Human polymorphonuclear leukocytes (PMNs) were purified from heparinized blood of healthy human donors using a standard method.22 All studies with human PMNs were performed in accordance with a protocol (01-I-N055) approved by the Institutional Review Board for human subjects, National Institute of Allergy and Infectious Diseases (Hamilton, MT). The studies conformed to the principles set forth in the Declaration of Helsinki. Healthy volunteers gave written informed consent before participation in the study. Parental strain MGAS2221 and four isogenic mutant derivatives were cultured to midexponential phase of growth (OD600 = 0.170 to 0.250) in THY broth, and washed once with phosphate-buffered saline. A total of 106 CFUs of each strain were combined with 106 PMNs in a 1.7-mL tube containing 10% autologous human serum in HEPES-buffered RPMI 1640 medium (600 μL total volume). Assay mixtures were rotated gently at 37°C for 1 or 3 hours, at which time saponin (0.1% final concentration) was added to each assay mixture and the tubes were placed on ice for 15 minutes. An aliquot of each was diluted and grown overnight on THY agar, and CFUs were counted. The percentage bacterial survival was determined by comparing CFUs from assays with PMNs to data from control assays without PMNs. Data were analyzed with a repeated-measures one-way analysis of variance and Tukey's post test to correct for multiple comparisons (GraphPad Prism software version 6.05; GraphPad Software, Inc.).
Results
In Vitro Phenotypes of Strain MGAS2221 and Its Isogenic Mutant Derivatives
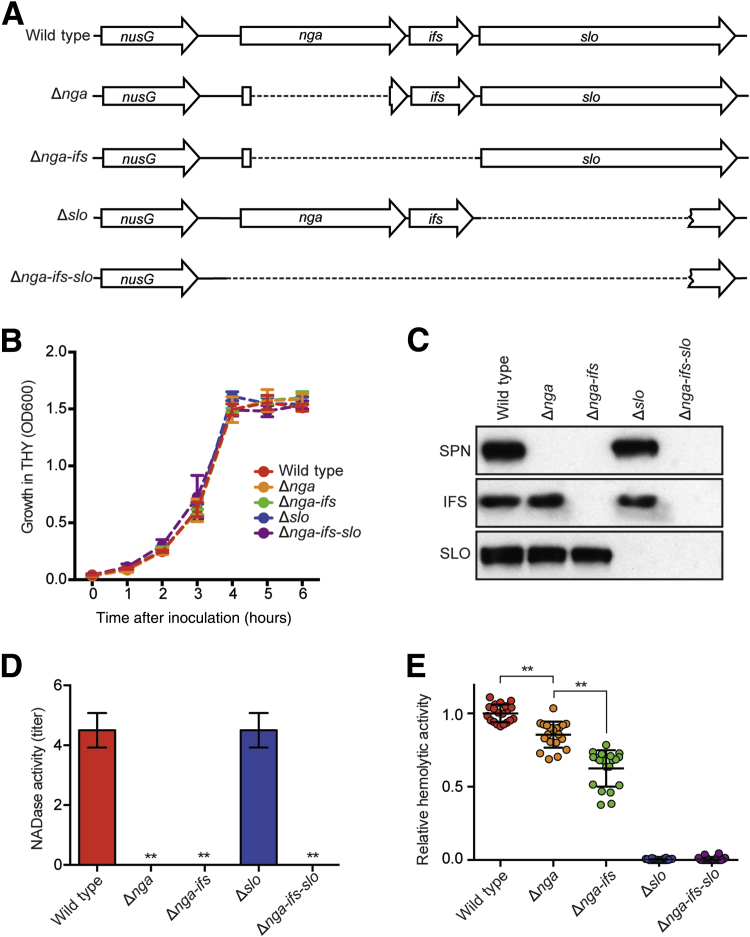
Isogenic mutant strains with deletion of nga, slo, or both genes were derived from wild-type serotype M1 strain MGAS2221 (Figure 1A). Compared to parental strain MGAS2221, none of the four isogenic mutant strains had a growth defect when cultured in THY broth (Figure 1B). Western immunoblot analysis was performed to determine the SPN, IFS, and SLO production phenotypes of the parental and mutant strains (Figure 1C). As expected, all three proteins (SPN, IFS, and SLO) were made by wild-type strain MGAS2221. Strain Δnga lacked SPN but produced IFS and SLO, whereas strain Δnga-ifs was deficient in both SPN and IFS, but produced SLO. Strain Δslo lacked SLO production but produced SPN and IFS. Strain Δnga-ifs-slo failed to produce all three proteins, as expected.
Figure 1.
Characteristics of serotype M1 parental strain MGAS2221 and isogenic mutant derivatives with different deletions in the nga-ifs-slo region. A: Schematic showing the nga-ifs-slo region of MGAS2221 and the four isogenic mutant derivatives. B: Growth curves of the five strains cultured in Todd-Hewitt broth supplemented with 0.2% yeast extract (THY). C: Western immunoblot of midexponential cultures showing production of SPN, IFS, and SLO by wild-type strain MGAS2221 and isogenic mutant derivative strains. D and E: NADase activity and SLO hemolytic activity of the five strains studied. Data are expressed as means ± SD (D and E). n = 4 (D); n = 20 (E). ∗∗P < 0.01 [versus the wild-type strain using one-way analysis of variance and Dunnett's post test (D), and using one-way analysis of variance and Dunnett's post test (E)].
To quantitatively characterize the SPN and SLO production phenotypes of the isogenic mutant strains, we measured the NADase enzymatic activity and SLO hemolytic activity present in midexponential culture supernatants obtained from the parental and four mutant strains (Figure 1D). As expected, strains Δnga, Δnga-ifs, and Δnga-ifs-slo lacked NADase activity. Strain Δslo produced a wild-type level of NADase activity, a result indicating that deletion of slo had no significant effect on the level of SPN production. Of interest, compared to parental strain MGAS2221, strain Δnga had significantly lower SLO activity (Figure 1E). This result is consistent with a previous report that SPN augments SLO-mediated hemolysis and cytotoxicity.4 Unexpectedly, strain Δnga-ifs had a further decrease in SLO activity compared to Δnga.
Virulence Features of MGAS2221 and Its Isogenic Mutant Derivatives in Three Mouse Models of Invasive Infection
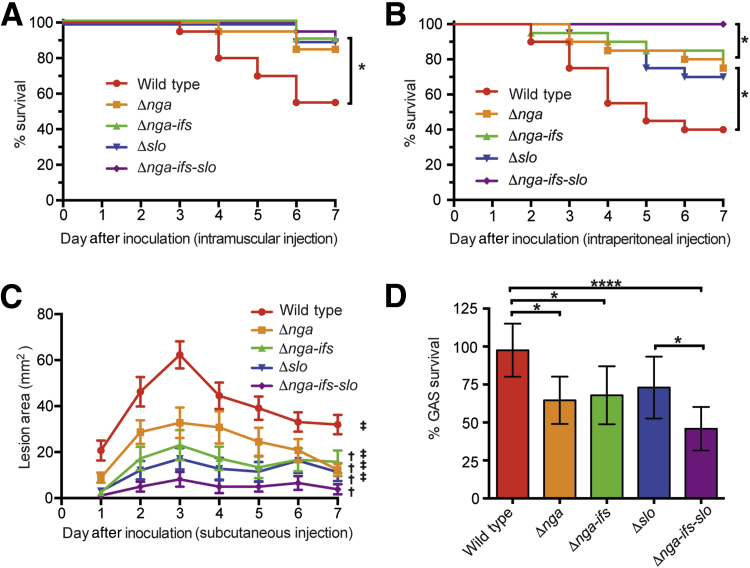
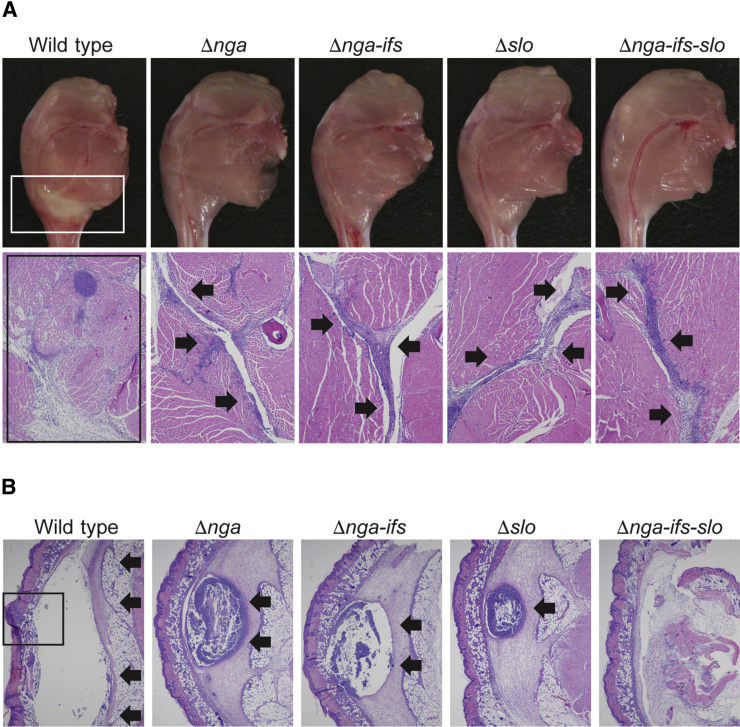
To study the relative contribution of SPN and SLO toxins to pathogenesis, we compared the virulence of strain MGAS2221 and its isogenic mutant derivatives deficient in SPN, SLO, and both, using three distinct mouse infection models (Figure 2). In a mouse model of necrotizing myositis, compared to the wild-type strain MGAS2221, SPN-deficient strains (Δnga and Δnga-ifs), SLO-deficient strain Δslo, and SPN-SLO double-deficient strain (Δnga-ifs-slo) caused significantly less near mortality (Figure 2A). The magnitude of virulence attenuation among the four mutant strains was similar. We also compared the extent of tissue damage caused by the five assayed strains. Visual examination of infected limbs showed that the wild-type strain MGAS2221 caused large lesions with massive tissue destruction (Figure 3A). The four isogenic mutant strains caused markedly smaller lesions (Figure 3A). In addition, microscopic examination of the limb lesions demonstrated that the wild-type strain caused a large area of fascial and myocyte necrosis (Figure 3A). In comparison, the four isogenic mutant strains caused a mild fascial infection (Figure 3A) that did not breach the fascial plane to cause myonecrosis. These results indicate that in this mouse infection model, SPN and SLO contribute significantly to the virulence of MGAS2221, and the consequence of lacking production of one toxin (SPN or SLO) is closely similar to the consequence of lacking production of both toxins (SPN and SLO).
Figure 2.
Strain virulence in mouse infection models and resistance to killing by polymorphonuclear leukocytes (PMNs). A: Ability of wild-type and mutant strains to cause near mortality in a mouse model of necrotizing fasciitis. B: Ability of wild-type and mutant strains to cause near mortality in a mouse model of bacteremia. Near mortality is expressed as Kaplan-Meier survival curves, and statistical differences are determined using the log-rank test. C: Ability of wild-type and mutant strains to cause skin lesions in a mouse model of skin and soft tissue infection. Lesion areas are expressed. P values were determined by repeated measures of analysis of variance. D: Resistance of the five strains to the bactericidal activity of purified human PMNs. Bacterial survival data are expressed as % GAS survival. Statistical differences are determined using one-way analysis of variance and Tukey's post test. Data are expressed as means ± SEM (C and D). n = 6 (D). ∗P < 0.05, ∗∗∗∗P < 0.0001; †P < 0.05 relative to wild-type; ‡P < 0.05 relative to Δnga-ifs-slo.
Figure 3.
Mutant strains lacking production of SPN, SLO, or both toxins are attenuated in ability to cause tissue destruction in mouse models of necrotizing myositis and skin and soft tissue infection. A: Visual and microscopic examination of muscle lesions collected at 72 and 24 hours, respectively, after inoculation. The wild-type parental strain MGAS2221 causes large lesions (white boxed region, top left panel) with extensive destruction of the fascial tissues and myocytes (black boxed region, bottom left panel). Each isogenic mutant strain caused markedly smaller lesions (top panels) with less severe tissue damage that is confined to the fascial planes (bottom panels, inward facing arrows). B: Microscopic examination of skin lesions collected 72 hours after inoculation. The wild-type parental strain MGAS2221 caused large abscesses extending beyond the field of view (arrows) and ruptured through the skin surface (boxed region, left panel). Each single-gene isogenic mutant strain caused markedly smaller abscesses confined to the underlying soft tissue (arrows). The double-toxin deficient strain caused soft tissue infections without a discrete abscess. Hematoxylin and eosin staining was used. Original magnifications: ×4 (A); ×2 (B).
To test the hypothesis that SPN and SLO toxins contribute to virulence in a second infection model, we compared the same five strains in a mouse model of bacteremia. Similar to the findings in the necrotizing myositis model, all four mutant strains were significantly impaired in ability to cause near mortality compared to the wild-type strain (Figure 2B). Interestingly, we also discovered that the isogenic mutant strain lacking ability to produce SPN and SLO (Δnga-ifs-slo) was significantly less virulent than each single-toxin deficient strain (Δnga, Δnga-ifs, and Δslo). That is, in this mouse bacteremia model, we observed a hierarchy of virulence: wild-type parental strain > single-toxin deficient strains > double-toxin deficient strain. Thus, SPN and SLO have an additive effect on virulence in this model.
We next compared the virulence of wild-type reference strain MGAS2221 and the four isogenic mutants using a well-described mouse model of skin and soft tissue infection. Compared to the wild-type parental strain, all four isogenic mutant strains caused significantly smaller lesions characterized by smaller abscesses and less soft tissue destruction (Figure 2C and Figure 3B), indicating that SPN and SLO also play important roles in virulence in this model. Similar to our findings in the bacteremia model, the double-toxin deficient strain (Δnga-ifs-slo) was significantly attenuated in virulence relative to either of the single-toxin deficient mutant strains.
SPN and SLO Contribute to Evasion of Killing by Human PMNs
PMNs are an essential part of innate immunity against bacterial infections.23 We previously showed that increased production of SPN and SLO renders group A Streptococcus more resistant to clearance by PMNs.8 Herein, we evaluated the relative contribution of SPN and SLO to circumvent the bactericidal effects of human PMNs. The results demonstrated that compared to the parental strain, the SPN-deficient strains (Δnga and Δnga-ifs) are significantly impaired in ability to resist PMN-mediated killing (P = 0.01 and P = 0.023, respectively) (Figure 2D). The attenuation of SLO-deficient strain (Δslo) did not reach statistical significance (P = 0.077). The mutant strain lacking production of both SPN and SLO (Δnga-ifs-slo) was severely attenuated (P < 0.0001) (Figure 2D). These results are consistent with our findings using mouse models of bacteremia as well as skin and soft tissue infection (Figure 2, B and C) that the mutant lacking production of both toxins is the most attenuated in virulence.
Discussion
SPN and SLO were among the earliest investigated group A Streptococcus virulence factors. In 1956, Bernheimer et al24 reported that the partially purified streptococcal fraction containing SLO inhibits the oxidative metabolism of mitochondria via affecting coenzyme NAD+. The responsible agent was subsequently identified to be an extracellular NADase (SPN).25 In 1957, by studying multiple streptococcal strains with and without NADase activity, Bernheimer et al26 proposed a possible association between NADase production and streptococcal leukotoxicity. SLO, an oxygen-sensitive hemolysin, was first described by Neil and Mallory in 1928 and subsequently confirmed by Herbert and Todd in 1941.27 Multiple roles in virulence have been attributed to SLO. Van Epps and Andersen28 showed that SLO inhibits neutrophil chemotaxis and mobility. Studies in the recent decade demonstrate SPN and SLO play important roles in circumventing the host immune system via i) inhibiting group A Streptococcus internalization by host cells,3 ii) promoting apoptosis of macrophages,29 iii) preventing the bactericidal effects of neutrophils,7, 8 iv) disrupting maturation of the phagosome, and v) promoting intracellular bacterial survival.5, 6, 30 Recent advances in S. pyogenes population genomics and pathogenesis further underlined the important roles of SPN and SLO in virulence. We previously showed that polymorphisms in the nga upstream region result in enhanced production of SPN and SLO, which is strongly associated with increased transmission and virulence of epidemic M1 and M89 clones.8, 15, 16, 17 In the present work, we performed a systematic deletion study on the nga-ifs-slo operon to evaluate the relative contribution of SPN and SLO toxins to the virulence of an epidemic M1 strain.
In vitro features of the mutant strains demonstrate that when grown in THY, deleting slo has no major effect on SPN production or NADase activity (Figure 1, C and D). However, deleting nga results in a significant reduction of SLO hemolytic activity (Figure 1E). This observation is consistent with a previous report that NADase activity augments the hemolytic activity and cytotoxicity of SLO.3 We unexpectedly found that deleting both the nga and ifs genes leads to a further reduction of SLO hemolytic activity (Figure 1E). Although this further reduction in SLO activity is modest and does not have significant consequence in bacterial virulence in the animal infection models used (Figures 2 and 3), it is significant and reproducible in THY (Figure 1E). There are multiple potential explanations for this observation. First, although slo is transcribed primarily from the nga promoter,13 it has been demonstrated that a weak second slo promoter is located in the ifs coding region.31 Deletion of the ifs gene removes this weak promoter, which may result in slightly decreased slo expression. However, our quantitative RT-PCR studies failed to demonstrate that deleting ifs results in significant decrease of slo expression (Supplemental Figure S1). Because of the resolution of this assay, a modest change in gene expression may not be precisely determined by quantitative RT-PCR. Another possibility is that IFS plays a role in post-transcriptional regulation, or the secretion of SLO.
We used three mouse invasive infection models to systematically evaluate the contribution of SPN and SLO to the virulence of epidemic M1 strain MGAS2221. The results clearly show that mutant strains lacking either SPN or SLO are severely impaired in ability to cause necrotizing myositis, bacteremia, and soft tissue infections (Figure 2). SPN-deficient mutants and SLO-deficient mutants are attenuated to similar levels (Figure 2). Interestingly, a previous study showed that although a SPN-deficient mutant is severely attenuated, SLO-deficient strains are either modestly attenuated or not attenuated.32, 33, 34 The difference between our finding and the previous observation suggests that the relative significance of SPN and SLO in pathogenesis might be strain dependent. However, it has been established that SPN and SLO are functionally associated. Translocation of SPN into the host cells requires SLO.2 SPN binding to the host cell membrane promotes SLO-mediated membrane pore formation.12 Furthermore, the NADase activity of SPN augments the cytotoxicity and the hemolytic activity of SLO.4 These findings suggest the virulence function of SPN is (at least partially) dependent on SLO, and vice versa. We therefore are more inclined to believe that in epidemic M1 strains, the SPN-deficient mutant and the SLO-deficient mutants are attenuated to similar levels in animal models of invasive infection.
If the virulence function of SPN is entirely dependent on SLO, and vice versa, one would expect that inactivating SPN, or SLO, or both toxins would have the same consequence in virulence. It has been demonstrated that SPN mutant, SLO mutant, and SPN-SLO double mutant are attenuated to similar levels in cytotoxicity on keratinocytes.3 Our virulence results also show that mutant strains lacking SPN, SLO, or both toxins are equally impaired in ability to cause near mortality in the mouse model of necrotizing myositis (Figure 2A). These results confirmed the intimate functional association between SPN and SLO. However, our results also showed that the mutant lacking both SPN and SLO is more attenuated compared to strains lacking SPN, or SLO, in ability to cause bacteremia or soft tissue infections, and to resist the bactericidal effects of human PMNs (Figure 2, B–D). These observations suggest the virulence function of SPN is not entirely dependent on SLO, and vice versa. We speculate that SPN may retain a function in virulence in the absence of SLO. This hypothesis is supported by a previous observation that the translocation of SPN into the host cell by a SLO-deficient mutant is reduced, but not abolished.6 Similarly, SLO might have the potential to confer virulence without the assistance of SPN, presumably because of its ability to destroy host cell membranes and cause cell injuries. Many SLO-like cholesterol-dependent cytolysins made by other streptococcal pathogens (eg, pneumolysin in Streptococcus pneumoniae and suilysin in Streptococcus suis) are known virulence factors,35, 36 but they are not coupled with NADase. In summary, our mouse virulence data confirmed the functional association between SPN and SLO. More important, our results suggest virulence function of SPN is not entirely dependent on SLO, and vice versa.
In summary, using a genetically representative epidemic M1 strain, and a set of isogenic mutant strains deficient in SPN, SLO, and both, we evaluated the relative contribution of SPN and SLO to cause invasive infections in mice, and to resist the bactericidal effects of human PMNs. We conclude that both SPN and SLO contribute significantly to S. pyogenes pathogenesis. Our findings improve understanding of the pathogenesis of the epidemic M1 clone, and have implications for translational research on a pathogen that has affected millions of people.
Acknowledgments
We thank Matthew Ojeda Saavedra and Concepcion Cantu for assistance with the mouse experiments. All authors have read and approved the final manuscript.
Footnotes
Supported by the Fondren Foundation (J.M.M.) and in part by the National Institute of Allergy and Infectious Diseases, NIH, Intramural Research Program (F.R.D.). J.D.L. was supported by the Rice University Century Scholar program.
Disclosures: None declared.
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.ajpath.2016.11.003.
Supplemental Data
Supplemental Figure S1.
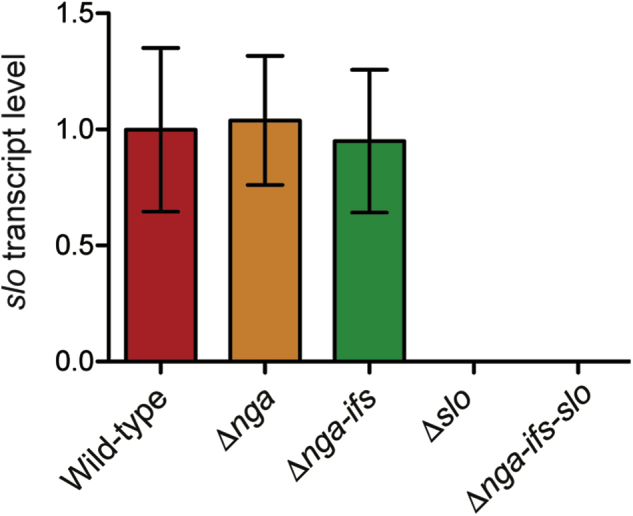
Relative transcript level of slo in MGAS2221 and four isogenic mutants. Transcript level of slo was normalized with the transcript level of housekeeping gene rpsL. Each experiment was performed in triplicate. Statistical differences between strains were determined using one-way analysis of variance and Dunnett's post test. Data are expressed as means ± SD.
References
- 1.Walker M.J., Barnett T.C., McArthur J.D., Cole J.N., Gillen C.M., Henningham A., Sriprakash K.S., Sanderson-Smith M.L., Nizet V. Disease manifestations and pathogenic mechanisms of group A streptococcus. Clin Microbiol Rev. 2014;27:264–301. doi: 10.1128/CMR.00101-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Madden J.C., Ruiz N., Caparon M. Cytolysin-mediated translocation (CMT): a functional equivalent of type III secretion in gram-positive bacteria. Cell. 2001;104:143–152. doi: 10.1016/s0092-8674(01)00198-2. [DOI] [PubMed] [Google Scholar]
- 3.Bricker A.L., Cywes C., Ashbaugh C.D., Wessels M.R. NAD+-glycohydrolase acts as an intracellular toxin to enhance the extracellular survival of group A streptococci. Mol Microbiol. 2002;44:257–269. doi: 10.1046/j.1365-2958.2002.02876.x. [DOI] [PubMed] [Google Scholar]
- 4.Michos A., Gryllos I., Hakansson A., Srivastava A., Kokkotou E., Wessels M.R. Enhancement of streptolysin O activity and intrinsic cytotoxic effects of the group A streptococcal toxin, NAD-glycohydrolase. J Biol Chem. 2006;281:8216–8223. doi: 10.1074/jbc.M511674200. [DOI] [PubMed] [Google Scholar]
- 5.Bastiat-Sempe B., Love J.F., Lomayesva N., Wessels M.R. Streptolysin O and NAD-glycohydrolase prevent phagolysosome acidification and promote group A Streptococcus survival in macrophages. MBio. 2014;5:e01690–e01714. doi: 10.1128/mBio.01690-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O'Seaghdha M., Wessels M.R. Streptolysin O and its co-toxin NAD-glycohydrolase protect group A Streptococcus from xenophagic killing. PLoS Pathog. 2013;9:e1003394. doi: 10.1371/journal.ppat.1003394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uchiyama S., Dohrmann S., Timmer A.M., Dixit N., Ghochani M., Bhandari T., Timmer J.C., Sprague K., Bubeck-Wardenburg J., Simon S.I., Nizet V. Streptolysin O rapidly impairs neutrophil oxidative burst and antibacterial responses to group A streptococcus. Front Immunol. 2015;6:581. doi: 10.3389/fimmu.2015.00581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu L., Olsen R.J., Nasser W., Beres S.B., Vuopio J., Kristinsson K.G., Gottfredsson M., Porter A.R., DeLeo F.R., Musser J.M. A molecular trigger for intercontinental epidemics of group A Streptococcus. J Clin Invest. 2015;125:3545–3559. doi: 10.1172/JCI82478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fehrenbach F.J. Identity of streptolysin-O and NAD-glycohydrolase (EC 3.2.2.5) Z Naturforsch B. 1971;26:1336–1338. doi: 10.1515/znb-1971-1224. [DOI] [PubMed] [Google Scholar]
- 10.Fehrenbach F.J. Purification and crystallization of NAD glycohydrolase of C streptococciEur J Biochem. 1971;18:94–102. doi: 10.1111/j.1432-1033.1971.tb01219.x. German. [DOI] [PubMed] [Google Scholar]
- 11.Shany S., Grushoff P.S., Bernheimer A.W. Physical separation of streptococcal nicotinamide adenine dinucleotide glycohydrolase from streptolysin O. Infect Immun. 1973;7:731–734. doi: 10.1128/iai.7.5.731-734.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mozola C.C., Caparon M.G. Dual modes of membrane binding direct pore formation by streptolysin O. Mol Microbiol. 2015;97:1036–1050. doi: 10.1111/mmi.13085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kimoto H., Fujii Y., Yokota Y., Taketo A. Molecular characterization of NADase-streptolysin O operon of hemolytic streptococci. Biochim Biophys Acta. 2005;1681:134–149. doi: 10.1016/j.bbaexp.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 14.Meehl M.A., Pinkner J.S., Anderson P.J., Hultgren S.J., Caparon M.G. A novel endogenous inhibitor of the secreted streptococcal NAD-glycohydrolase. PLoS Pathogens. 2005;1:e35. doi: 10.1371/journal.ppat.0010035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu L., Olsen R.J., Nasser W., de la Riva Morales I., Musser J.M. Trading capsule for increased cytotoxin production: contribution to virulence of a newly emerged clade of emm89 Streptococcus pyogenes. MBio. 2015;6:e01378–e01415. doi: 10.1128/mBio.01378-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beres S.B., Kachroo P., Nasser W., Olsen R.J., Zhu L., Flores A.R., de la Riva I., Paez-Mayorga J., Jimenez F.E., Cantu C., Vuopio J., Jalava J., Kristinsson K.G., Gottfredsson M., Corander J., Fittipaldi N., Di Luca M.C., Petrelli D., Vitali L.A., Raiford A., Jenkins L., Musser J.M. Transcriptome remodeling contributes to epidemic disease caused by the human pathogen Streptococcus pyogenes. MBio [Internet] 2016;7(3) doi: 10.1128/mBio.00403-16. e00403-16, doi: 10.1128/mBio.00403-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nasser W., Beres S.B., Olsen R.J., Dean M.A., Rice K.A., Long S.W., Kristinsson K.G., Gottfredsson M., Vuopio J., Raisanen K., Caugant D.A., Steinbakk M., Low D.E., McGeer A., Darenberg J., Henriques-Normark B., Van Beneden C.A., Hoffmann S., Musser J.M. Evolutionary pathway to increased virulence and epidemic group A Streptococcus disease derived from 3,615 genome sequences. Proc Natl Acad Sci U S A. 2014;111:E1768–E1776. doi: 10.1073/pnas.1403138111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sumby P., Porcella S.F., Madrigal A.G., Barbian K.D., Virtaneva K., Ricklefs S.M., Sturdevant D.E., Graham M.R., Vuopio-Varkila J., Hoe N.P., Musser J.M. Evolutionary origin and emergence of a highly successful clone of serotype M1 group a Streptococcus involved multiple horizontal gene transfer events. J Infect Dis. 2005;192:771–782. doi: 10.1086/432514. [DOI] [PubMed] [Google Scholar]
- 19.Olsen R.J., Sitkiewicz I., Ayeras A.A., Gonulal V.E., Cantu C., Beres S.B., Green N.M., Lei B., Humbird T., Greaver J., Chang E., Ragasa W.P., Montgomery C.A., Cartwright J., Jr., McGeer A., Low D.E., Whitney A.R., Cagle P.T., Blasdel T.L., DeLeo F.R., Musser J.M. Decreased necrotizing fasciitis capacity caused by a single nucleotide mutation that alters a multiple gene virulence axis. Proc Natl Acad Sci U S A. 2010;107:888–893. doi: 10.1073/pnas.0911811107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Committee for the Update of the Guide for the Care and Use of Laboratory Animals. National Research Council . National Academies Press; Washington, DC: 2011. Guide for the Care and Use of Laboratory Animals: Eighth Edition. [Google Scholar]
- 21.Sanson M., O'Neill B.E., Kachroo P., Anderson J.R., Flores A.R., Valson C., Cantu C.C., Makthal N., Karmonik C., Fittipaldi N., Kumaraswami M., Musser J.M., Olsen R.J. A naturally occurring single amino acid replacement in multiple gene regulator of group A Streptococcus significantly increases virulence. Am J Pathol. 2015;185:462–471. doi: 10.1016/j.ajpath.2014.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kobayashi S.D., Voyich J.M., Buhl C.L., Stahl R.M., DeLeo F.R. Global changes in gene expression by human polymorphonuclear leukocytes during receptor-mediated phagocytosis: cell fate is regulated at the level of gene expression. Proc Natl Acad Sci U S A. 2002;99:6901–6906. doi: 10.1073/pnas.092148299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Segal A.W. How neutrophils kill microbes. Annu Rev Immunol. 2005;23:197–223. doi: 10.1146/annurev.immunol.23.021704.115653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bernheimer A.W., Carlson A.S., Kellner A. Selective inhibition by preparations of streptococcal filtrates of the oxidative metabolism of mitochondria procured from rabbit myocardium. J Exp Med. 1956;104:577–587. doi: 10.1084/jem.104.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carlson A.S., Kellner A., Bernheimer A.W., Freeman E.B. A streptococcal enzyme that acts specifically upon diphosphopyridine nucleotide: characterization of the enzyme and its separation from streptolysin O. J Exp Med. 1957;106:15–26. doi: 10.1084/jem.106.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bernheimer A.W., Lazarides P.D., Wilson A.T. Diphosphopyridine nucleotidase as an extracellular product of streptococcal growth and its possible relationship to leukotoxicity. J Exp Med. 1957;106:27–37. doi: 10.1084/jem.106.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herbert D., Todd E.W. Purification and properties of a haemolysin produced by group A haemolytic streptococci (streptolysin O) Biochem J. 1941;35:1124–1139. doi: 10.1042/bj0351124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Epps D.E., Andersen B.R. Streptolysin O inhibition of neutrophil chemotaxis and mobility: nonimmune phenomenon with species specificity. Infect Immun. 1974;9:27–33. doi: 10.1128/iai.9.1.27-33.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Timmer A.M., Timmer J.C., Pence M.A., Hsu L.C., Ghochani M., Frey T.G., Karin M., Salvesen G.S., Nizet V. Streptolysin O promotes group A Streptococcus immune evasion by accelerated macrophage apoptosis. J Biol Chem. 2009;284:862–871. doi: 10.1074/jbc.M804632200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharma O., O'Seaghdha M., Velarde J.J., Wessels M.R. NAD+-glycohydrolase promotes intracellular survival of group A Streptococcus. PLoS Pathog. 2016;12:e1005468. doi: 10.1371/journal.ppat.1005468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Savic D.J., McShan W.M., Ferretti J.J. Autonomous expression of the slo gene of the bicistronic nga-slo operon of Streptococcus pyogenes. Infect Immun. 2002;70:2730–2733. doi: 10.1128/IAI.70.5.2730-2733.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Limbago B., Penumalli V., Weinrick B., Scott J.R. Role of streptolysin O in a mouse model of invasive group A streptococcal disease. Infect Immun. 2000;68:6384–6390. doi: 10.1128/iai.68.11.6384-6390.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sierig G., Cywes C., Wessels M.R., Ashbaugh C.D. Cytotoxic effects of streptolysin o and streptolysin s enhance the virulence of poorly encapsulated group a streptococci. Infect Immun. 2003;71:446–455. doi: 10.1128/IAI.71.1.446-455.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bricker A.L., Carey V.J., Wessels M.R. Role of NADase in virulence in experimental invasive group A streptococcal infection. Infect Immun. 2005;73:6562–6566. doi: 10.1128/IAI.73.10.6562-6566.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rossjohn J., Gilbert R.J., Crane D., Morgan P.J., Mitchell T.J., Rowe A.J., Andrew P.W., Paton J.C., Tweten R.K., Parker M.W. The molecular mechanism of pneumolysin, a virulence factor from Streptococcus pneumoniae. J Mol Biol. 1998;284:449–461. doi: 10.1006/jmbi.1998.2167. [DOI] [PubMed] [Google Scholar]
- 36.Tenenbaum T., Seitz M., Schroten H., Schwerk C. Biological activities of suilysin: role in Streptococcus suis pathogenesis. Future Microbiol. 2016;11:941–954. doi: 10.2217/fmb-2016-0028. [DOI] [PubMed] [Google Scholar]