Abstract
The National Cancer Institute–Molecular Analysis for Therapy Choice (NCI-MATCH) trial is a national signal-finding precision medicine study that relies on genomic assays to screen and enroll patients with relapsed or refractory cancer after standard treatments. We report the analytical validation processes for the next-generation sequencing (NGS) assay that was tailored for regulatory compliant use in the trial. The Oncomine Cancer Panel assay and the Personal Genome Machine were used in four networked laboratories accredited for the Clinical Laboratory Improvement Amendments. Using formalin-fixed paraffin-embedded clinical specimens and cell lines, we found that the assay achieved overall sensitivity of 96.98% for 265 known mutations and 99.99% specificity. High reproducibility in detecting all reportable variants was observed, with a 99.99% mean interoperator pairwise concordance across the four laboratories. The limit of detection for each variant type was 2.8% for single-nucleotide variants, 10.5% for insertion/deletions, 6.8% for large insertion/deletions (gap ≥4 bp), and four copies for gene amplification. The assay system from biopsy collection through reporting was tested and found to be fully fit for purpose. Our results indicate that the NCI-MATCH NGS assay met the criteria for the intended clinical use and that high reproducibility of a complex NGS assay is achievable across multiple clinical laboratories. Our validation approaches can serve as a template for development and validation of other NGS assays for precision medicine.
Precision medicine attempts to direct treatment for a patient based on molecular alterations known to exist in the patient's disease. The treatment of patients with cancer has been at the center of the evolution for precision medicine studies. Many recently developed treatments, including those approved by the US Food and Drug Administration (FDA), target specific genetic defects known to drive or significantly contribute to the cancer phenotype. Well-defined, reproducible, and robust molecular assays are therefore required that can efficiently assess tumor tissue to identify defects for which a treatment exists. Such assays play a pivotal role in the success of precision medicine.
The National Cancer Institute (NCI) initiated a large national precision medicine trial called the Molecular Analysis for Therapy Choice (referred to as NCI-MATCH) that is conducted through the National Clinical Trial Network and National Clinical Oncology Research Program and led by the Eastern Cooperative Oncology Group–American College of Radiology Imaging Network (ECOG-ACRIN) Cancer Research Group. The goal of this trial is to screen thousands of patients recruited from up to 2400 National Clinical Trial Network clinical sites who have relapsed or refractory solid tumors and lymphomas after standard systemic treatment for their cancer and then to assign the patients to a treatment appropriately matched to their cancer genotype. Details of the trial and protocol can be found at the NCI website (http://cancer.gov/nci-match, last accessed October 13, 2016).1, 2, 3
Targeted next-generation sequencing (NGS) panels can identify mutations in key genes with predictive value for approved or investigational cancer treatments.4, 5, 6, 7 Although NGS technology is a powerful tool, it is also new and not yet standardized among clinical laboratories. Different sample collection and processing methods, sequencing chemistries, instruments, protocols, and data analysis methods are known to affect NGS assay results.8, 9 In addition, regulatory compliance, such as the Code of Federal Regulations title 21 part 812 for investigational device exemption (Code of Federal Regulations, http://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfcfr/CFRsearch.cfm?CFRPart=812, last accessed October 13, 2016) for the FDA must be achieved for use of assays in clinical trials.
To provide a robust, standardized, and reproducible assay to support the NCI-MATCH trial and accommodate potentially large numbers of specimens, four clinical molecular diagnostics laboratories accredited through the Clinical Laboratory Improvement Amendments (CLIA) program formed a network to synergistically develop and validate the targeted NGS assay system with input from regulatory agencies. These four laboratories are located at Frederick National Laboratory for Cancer Research (FNLCR), Massachusetts General Hospital (MGH), the University of Texas MD Anderson Cancer Center (MDACC), and the Yale School of Medicine (YSM). We report on the methods used and the results obtained during the analytical performance testing and validation for analytical sensitivity, specificity, reproducibility, and limit of detection in each of the four laboratories and the overall combined data set and performance of the four laboratories. We believe our validation approach can serve as a template for development and validation of other clinical applications of NGS in support of precision medicine trials.
Materials and Methods
Tumor Specimens and Cell Lines
For evaluating analytical performance of this assay, archived formalin-fixed, paraffin-embedded (FFPE) clinical tumor specimens with various histopathologic diagnoses from the four network laboratories were chosen as samples of convenience to include a wide variety of known somatic variants encompassing all five variant types: single-nucleotide variants (SNVs), small insertions/deletions (indels), large indels (gap ≥4 bp), copy number variants (CNVs), and gene fusions. These variants were originally identified by orthogonal analytically validated assays (eg, digital PCR, Sanger sequencing, and fluorescent in situ hybridization [FISH]) in the CLIA-accredited laboratories. Tumor content for the specimens was assessed by board-certified pathologists.
Although every effort was made to include informative FFPE clinical specimens, a few FFPE cell line pellets were also included in this assay validation study because of the scarcity of specific variant types in available clinical specimens. Cell lines (Supplemental Table S1) were obtained from the Frederick National Laboratory for Cancer Research (Frederick, MD), American Type Culture Collection (Manassas, VA), and Coriell Institute for Medical Research (Camden, NJ) and were cultured using vendor-recommended conditions. Cultured cells were harvested and pelleted by centrifugation, fixed overnight in 10% neutral buffered formalin, and embedded in paraffin blocks.
Sections were cut from tumor and cell line FFPE blocks, and the relevant regions were collected for nucleic acid extraction. Numbers of specimens sequenced in each assay performance assessment in the validation study are summarized in Table 1, and the complete list of the specimens is provided in Supplemental Table S1.
Table 1.
Summary of Specimens Sequenced in Analytical Validation Study for the NCI-MATCH NGS Assay
| Institution | Sensitivity, % | Specificity, % | Reproducibility, % | Limit of detection, % |
|---|---|---|---|---|
| FNLCR | 56 | 20 | 64 | 10 |
| MDACC | 45 | 20 | 64 | 10 |
| MGH | 58 | 20 | 64 | 10 |
| YSM | 56 | 20 | 64 | 10 |
| Total | 215 | 80 | 256 | 40 |
| Total unique specimens∗ | 198 | 5 | 16 | 3 |
| Total unique clinical specimens | 186 | 0 | 16 | 1 |
| Total unique cell lines | 12 | 5 | 0 | 2 |
FNLCR, Frederick National Laboratory for Cancer Research; MDACC, MD Anderson Cancer Center; MGH, Massachusetts General Hospital; YSM, Yale University School of Medicine.
Some clinical specimens are used more than once because of scarcity of the variant type. Please refer to Supplemental Table S1 for complete details of all specimens sequenced.
Assay System and Content
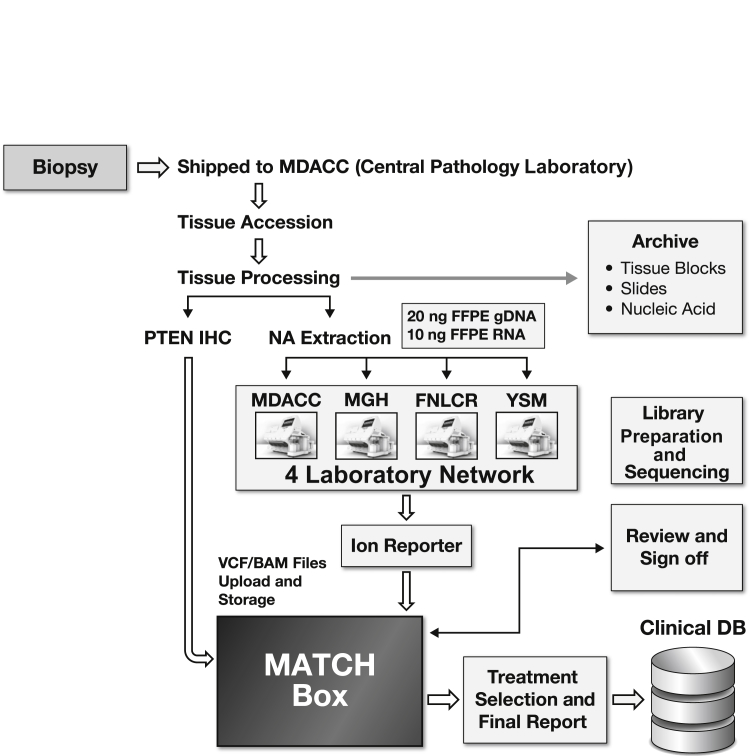
The overall laboratory workflow and components in the NCI-MATCH assay system are depicted in Figure 1. Clinical biopsy samples were sent overnight to the central pathology laboratory of the MDACC for preanalytical histologic assessment followed by extraction of nucleic acids (DNA and RNA). Nucleic acid specimens from each patient's biopsy sample were shipped to one of the four clinical laboratories where the NCI-MATCH NGS assay was performed using locked standard operating procedures (SOPs) and validated personal genome machine (PGM) instruments. The work used a locked data analysis pipeline, Torrent Suite version 4.4.2 (Thermo Fisher Scientific, Waltham, MA), and Ion Reporter version 4.4.2 (Thermo Fisher Scientific). A variant calling format (VCF) file generated from this locked pipeline was uploaded by each network laboratory to a centralized data processing system, termed MATCHBox, a rules engine designed to analyze all relevant data, including disease histologic and test findings from assays other than NGS (eg, immunohistochemistry for PTEN expression; see below) together with the results of the NCI-MATCH NGS assay to assign patients to trial arms and to generate clinical reports returned to the referring oncologist. MATCHBox is considered outside the assay system and will not be further discussed here.
Figure 1.
Workflows and components in the NCI-MATCH assay system. Four core needle specimens biopsied from a patient are shipped to central pathology laboratory at MD Anderson Cancer Center (MDACC). Tissue specimens are registered, and the preanalytic processes include formalin fixation and paraffin embedding (FFPE), hematoxylin and eosin staining, and tumor content assessment. The content of tumor cellularity of a specimen is required to be ≥70%, and enrichment by manual microdissections is performed on all specimens to attempt to achieve >70% tumor cellularity before nucleic acid (NA) extraction. A PTEN immunohistochemistry (IHC) stain is also performed as an inclusion biomarker for one treatment arm in the NCI-MATCH study. All residual tissue and nucleic acid samples are achieved in the ECOG-ACRIN Central Biorepository and Pathology Facility at the MDACC. One patient's genomic DNA (gDNA) and cDNA (reverse transcribed from total RNA) are shipped to one of four clinical laboratories to perform the NCI-MATCH next-generation sequencing (NGS) assay. The NGS data are analyzed by Ion Reporter (Torrent Suite version 4.4.2 Oncomine workflow) and the variant calling format (VCF) file and binary alignment map (BAM) file are automatically uploaded to MATCHBox, the centralized data processing system. MATCHBox identifies the actionable mutations of interest and assigns the treatment. The variant report and treatment assignment are reviewed by a group of experts composed of bioinformaticans, molecular biologists, statisticians, oncologists, and pathologists before sign off of the final report. The final results are sent to the enrolling physicians and patients and to a clinical medical database (DB) for long-term storage. The overall turnaround time from biopsy to final report is intended to be between 11 and 14 days. FNLCR, Frederick National Laboratory for Cancer Research; MGH, Massachusetts General Hospital; YSM, Yale University School of Medicine.
The Oncomine Cancer Panel assay10 using AmpliSeq chemistry and the PGM sequencer was selected and optimized for the NCI-MATCH NGS assay. The NCI-MATCH NGS assay system was designed to detect and report 4066 predefined genomic variations, encompassing 3259 SNVs, 114 indels, 435 large indels, 75 CNVs, and 183 gene fusions (Supplemental Table S2) across 143 unique genes (after removal of genes duplicated in more than one variant type) (Supplemental Table S3) in the Oncomine Cancer Research Panel (Thermo Fisher Scientific). Although other NGS assays attempt to detect gene fusions by sequencing intronic DNA to identify breakpoints, the NCI-MATCH NGS assay detects the targeted gene fusions by sequencing cDNA converted directly from specific targeted RNA transcripts. In addition, the assay reports novel nonsense or frameshift variants in 26 tumor suppressor genes, in-frame deletions in exon 19 of EGFR, in-frame insertions in exon 20 of ERBB2, and in-frame insertions and deletions in exons 9 and 11 of KIT. These reportable variants are collectively termed mutations of interest (MOIs). A subset of these MOIs considered actionable are used for treatment selection in the NCI-MATCH clinical trial and are identified based on the assumption of three levels of evidence suitable for assigning patients to treatment with corresponding drugs present in the trial arms. As stated in the protocol, level 1 evidence is accorded to gene variants that are credentialed for selection of an FDA-approved drug (eg, BRAF V600E and vemurafenib in melanoma) in any tissue; level 2a evidence is accorded to gene variants that are eligibility criteria for an ongoing clinical trial; level 2b evidence is accorded to gene variants that have been identified as a target in an N-of-1 response (eg, TSC1 mutations for everolimus11); and level 3 evidence is based on preclinical inferential data (in vitro or in vivo models) supporting use of the variants for treatment selection (eg, models in which cells or tissues containing a variant respond to the drug, whereas those without the variant do not). Two types of variants with level 3 evidence are gain-of-function variants, such as activating mutations in kinases found in a preclinical model (eg, D769H variant of ERBB2 results in increased tyrosine kinase–specific activity), and loss-of-function variants in tumor suppressor genes or pathway inhibitors (eg, NF1), producing a stop codon, frameshift, or a demonstrated loss of function of the resultant protein in a preclinical model.
The content of the Oncomine panel will be reversioned and revalidated to match the targets of additional treatment arms in the future.
Overview of Assay Validation Process
The four CLIA-accredited laboratories first defined the general assay system and undertook feasibility testing. The staff from the four laboratories then met face to face to harmonize the experimental conditions, and to edit and finalize common SOPs. The overall process used for this effort was consistent with a quality system approach and followed document design control. This process began with defining and documenting the intended use of the NGS assay. The description of intended use included the details of the assay system, beginning with the details of the clinical specimens and preanalytic processing, through nucleic acid extraction, NGS, and data analysis. Document control included requiring signatures of essential personnel on the documents and SOPs, and reversioning of SOPs occurred whenever changes were made. On the basis of the intended use and assay system feasibility testing, a validation plan was developed to include required minimum assay performance metrics to be tested during the analytical validation. At this point, presubmission discussions were held with the Center for Devices and Radiologicalal Health at the FDA to assess the validation plan and define risks involved in the use of the assay for its clinical intended use as part of the NCI-MATCH trial. Input and suggestions from the FDA were used to modify the validation plan, whereupon locked SOPs were used to test the analytical performance of the assay in each of the four laboratories.
Preanalytical Considerations
To test a broad spectrum of known and clinically relevant variants, it was necessary to use specimens collected and processed in a variety of laboratories using different methods of preparation, not following the NCI-MATCH preanalytical SOPs. Although the specimens used for the assay validation studies were not processed using standard and uniform preanalytic procedures, data quality was high and failure rate as detailed below was very low. For the processing of actual patient samples in the NCI-MATCH trial, a set of standard and uniform procedures were subsequently established [NCI: NCI-Molecular Analysis for Therapy Choice (NCI-MATCH) Trial, http://www.cancer.gov/about-cancer/treatment/clinical-trials/nci-supported/ncimatch#3, last accessed October 13, 2016]. The full-system fit-for-purpose pre-analytic specimen protocol was then tested in the CLIA-accredited surgical pathology laboratory and Tissue Qualification Laboratory of The University of Texas MD Anderson Cancer Center with approval of that institution's IRB. All tumor resection specimens received in the surgical pathology laboratory with <30 minutes of recorded warm ischemia time were evaluated immediately for use. The supervising staff pathologist first determined if sufficient tumor tissue was present to permit pathological assessment for clinical care based on standard-of-care using College of American Pathologists Cancer Protocols (http://www.cap.org/web/oracle/webcenter/portalapp/pagehierarchy/cancer_protocol_templates.jspx?_afrLoop=590901100057455#!%40%40%3F_afrLoop%3D590901100057455%26_adf.ctrl-state%3Dj1cn7h2lw_4, last accessed October 13, 2016) in routine use. Resection specimens were excluded from use for the preanalytic fit-for-purpose testing if collection of tissue could compromise clinical care because of tumor location or small size. Up to four core needle biopsy specimens with a 16-gauge needle biopsy apparatus were collected from each suitable resection specimen. The cores were fixed in 10% neutral buffered formalin as with routine clinical specimens, placed in the NCI-MATCH specimen collection and shipping kit with temperature control and monitoring devices, shipped overnight to the Tissue Qualification Laboratory in the Department of Pathology at the MDACC, and tracked in the ECOG-ACRIN Central Biorepository and Pathology Facility database, according to the sample collection protocol of the NCI-MATCH trial.
The received fixed core biopsy tissue was processed in up to four cassettes to conserve tissue and embedded in paraffin blocks within 36 hours after collection. Quality control and quality assurance evaluation was performed by one of the trial pathologists using a section (4 μm thick) cut from each block and stained with hematoxylin and eosin. The block with the largest amount of intact tumor and the least stroma and inflammation was selected, and the region(s) with viable tumor were demarcated. The tumor content of the demarcated region was estimated as the percentage of the total cell nuclei, and approximately 10 unstained sections were cut for use in microdissection with a No. 11 scalpel under a dissecting microscope with the demarcated slide as a guide, followed by extraction of nucleic acids from the scraped tissue. The goal for tumor enrichment was a minimum of 70% tumor nuclei. Unstained slides for immunohistochemistry were also prepared. Any sample that could not be enriched to the minimum goal was annotated, and its results were reported with a disclaimer.
DNA and RNA Extraction and Preparation
For sensitivity assessment, nucleic acid samples that had been previously processed and analyzed by orthogonal methods in the four laboratories were used. These archived samples had undergone different nucleic extraction methods: FNLCR used Qiagen AllPrep DNA/RNA FFPE kit (Qiagen, Valencia, CA); MDACC extracted DNA using the PicoPure DNA Extraction Kit (Arcturus, Mountain View, CA) and extracted RNA using the Agencourt AMPureXP Kit (Agencourt Biosciences, Beverly, MA); MGH extracted total nucleic acids using the Agencourt FormaPure kit; and YSM extracted DNA using the QIAamp DNA FFPE Tissue Kit and RNA using the Qiagen RNeasy FFPE kit. Nucleic acid samples used in other assessments and the fit-for-purpose study (and subsequently for the NCI-MATCH trial tumor specimen) were extracted by Qiagen AllPrep DNA/RNA FFPE kit. All nucleic acid samples were quantitated by Qubit fluorometer (Thermo Fisher Scientific) before use in the NGS assay.
Library and Template Preparation for NGS Sequencing
The assay required 20 ng of genomic DNA and 10 ng of RNA from FFPE specimens. RNA was reverse transcribed to cDNA using the VILO cDNA Synthesis Kit (Thermo Fisher Scientific). Libraries were then prepared for each sample by setting up three PCR reactions, one for each of the three primer pools (two pools for DNA and one pool for RNA) in the Oncomine Cancer Panel, using either 10 ng of DNA or 10 μL of the cDNA generated for each sample. Thermal cycling was performed according to the manufacturer's recommendations and consisted of 18 cycles of amplification for the DNA reactions and 30 cycles of amplification for the cDNA reactions. After thermal cycling, the two DNA PCR reactions mixtures were combined, and both the DNA and RNA components were treated with the FuPa enzyme to digest the primer regions, ligated with IonXpress barcode oligonucleotides, purified, and quantified using the Ion Library Quantification Kit (Thermo Fisher Scientific).
A 10-pmol/L pool of DNA and RNA libraries in a 5:1 ratio (DNA to RNA) was used to prepare templated ion sphere particles with the Ion PGM Template OT2 200 Kit and the Ion OneTouch 2 instrument (Thermo Fisher Scientific). The templated ion sphere particles were enriched on the Ion OneTouch ES instrument. The Ion PGM Sequencing 200 Kit version 2, Ion 318 version 2 chips, and Ion Torrent PGM were used to sequence the enriched, templated ion sphere particles. All wet laboratory procedures were performed following SOPs based on the manufacturer's instructions.
Bioinformatics and Data Analysis Pipeline
Sequencing data analysis was performed using Torrent Suite version 4.4.2 and Ion Reporter version 4.4.2. This workflow was created by adding the custom hotspots BED (Browser Extensible Data) file (Supplemental Table S4) to report the MOIs and a custom CNV baseline (described in the next paragraph) to the default manufacturer's workflow (named the Oncomine Cancer Research Panel version 1.2–DNA and Fusions–Single Sample). This workflow could report SNVs and indels as low as 3% variant allele fraction (VAF), but based on the results of the feasibility study, a threshold was established at ≥5% VAF. For novel deleterious indels that create frameshifts in protein codon in tumor suppressor genes, a threshold was established at ≥10%. Detected SNVs and indels also require at least 25 variant-containing reads to be reported as positive. For example, an MOI reported at 5% VAF with a 200 read depth was considered negative because the product of VAF and reads (0.05 × 200) was <25. This approach combined read depth with allele frequency to ensure confidence in variant calls with either low read depth and/or low allele frequency.
Copy number analysis was performed using the copy number module within the aforementioned workflow of the Ion Reporter system. To create the baseline for CNV calling, nine replicates of a previously generated normal hapmap (haplotype map) data set (triplicates of each of Centre d'Etude du Polymorphisme Humain NA12878, Chinese female NA18526, and Yoruban NA18507) were used at each of the four network laboratories. In the analytical sensitivity study, copy numbers of four or greater were considered concordant if the orthogonal assay also reported a copy number of four or greater for the target genes. Only gene amplification–type CNVs were validated in this study.
Fusions were detected using the fusion detection module within the Ion Reporter workflow. This pipeline only reported previously annotated fusions as defined in a reference file that was preloaded into the workflow. All fusions with a read count ≥25 reads, except EGFRvIII mutation (exons 2 to 7 deletion in transcript), were considered positive. A higher threshold with a read count ≥1000 was established for EGFRvIII based on the level of background noise detected in the feasibility study.
Assay Quality Metrics
To ensure the quality of results obtained from the NCI-MATCH NGS assay and to set quality control criteria for reportable results, quality systems were developed in the four CLIA-accredited laboratories. Thresholds listed in Table 2 were set for library yields, number of DNA reads, number of RNA reads, RNA read length, uniformity, and percentage of amplicons with at least 100× coverage. These criteria were applied to all validation samples. If a sample failed because of poor sequencing quality (determined by any of the aforementioned metrics with the exception of final library yield), it was reprocessed from template preparation. All other sample failures were repeated from library preparation. A flagged sample proceeded to data review, which included scrutinizing the variant calls using the quality metrics within the VCF file, at which point it could be accepted or rejected. The thresholds of quality control metrics were determined empirically using the data from feasibility study and input from the vendor.
Table 2.
NGS QC Metrics and Results in the Analytical Validation Study for NCI-MATCH NGS Assay
| NGS QC metrics | Threshold, call | Failed samples, n | Flagged samples, n | Mean (range) |
||||
|---|---|---|---|---|---|---|---|---|
| FNLCR | MDACC | MGH | YSM | Four laboratories | ||||
| DNA library yield (pmol/L) | ≥20, Fail | 0 | NA | 1966 (112–7720) | 1638 (36–13,786) | 1556 (90–4994) | 1734 (117–9306) | 1724 (36–13,786) |
| RNA library yield (pmol/L) | ≥20, Fail | 0 | NA | 2443 (192–7892) | 1827 (137–12,621) | 2099 (198–5195) | 2408 (188–8680) | 2197 (137–12,621) |
| DNA total reads (million counts) | ≥3, Flag | NA | 8 | 4.20 (0.90–5.51) | 4.30 (0.91–6.10) | 4.37 (0.67–5.17) | 5.04 (1.82–6.22) | 4.46 (0.67–6.22) |
| DNA uniformity (%)∗ | ≥80, Fail; ≥90, flag | 2 | 52 | 93.70 (85.65–96.61) | 93.99 (80.98–98.70) | 95.31 (84.81–98.00) | 91.75 (67.66–97.44) | 93.67 (67.66–98.70) |
| DNA percentage of amplicons 100× (%) | ≥90, Fail | 6† | NA | 97.80 (81.00–99.05) | 98.39 (85.93–99.49) | 98.63 (85.22–99.45) | 97.78 (85.89–99.21) | 98.15 (81.00–99.49) |
| DNA MAPD‡ | ≤0.9, Fail | 0 | NA | 0.277 (0.14–0.574) | 0.403 (0.196–0.762) | 0.27 (0.134–0.534) | 0.385 (0.225–0.691) | 0.333 (0.134–0.762) |
| RNA total reads (thousand counts) | ≥100, Flag | NA | 0 | 918 (405–1313) | 925 (186–2266) | 1037 (510–2725) | 1112 (182–2884) | 998 (182–2884) |
| RNA read length (bp) | ≥40, Flag | NA | 0 | 93 (77–99) | 98 (74–112) | 98 (84–116) | 96 (79–104) | 96 (74–116) |
FNLCR, Frederick National Laboratory for Cancer Research; MAPD, median of the absolute values of all pairwise differences; MDACC, MD Anderson Cancer Center; MGH, Massachusetts General Hospital; NA, not applicable; QC, quality control; YSM, Yale University School of Medicine.
Uniformity is the percentage of bases in all amplicon-targeted regions covered by at least 0.2× the mean base read depth.
Two of 6 samples failed by percentage of amplicon >100× also failed by uniformity.
The MAPD between log2 ratios of two adjacent amplicons (except at the beginning and end of a chromosome).
Assay Performance Characteristics and Statistics
As detailed in Results, the laboratory network established the following performance characteristics for the NCI-MATCH NGS assay: sensitivity, specificity, reproducibility, limit of detection, and fit for purpose.
Assay Sensitivity
A true-positive result was defined as an MOI variant that was identified by prior testing in network laboratories and also reported by the NCI-MATCH NGS assay. A false-negative result was defined as an MOI variant previously identified by prior testing in network laboratories or reported by Catalogue of Somatic Mutations in Cancer (COSMIC) database in cell lines but not detected by the NCI-MATCH NGS assay. The assay sensitivity was expressed by the percentage of MOIs detected by the NCI-MATCH NGS assay in the total known MOIs according to the formula: (True Positive)/(True Positive + False Negative).12 The 95% CI was estimated using the Clopper and Pearson method (cran.r-project.org, last accessed October 14, 2016).13 The analytical sensitivity was calculated for each variant type (SNV, indel, large indel, CNV, gene fusion) and all types together in each laboratory and in the four laboratories combined together. The acceptable sensitivity was prespecified as ≥95% for SNVs and ≥90% for each of the other variant types within each clinical laboratory and across the four laboratories combined.
Assay Specificity
Specificity was estimated based on the total correct calls of the normal sequence within the total reportable MOIs (a total of 4066 predefined hotspots listed in Supplemental Table S2 plus nonhotspots in 26 tumor suppressor genes). A false-positive result was defined as an MOI reported by the NCI-MATCH NGS assay at a locus that had been previously reported as wild-type sequence in the 1000 Genome Project Consortium.14 A true-negative result was defined as the absence of an MOI by the NCI-MATCH assay that is in agreement with the previously reported results. Specificity was expressed by the percentage of total correct calls of the normal sequence within the full set of reportable MOIs according to the formula (True Negative)/(True Negative + False Positive).12 The 95% CI for each MOI was first estimated according the true negative detection rate across all replicates (20 replicates per laboratory and 80 replicates for four laboratories) in five cell lines using the Clopper and Pearson method (cran.r-project.org).13 Averaged values of low and high 95% confidence limits across all reportable MOIs were used to represent the interval range for the assay.
The analytical specificity was assessed in each variant type and overall for each laboratory, as well as for the four laboratories combined. The target for acceptable specificity was ≥99.9% for SNVs, ≥99.0% for indels and large indels, ≥97.0% for CNVs, and ≥99.0% for gene fusions within each laboratory. The acceptable mean number of false-positive variants per tested sample was prespecified to be 0.10 for each laboratory (1 false-positive MOI per 10 tested samples).
Assay Reproducibility
Reproducibility was assessed by calculating pairwise concordance within (intra-) and between (inter-) operators for all assay-reportable variants in each of the 16 nucleic acid specimens separately for DNA and RNA. Positive pairwise concordance was defined as the mean agreement in positive variant calls (MOI variants detected and reported by the NCI-MATCH NGS assay) between two replicates (number of positive variants called in both replicates/the total number of unique positive calls between the two runs). The overall pairwise concordance was defined as the number of MOIs in agreement, positive or negative, between both replicates divided by the number of unique positive and negative calls between the two replicates. Acceptable mean intraoperator and interoperator overall concordance were prespecified to be ≥99% in each of the four laboratories, as well as combined.
Results
Overall NGS Data Quality
A total of 455 sequencing runs of the NCI-MATCH NGS assay were performed in the four CLIA-accredited laboratories for this analytical validation study. The quality metrics of the assays performed in each individual laboratory and in all four laboratories combined are summarized in Table 2. During the entire validation study, six samples failed to meet sequencing quality control metrics in the first attempted analysis, but all six samples passed in the repeated analysis.
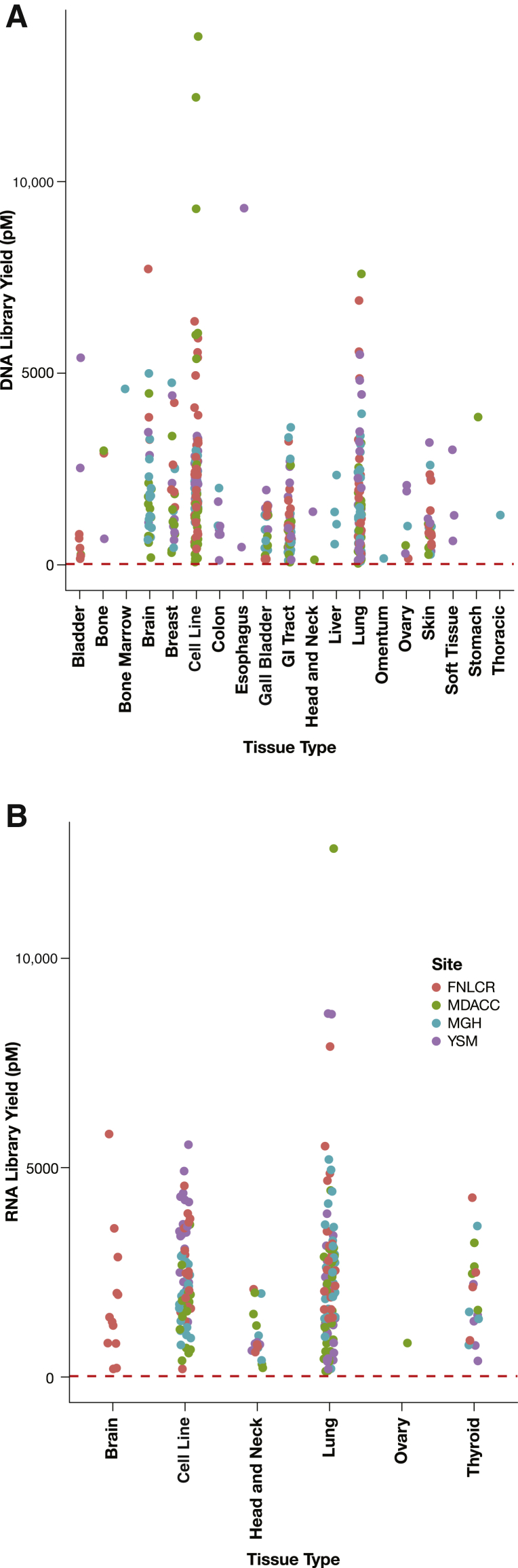
Nucleic acid specimens were extracted from 18 tissue types plus cell lines for DNA and five tissue types for RNA. Library yields of both DNA samples (Figure 2A) and RNA samples (Figure 2B) passed the minimal requirement (20 pmol/L) with all tissue types, indicating that successful library preparation was feasible from all tissue types studied.
Figure 2.
Tissue type has no effect on library yield. Library yields are plotted against the tissue types from which DNA samples (A) and RNA samples (B) were extracted. Each dot represents a sample, and each color code represents the laboratory site where the NCI-MATCH NGS assay was performed. The red dashed lines represent a minimal yield of 20 pmol/L required for template preparation. FNLCR, Frederick National Laboratory for Cancer Research; MDACC, MD Anderson Cancer Center; MGH, Massachusetts General Hospital; YSM, Yale University School of Medicine.
Sensitivity Assessment
To assess assay sensitivity, a total of 198 specimens (12 FFPE cell lines and 186 clinical specimens) were used to test 265 variants previously verified by orthogonal analytically validated assays or identified in the COSMIC database for well-characterized cell lines (Supplemental Figure S1 and Supplemental Table S5). Because of the scarcity of clinical specimens harboring specific variant types (especially indel and gene fusion variants), some specimens were tested in more than one laboratory.
The sensitivity of the NCI-MATCH NGS assay as determined from the combined results in the four laboratories was 99.02% for SNVs, 95.00% for indels, 97.50% for large indels, 92.50% for CNVs, 97.67% for gene fusions, and 96.98% overall for 265 variants combined. The sensitivities for all variants used in each laboratory were 95.59% for FNLCR, 100.00% for MDACC, 98.50% for YSM, and 93.85% for MGH. These results met the acceptance criteria of ≥95% for SNVs and ≥90% for each of the other variant types within each clinical laboratory and across the four laboratories combined (Table 3). Detailed information for all 265 known variants used in sensitivity validation is listed in Supplemental Table S5.
Table 3.
Analytical Sensitivity of the NCI-MATCH Assay
| Institution | SNV, % (n, 95% CI) | Indel l, % (n, 95% CI) | Large indel, % (n, 95% CI) | CNV, % (n, 95% CI) | Fusion, % (n, 95% CI) | All, % (n, 95% CI) |
|---|---|---|---|---|---|---|
| FNLCR | 96.00 (25, 79.64–99.89) | 90.00 (10, 55.49–99.74) | 100.00 (10, 69.15–100.00) | 90.00 (10, 55.49–99.74) | 100.00 (13, 75.29–100.00) | 95.59 (68, 86.64–99.08) |
| MDACC | 100.00 (25, 86.28–100.00) | 100.00 (10, 69.15–100.00) | 100.00 (10, 69.15–100.00) | 100.00 (10, 69.15–100.00) | 100.00 (10, 69.15–100.00) | 100 (65, 94.48–100.00) |
| MGH | 100.00 (25, 86.28–100.00) | 100.00 (10, 69.15–100.00) | 90.00 (10, 55.49–99.74) | 80.00 (10, 44.39–97.47) | 90.00 (10, 55.49–99.74) | 93.85 (65, 84.98–98.29) |
| YSM | 100.00 (27, 87.22–100.00) | 90.00 (10, 55.49–99.74) | 100.00 (10, 69.15–100.00) | 100.00 (10, 69.15–100.00) | 100.00 (10, 69.15–100.00) | 98.5 (67, 91.96–99.96) |
| Four laboratories | 99.02 (102, 94.65–99.97) | 95.00 (40, 83.08–99.38) | 97.50 (40, 86.84–99.93) | 92.50 (40, 79.61–98.42) | 97.67 (43, 87.71–99.94) | 96.98 (265, 94.13–98.68) |
| Acceptance criteria | 95.00 | 90.00 | 90.00 | 90.00 | 90.00 | 90.00 |
CNV, copy number variant; FNLCR, Frederick National Laboratory for Cancer Research; indel, insertion/deletion; MDACC, MD Anderson Cancer Center; MGH, Massachusetts General Hospital; SNV, single-nucleotide variant; YSM, Yale University School of Medicine.
The NCI-MATCH NGS assay detected a total of 257 variants of the 265 previously identified (Supplemental Table S5) variants combined among all four laboratories. The eight false-negative variants included one SNV (c.3140A>G in PIK3CA), two indels (c.821delG in PTEN and c.233_234insT in TP53); one large indel (c.2236_2248delGAATTAAGAGAAGinsCAAC in EGFR); three CNVs in ERBB2, PDGFRA, and KRAS; and one gene fusion (EML4-ALK E20A20). The allele frequency of SNV c.3140A>G in PIK3CA was seen at 2.8% in the VCF file, but this percentage was below the NCI-MATCH assay reportable threshold of 5% VAF. Because we would have filtered out this call because of the allele frequency below our threshold, this call was counted as a false-negative result. The indel c.821delG in PTEN was not detected because of strand bias (ie, lack of coverage from both strands). A long homopolymeric region upstream from the variant likely blocked the efficient extension of the strand by the polymerase (Supplemental Figure S2A). For the indel c.233_234insT in TP53, we did not observe any evidence of the reported variant allele. Because the orthogonal NGS assay detected this variant at 15% VAF, this variant was determined to be a false-negative result (Supplemental Figure S2B). The large indel c.2236_2248delGAATTAAGAGAAGinsCAAC in EGFR is a complex variant with a large (approximately 9 bp) deletion in combination with two single-base substitutions. Although the sequencing reads carrying the previously reported (Supplemental Table S5) variant allele were observed, it was not reported by the data analysis pipeline (Supplemental Figure S2C). The CNVs were not considered to be detected because the copy numbers reported by the assays were below the established threshold of 4: ERBB2 copy number was 2.8 as contrasted with 5.8 in the FISH-based orthogonal assay, PDGFRA copy number was 2.9 as contrasted with >25:1 amplification with FISH, and KRAS copy number was 3.5 as contrasted with 6.7 for FISH.
Specificity Assessment
For specificity analysis, five normal hapmap cell lines [Finnish (HG00272), Han Chinese (HG00403), Puerto Rican (HG00640), Japanese (NA18950), and African Ancestry (NA19701)] were cultured and prepared as FFPE cell pellets before DNA and RNA extraction. Each of the five hapmap cell lines was characterized by the NCI-MATCH NGS assay in quadruplicate at each laboratory.
The NCI-MATCH assay reported four false-positive variants in 80 total assay runs of the hapmap samples across the four laboratories (Supplemental Table S6). Although each of five hapmap samples was sequenced 16 times across the four laboratories, each of the four false-positive variants was reported in only one replicate. The resulting estimated number of false-positive variants per sample was 4 of 80 (0.05), meeting the acceptance criteria of <0.1 false-positive variant per sample. The overall specificity of the NCI-MATCH NGS assay across the four laboratories was 99.99% for SNVs, 99.98% for indels, 100.00% for large indels, 100.00% for CNVs, 99.99% for gene fusions, and 99.99% for all reportable variants. These results met the acceptance criteria of ≥99.9% for SNVs; ≥99% for indels, large indels, and gene fusions; ≥97% for CNV; and overall acceptance criteria of ≥99% (Table 4).
Table 4.
Analytical Specificity of the NCI-MATCH Assay
| Institution | SNV, % (n, 95% CI) | Indel, % (n, 95% CI) | Large indel, % (n, 95% CI) | CNV, % (n, 95% CI) | Fusion, % (n, 95% CI) | All, % (n, 95% CI) |
|---|---|---|---|---|---|---|
| FNLCR | 100.00 (3258, 83.16–100.00) | 100.00 (114, 83.16–100.00) | 100.00 (435, 83.16–100.00) | 100.00 (75, 83.16–100.00) | 100.00 (183, 83.16–100.00) | 100.00 (4065, 83.16–100.00) |
| MDACC | 100.00 (3258, 83.16–100.00) | 100.00 (114, 83.16–100.00) | 100.00 (435, 83.16–100.00) | 100.00 (75, 83.16–100.00) | 99.97 (183, 83.11–99.99) | 99.99 (4065, 83.16–99.99) |
| MGH | 100.00 (3258, 83.16–100.00) | 99.96 (115, 83.09–99.99) | 100.00 (435, 83.16–100.00) | 100.00 (75, 83.16–100.00) | 100.00 (183, 83.16–100.00) | 99.99 (4066, 83.16–99.99) |
| YSM | 99.99 (3259, 83.15–99.99) | 99.96 (115, 83.09–99.99) | 100.00 (435, 83.16–100.00) | 100.00 (75, 83.16–100.00) | 100.00 (183, 83.16–100.00) | 99.99 (4067, 83.15–99.99) |
| Four laboratories | 99.99 (3259, 95.49–99.99) | 99.98 (116, 95.42–99.9) | 100.00 (435, 95.49–100.00) | 100.00 (75, 95.49–100.00) | 99.99 (183, 95.48–99.99) | 99.99 (4068, 95.49–99.99) |
| Acceptance criteria | 99.90 | 99.00 | 99.00 | 97.00 | 99.00 | 99.00 |
CNV, copy number variant; FNLCR, Frederick National Laboratory for Cancer Research; indel, insertion/deletion; MDACC, MD Anderson Cancer Center; MGH, Massachusetts General Hospital; SNV, single-nucleotide variant; YSM, Yale University School of Medicine.
The four false-positive variants were one SNV variant (c.1207G>T, p.Glu403Ter in APC in HG00640), two indel variants (c.3909_3910insC, p.Arg1304fs in PTCH1 in HG00640 and c.1309_1310insT, p.Thr438fs in MSH2 in HG00272), and one gene fusion (FGFR3-TACC3.F15T1 in HG00403). The four variants reported by the NCI-MATCH NGS assay were false-positive variants because no such variants have been reported in corresponding hapmap cell lines in the 1000 Genomes Project. Gene fusion FGFR3-TACC3.F15T1 was detected with 440 read counts only once in 16 replicate analyses of the HG00403 hapmap cell lines across all four laboratories. Further examination of the binary alignment map (BAM) file using the Integrative Genomics Viewer15 revealed a rare mispriming event that was identified as a series of six homozygous variants in one of the fusion partners, which could be mapped to a different region of the hg19 reference genome (Supplemental Figure S3). To reduce potential false reporting of fusion variants in clinical samples by the NCI-MATCH NGS, the SOP for evaluation of all gene fusions requested examining the BAM file in the Integrative Genomics Viewer to eliminate the false-positive variants that resulted from mispriming.
Four other variants (SNV c.5557G>A in ATM in HG00272, SNV c.1124A>G in MET in HG00403, and SNV c.215C>G in TP53 in all five hapmap cell lines and large indel c.6859_6863delAGTCA in NF1 in HG00272) reported by the NCI-MATCH NGS assay in all replicates analyses of these hapmap cell lines were confirmed as true-positive variants because they were previously reported at similar allele frequency in the 1000 Genome Project.14
Reproducibility Assessment
Reproducibility was evaluated on nine DNA and seven RNA samples extracted from 16 different clinical specimens. There were 15 known sample-specific variants: five SNVs, one indel, one large indel, and one CNV in nine DNA samples and seven gene fusions in seven RNA samples. Aliquots of the same nucleic acid samples were characterized by the NCI-MATCH NGS assay in four laboratories. In each laboratory, each of the samples was analyzed by different operators, and each operator performed the assay twice. The assay replicates were generated by construction of new library preparations proceeding through sequencing and data analysis of the specimen. Two different instruments were used by each operator for each of their replicates. Each laboratory had two operators except YSM, which included a third operator in the analysis (Supplemental Table S1).
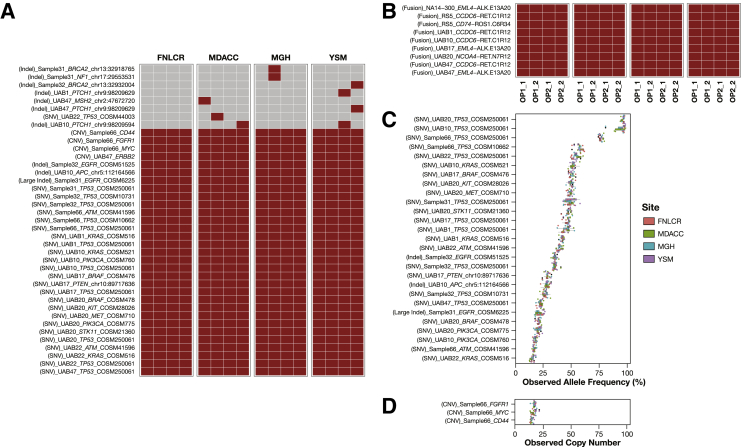
The NCI-MATCH NGS assay reported a total of 45 variants: 25 SNVs, 9 indels, 1 large indel, 3 CNVs, and 7 gene fusions in 16 nucleic acid samples analyzed in the four clinical laboratories. Of the 45 reported variants, 15 (5 SNVs, 1 indel, 1 large indel, 1 CNV, and 7 gene fusions) were identified previously, and 30 (20 SNVs, 8 indels, and 2 CNVs) were previously unknown. The assay identified all expected variants in 16 replicates, which revealed 100% concordance for pairwise intraoperator and interoperator comparisons. A total of 37 of the 45 reported variants were concordantly called by the NCI-MATCH NGS assay, but 7 variants (representing 6 unique variants) were detected only once, and 1 indel variant was detected twice among the 16 technical replicates in the four network laboratories. Those seven discordantly detected unique variants were an indel in BRCA2 (chr13: 32918765, A insertion), an indel in BRCA2 (chr13: 32932004, G deletion), an indel in MSH2 (chr2: 47672720, T insertion), an indel in NF1 (chr17:29553531, T insertion), an indel in PTCH1 (chr9: 98209629, G insertion) detected in two separated samples; an indel in PTCH1 (chr9: 98209594, A insertion), and an SNV c.452C>G in TP53. The detection results of 45 reported variants across 16 technical replicates in the four laboratories are depicted in tile plots (Figure 3, A and B), and the reported variants in reproducibility assessment are listed in Supplemental Table S7. The intralaboratory and interlaboratory and operator pairwise concordances over 45 detected variants (positive concordance) and all reportable variants (overall concordance) were calculated and summarized in Table 5. The mean positive concordances for all intraoperative and interoperator pairwise comparisons over 45 reported variants were >96% in nine DNA samples and 100% in seven RNA samples. The mean overall concordances over all MOI loci were ≥99.99% among all intraoperator and interoperator pairwise comparisons. The reproducibility of the NCI-MATCH NGS assay met the required acceptance criteria of 99% for overall concordance. The reproducibility in variant allele frequency (Figure 3C) or copy number (Figure 3D) of 30 commonly detected DNA variants are depicted in box plots.
Figure 3.
Reproducibility assessment across 16 technical replicates in four network laboratories. A and B: Reproducibility of 45 positive variants consisting of 38 variants detected in 9 DNA samples (A) and 7 fusion variants in 7 RNA samples (B) that were called at least once in 16 technical replicates across 4 network laboratories. Each row represents a variant, and the type of variant, sample, gene, Catalogue of Somatic Mutations in Cancer(COSMIC) identification or position (hg19) for variants without COSMIC identification are labeled. Each column represents a replicate; the laboratory and operator and replicate are indicated. The color code represents a variant call by the NCI-MATCH NGS assay (gray, no call; brown, a call). A: Eight DNA variants discordantly called (top) include 7 variants (6 indels and 1 SNV) detected once and 1 variant (Indel) detected only twice in 16 technical replicates. To simplify the presentation, replicates generated from OP2 and OP3 at the Yale University School of Medicine (YSM) are grouped and indicated as OP2. C and D: Allele frequencies of 27 variants (C) and copy numbers of 3 variants (D) were concordantly detected in 16 technical replicates and are plotted as box plot. Each row represents a variant, and the allele frequency copy number detected in 16 replicates are grouped by laboratory and color-coded as indicated in the legends. FNLCR, Frederick National Laboratory for Cancer Research; indel, insertion and deletion; MDACC, MD Anderson Cancer Center; MGH, Massachusetts General Hospital; OP, operator.
Table 5.
Analytical Reproducibility of the NCI-MATCH Assay
| Reproducibility across 16 specimens | Nucleic acid | Type | Pairwise concordances, n | Mean of concordances | Median of concordances | Range of concordances |
|---|---|---|---|---|---|---|
| Positive concordance | DNA | Intraoperator | 72 | 96.204 | 100 | 50.00–100.00 |
| RNA | Intraoperator | 56 | 100.000 | 100 | 100.00–100.00 | |
| DNA | Within-laboratory interoperator | 144 | 96.204 | 100 | 50.00–100.00 | |
| RNA | Within-laboratory interoperator | 112 | 100.000 | 100 | 100.00–100.00 | |
| DNA | Cross lab InterOperator | 864 | 96.289 | 100 | 33.33–100.00 | |
| RNA | Cross-laboratory interoperator | 672 | 100.000 | 100 | 100.00–100.00 | |
| Overall concordance | DNA | Intraoperator | 72 | 99.997 | 100 | 99.95–100.00 |
| RNA | Intraoperator | 56 | 100.000 | 100 | 100.00–100.00 | |
| DNA | Within-laboratory interoperator | 144 | 99.997 | 100 | 99.95–100.00 | |
| RNA | Within-laboratory interoperator | 112 | 100.000 | 100 | 100.00–100.00 | |
| DNA | Cross-laboratory interoperator | 864 | 99.997 | 100 | 99.95–100.00 | |
| RNA | Cross-laboratory interoperator | 672 | 100.000 | 100 | 100.00–100.00 |
Limit-of-Detection Assessment
To assess the limit of detection of variant calling, two mixed DNA samples were used. Pool 1 consisted of an SNV (BRAF, c.1799T>A, p.V600E) at 50% allele frequency and a MYC amplification (CNV) at 60 copies. Pool 2 consisted of one large indel (RB1, c.346_349delACTT, p.T116fs*8) at 100% allele frequency and one indel (TP53, c.1023delC, p.R342fs*3) at 100% allele frequency. Each pool was diluted with FFPE Hapmap Centre d'Etude du Polymorphisme Humain (NA12878) DNA to obtain twofold serially diluted samples to estimate the lowest VAF or copy number detected by the NCI-MATCH NGS assay in SNV, indel, large indel, and CNV variant types. To assess the lowest detection variant calling, the rule of 5% VAF detection threshold and 25 sequencing reads with the variant was not applied.
A gene fusion variant was excluded from limit-of-detection assessment because the assay is designed to detect only gene fusions that are specifically targeted. Unlike the detection for DNA variants, there was no competition between the normal and fusion RNA transcripts for this fusion-specific PCR amplification. Therefore, the detection of gene fusion variants was considered as binary (ie, detected or not).
The detected VAFs of four variants in a dilution series across the four laboratories are depicted in a tile plot (Table 6 and Supplemental Table S8). As indicated in Table 6, the observed VAFs of detected SNV variants ranged from 45.00% to 2.80%, with an expected decrease in VAF with each dilution point. Similarly, observed copy numbers of detected MYC CNV ranged from 28.7 to 4 with an expected decrease in copy numbers with each dilution point. The higher than expected VAF for BRAF SNV mutations are likely the result of aneuploidy in chromosome 7 (six copies) in the source cell lines UACC62.16 For the pool consisting of an indel and large indel variants, both variants were detected at the first two dilution points in three laboratories, but MDACC detected at the first dilution points only for the large indels. The results of limit-of-detection studies are consistent with the default detection limit used in the data analysis pipeline.
Table 6.
Limit-of-Detection Assessment
| Site | Mutation | Fold of dilution |
||||
|---|---|---|---|---|---|---|
| 4 | 8 | 16 | 32 | 64 | ||
| FNLCR | CNV_MYC | 23.1 | 15.1 | 9.5 | 6.3 | 4.5 |
| SNV_BRAF_c.1799T>A | 41.8 | 26.9 | 13.5 | 7.2 | 4.0 | |
| Indel_TP53_c.1023delC | 27.9 | 10.7 | ND | ND | ND | |
| Large indel_RB1_c.346_349delACTT | 17.8 | 7.8 | ND | ND | ND | |
| MDACC | CNV_MYC | 23.1 | 16.1 | 9.6 | 6.0 | 4.1 |
| SNV_BRAF_c.1799T>A | 42.4 | 25.7 | 14.2 | 7.3 | 4.2 | |
| Indel_TP53_c.1023delC | 24.3 | 12.0 | ND | ND | ND | |
| Large indel_RB1_c.346_349delACTT | 16.3 | ND | ND | ND | ND | |
| MGH | CNV_MYC | 27.4 | 16.2 | 9.1 | 6.2 | 4.0 |
| SNV_BRAF_c.1799T>A | 42.7 | 25.4 | 13.6 | 6.7 | 4.4 | |
| Indel_TP53_c.1023delC | 23.6 | 10.7 | ND | ND | ND | |
| Large indel_RB1_c.346_349delACTT | 14.4 | 8.1 | ND | ND | ND | |
| YSM | CNV_MYC | 28.7 | 17.7 | 9.1 | 5.4 | 4.1 |
| SNV_BRAF_c.1799T>A | 45.4 | 24.8 | 11.4 | 7.6 | 2.8 | |
| Indel_TP53_c.1023delC | 23.8 | 10.5 | ND | ND | ND | |
| Large indel_RB1_c.346_349delACTT | 16.2 | 6.8 | ND | ND | ND | |
CNV, copy number variant; FNLCR, Frederick National Laboratory for Cancer Research; indel, insertion/deletion; MDACC, MD Anderson Cancer Center; MGH, Massachusetts General Hospital; ND, not detected; YSM, Yale University School of Medicine.
Fit-for-Purpose Assay System Assessment
To test the full assay system and informatics analysis pipeline performance from biopsy collection through the MATCHBox upload and reporting, the tumor in 22 prospective clinical surgical resection specimens was subjected to core needle biopsy as described above. The purpose of this effort was to reveal that all aspects of the NCI-MATCH assay system were fully functional and ready to accept specimens from patients. The criteria for evaluating if a specimen passes the fit-for-purpose testing included the NGS data passed the quality control metrics; the genomic results were successfully uploaded into MATCHBox; and variant calls and any treatment assignment were reported correctly.
The 22 samples consisted of 16 different tumor types (Supplemental Table S9). Of the 22 samples, 16 were analyzed once by FNLCR and 6 were analyzed either two or three times by the three other laboratories to test the reproducibility of reportable MOIs, resulting in a total of 32 runs of the assay. Although 29 runs passed the quality control metrics, three specimens analyzed by FNLCR failed to pass, yielding a 90.6% (n = 29/32) success rate. Two specimens (a squamous cell lung carcinoma and a hepatocellular carcinoma) failed to yield amplicon uniformity >80%, and the third specimen (a gastrointestinal stroma tumor) failed to generate a sufficient quantity of AmpliSeq library after two attempts (Supplemental Table S9). The VCF and BAM files of all 29 passing runs were successfully uploaded to MATCHBox. Of the six clinical specimens analyzed by MDACC, MGH, and YSM, two were tested in two laboratories, and the other four were tested in three laboratories. A total of 11 MOIs (8 SNVs, 1 indel, and 2 CNVs) were reported by MATCHBox for these six specimens. A CCDC6-RET.C1R12 fusion variant was detected in one of three laboratories and was considered a false-positive variant because of contamination. All 11 MOIs were detected at similar VAF or copy number in all replicates performed in different laboratories (Table 7), resulting in 100% mean cross-laboratory pairwise concordances for both positive and overall concordances.
Table 7.
Reproducibility of 11 Mutations of Interest in Six Clinical Specimens Used in Fit-for-Purpose Study Report
| Diagnose | Test sites, n | Type | Gene | Codon change | VAF (%) or CN reported by three laboratories |
||
|---|---|---|---|---|---|---|---|
| MDACC | MGH | YSM | |||||
| Adenocarcinoma, colon | 2 | SNV | APC | p.Glu582Ter | 0.42 | 0.50 | |
| Ependymoma, NOS | 3 | SNV | CTNNB1 | p.Ser45Pro | 0.07 | 0.11 | 0.25 |
| Papillary carcinoma, thyroid | 2 | Indel | VHL | p.Gly106fs | 0.38 | 0.43 | |
| SNV | IDH1 | p.Val178Ile | 0.51 | 0.52 | |||
| Adenocarcinoma lung, metastatic | 3 | CNV | CDK4 | 16.00∗ | 15.20∗ | 17.20∗ | |
| CNV | MDM2 | 21.00∗ | 21.30∗ | 22.80∗ | |||
| SNV | EGFR | p.Glu709Lys | 0.38 | 0.37 | 0.41 | ||
| SNV | EGFR | p.Leu858Arg | 0.35 | 0.36 | 0.40 | ||
| Adrenal tumor | 3 | SNV | CTNNB1 | p.Ser45Pro | 0.37 | 0.33 | 0.32 |
| SNV | KIT | p.Met541Leu | 0.51 | 0.49 | 0.51 | ||
| Adrenal cortical carcinoma | 3 | SNV | HRAS | p.Gln61Arg | 0.39 | 0.40 | 0.43 |
CN, copy number; CNV, copy number variant; indel, insertion/deletion; MDACC, MD Anderson Cancer Center; MGH, Massachusetts General Hospital; NOS, not otherwise specified; SNV, single-nucleotide variant; VAF, variant allele fraction; YSM, Yale University School of Medicine.
Copy number.
The fit-for-purpose study found that the NCI-MATCH NGS assay system was fully functional in all aspects and capable of supporting the NCI-MATCH clinical trial. The fact that 29 of 32 runs passed the quality control metrics (90.6% successful rate) was considered acceptable for clinical specimen performance.
Discussion
We outline an approach for clinical NGS assay development and analytical validation that was used successfully to prepare the NGS assay system for the NCI-MATCH clinical trial. Our validation involved testing cells and tumor tissues of multiple types in an effort to determine assay performance over a wide range of tumor specimens likely to be submitted for the NCI-MATCH NGS assay. We also focused on testing representative variants from each of the different variant types interrogated by the assay: SNV, indel, large indel, CNV and gene fusion. Not every reportable variant could be tested because the number of variants deemed reportable is very large, and the rarity of some variants made obtaining clinical specimens harboring such variants extremely challenging. However, an effort was made to represent a range of typical and frequent variants. As indicated by the results, the NGS assay system met the intended performance metrics.
Overall the assay produced robust results for all specimens, except for six samples that failed to pass quality metrics but passed when sequenced a second time. Furthermore, an effort was made to reveal that robust sequence results could be obtained from multiple tissue types. Eighteen different tissues and cell lines were evaluated for sequencing of DNA, and five different tissues were evaluated for RNA sequencing. These specimens represented tissues that are likely to be submitted for the NCI-MATCH trial screening and included tissues that may be difficult to sequence, such as pancreas, melanoma, skin, and bone. There were no detectable tissue-related failures when nucleic acid yields and sequencing quality metrics were achieved. This result suggests that DNA and RNA recovered from many different tissues can be successfully and accurately sequenced by this protocol.
Predefined metrics for sensitivity were met by each laboratory and across all laboratories (Table 3). Specificity results also met the high degree required (Table 4). Specificity was considered a critical component for the assay's intended use. Although it is important to minimize both false-positive and false-negative results, the NCI-MATCH leadership agreed that false-positive results would be of greater concern in this study because of the resultant drug assignment. The results observed in the limit-of-detection experiments indicated that the assay is capable of detecting variants at a level below the assay-defined limits (observed 2.8% for SNVs, 10.5% for indels, and 6.8% for large indels and four copies for gene amplification) (Table 6). Importantly, excellent reproducibility was found across the four laboratories (Table 5). These results are noteworthy because, to date, concerns have been widely expressed about the complexity, accuracy, and reproducibility of NGS for application in clinical trials.17, 18 Our results clearly indicate that locked and controlled procedures permit reliable, accurate, and reproducible use of NGS for clinical purposes. Reproducibility was found for variants known to exist in the specimen and across all reportable variants sites (MOIs), whether the allele was found or not on comparison to the human genome hg19 reference sequence. Assays developed independently of this particular assay may not indicate similar reproducibility to this specific assay, owing to the complexity of the assay systems. The topic of how well different clinical assays reproduce clinically relevant results is of great interest, and we hope that data addressing such comparisons will soon be available. This conclusion argues for continued development and implementation of assay standards, control materials, and proficiency testing that can be shared across laboratories in an effort to assess cross-assay performance comparisons. The standards and controls would complement traditional proficiency panels.
The discordant results observed between NGS and FISH assays for CNVs in our sensitivity study may have occurred as a result of heterogeneity in tumor samples and differences in measurement procedures. FISH assays rely on the signal number and a ratio of the locispecific probe to a chromosomal centromeric probe, whereas NGS assays rely on read depth or counts of amplicons across the gene compared with a normal genome baseline. The NGS assay sometimes reported fewer copies than FISH. Results of FISH focus exclusively on tumor cells, whereas NGS assays rely on read depth or counts of amplicons across the gene that are affected by the presence of nucleic acids from nonneoplastic cells. The SOPs used for the NCI-MATCH trial require tumor enrichment to achieve >70% viable tumor content. Nevertheless, each sample contained significant amounts of nontumor cells, which would artificially lower the observed CNV scores. Increasing the percentage of tumor cells in the samples by enrichment methods, such as laser capture microdissection, may provide more accurate copy numbers for the genes tested. In addition, discordance between NGS and FISH for CNV may arise in part from underestimations of aneuploidy within the tumor cells analyzed by FISH.
The detection of false-positive fusion variants by sequencing RNA in our fit-for-purpose study highlights the risks of cross-specimen contamination in high-throughput NGS assays that include gene fusions as targets. When a fusion-positive specimen is analyzed, a tremendous amount of uniform PCR product is generated, which entails the possibility of carrying minute amounts of these products forward in subsequent analyses. Consequently, the SOPs used for this assay note that great attention should be focused on samples processed in close temporal proximity to fusion-positive samples. If other specimens were found to contain the exact same gene fusion but with lower levels of detection, the laboratory repeated both suspected specimens. In addition, no-template controls (without the addition of nucleic acid) were tested periodically and immediately after suspected contamination was observed. For the trial, a laboratory will remain off line and not test patient specimens until the suspected contamination is resolved and no-template controls reveal the absence of fusion products. In general, good clinical laboratory and PCR practices should minimize such episodes of possible contamination.
Because it was not necessary for the specimens sequenced in analytical validation to be collected and processed following the NCI-MATCH pre-analytical SOPs, all fit-for-purpose specimens were processed consistent with the NCI-MATCH preanalytical SOPs. The three samples that failed to produce sequence in the fit-for-purpose study may have resulted from the process chosen to collect the specimens. The fit-for-purpose specimens were collected from surgical resection specimens; therefore, there were substantially longer intervals of warm ischemia than would be expected with core needle biopsy specimens or needle aspirates. The core needle biopsy specimens used for routine NCI-MATCH specimen collection will come directly from the tumor in the patient; therefore, less time will elapse before the tissue is placed in fixative.
In conclusion, the NCI-MATCH trial will provide an opportunity for cancer patients to be matched to treatments targeted to specific molecular defects based on the genomic analysis of their tumors. A targeted NGS assay has been developed and analytically validated to support this trial. We have described the process used for development and validation of this assay system.
This analytical validation study clearly found that the assay met the expected performance requirement for the intended use. In addition, the ability to accurately report results from multiple tissue types (including pancreas, melanoma, bone, and skin) suggests that nucleic acid specimens recovered from multiple tumor tissue types are acceptable. These validation data were submitted as an appendix to the NCI-MATCH trial Investigational New Drug documentation. This validation effort indicates that NGS assays can be robust and reproducible if the assay system is defined and standardized by locked SOPs. The process described within this article can serve as a template for other investigators who develop and validate NGS assay systems intended to measure biomarkers in CLIA laboratories to determine eligibility, assign treatment, or assess outcome in clinical trials (integral biomarkers).
Acknowledgments
We thank the members of the Eastern Cooperative Oncology Group–American College of Radiology Imaging Network (ECOG-ACRIN) Cancer Research Group and the NCI-MATCH Steering Committee; Drs. Liqiang Xi and Mark Raffeld (NCI) for providing clinical specimens; Mike Balco, Dr. Ignacio Wistuba, staff of the ECOG-ACRIN Central Biorepository and Pathology Facility, and faculty and staff in the Department of Pathology (The University of Texas MD Anderson Cancer Center) for assistance with prospective collection and processing of fit-for-purpose specimens; the MATCHBox team in the Biomedical Informatics and Information Technology group (NCI) for building MATCHBox; Dr. JoyAnn Philips Rohan, Lori Keisling, Erin Souhan, and Dr. Sanita Bharti for project management and administrative support; and the Oncomine team (Thermo Fisher Scientific) for their excellent support.
Concept and design: C.-J.L., S.R.H., J.L.S., A.J.I., B.A.C., A.P.C., H.D., P.M.W.; development of methods: C.-J.L., R.R.S., R.L., L.M.M., E.C.P., M.J.R., P.M.W., D.J.S., R.D.H., A.J.I., J.L.S.; acquisition of data: R.D.H., K.N.H., C.H.B., J.Y., K.R., S.C., H.R., A.R.; analysis and interpretation of data: C.-J.L., D.J.S., M.J.R., E.C.P.; writing, review, and revision of manuscript: C.-J.L., D.J.S., R.D.H., S.R.H., J.L.S., A.J.I., L.M.M., E.C.P., B.A.C., A.P.C., H.D., P.M.W.
Footnotes
See related Commentary on page 226
Supported by NIH grants HHSN261200800001E and NO1-CO-2008-00001, Eastern Cooperative Oncology Group–American College of Radiology Imaging Network (ECOG-ACRIN) Cancer Research Group Central Biorepository and Pathology Facility in the Division of Pathology and Laboratory Medicine at The University of Texas MD Anderson Cancer Center grant 1U24CA196172, and the ECOG-ACRIN Medical Research Foundation. S.R.H. is supported by the Frederick F. Becker Distinguished University Chair in Cancer Research from The University of Texas.
Disclosures: A.J.I. is a stockholder and scientific advisory board member of ArcherDx and is a consultant for Roche, Pfizer, DebioPharm, Chugai, and Constellation Pharmaceuticals. Other authors have no disclosures. This work does not express or represent the opinion of the National Cancer Institute, National Institutes of Health, or the US Department of Health and Human Services.
Current address of E.C.P., Department of Health Sciences Research, Mayo Clinic, Rochester, MN.
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.jmoldx.2016.10.007.
Supplemental Data
Variant distribution in sensitivity assessment. A: Number of variants in each type and total used in each laboratory. B: Number of unique variants in each type. n = 265 total variants; n = 149 unique variants. CNV, copy number variant; FNLCR, Frederick National Laboratory for Cancer Research; indel, insertion and deletion; MDACC, MD Anderson Cancer Center; MGH, Massachusetts General Hospital; SNV, single-nucleotide variant; YSM, Yale University School of Medicine.
Examination of variants not detected in sensitivity assessment by the Integrative Genomics Viewer (IGV). A: Indel in PTEN (c.821delG) in sample BT549; a strand bias is noted in the IGV in this amplicon, with reads from only the reverse strand (blue reads). This variant was verified by orthogonal assay and Sanger sequencing and also noted in the Catalogue of Somatic Mutations in Cancerdatabase. Thus, this variant was determined to be a false-negative variant. B: Indel in TP53 (c.233_234insT) in sample 297527; an insertion in TP53 c.233_234insT was not detected in sample 297527. The IGV image shows the low counts of the variant at the position indicated by the two lines. The orthogonal next-generation sequencing assay detected this variant at 15% variant allele fraction. Thus, this variant was determined to be a false-negative variant. C: Large indel in the EGFR gene (c.2236_2248 delGAATTAAGAGAAGinsCAAC) in sample 54; variant was not detected by the data analysis pipeline because this variant is composed of a large (approximately 9 bp) deletion and also the introduction of two single-base substitutions. indel, insertion and deletion.
Examination of variants not detected in specificity assessment by the Integrative Genomics Viewer. False-positive gene fusion FGFR3-TACC3.F15T1 detected in the HG00403 hapmap cell line showing the mispriming event, as denoted by the arrow.
References
- 1.Brower V. NCI-MATCH pairs tumor mutations with matching drugs. Nat Biotechnol. 2015;33:790–791. doi: 10.1038/nbt0815-790. [DOI] [PubMed] [Google Scholar]
- 2.Mullard A. NCI-MATCH trial pushes cancer umbrella trial paradigm. Nat Rev Drug Discov. 2015;14:513–515. doi: 10.1038/nrd4694. [DOI] [PubMed] [Google Scholar]
- 3.Conley B.A., Doroshow J.H. Molecular analysis for therapy choice: NCI MATCH. Semin Oncol. 2014;41:297–299. doi: 10.1053/j.seminoncol.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 4.Frampton G.M., Fichtenholtz A., Otto G.A., Wang K., Downing S.R., He J. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31:1023–1031. doi: 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh R.R., Patel K.P., Routbort M.J., Reddy N.G., Barkoh B.A., Handal B., Kanagal-Shamanna R., Greaves W.O., Medeiros L.J., Aldape K.D., Luthra R. Clinical validation of a next-generation sequencing screen for mutational hotspots in 46 cancer-related genes. J Mol Diagn. 2013;15:607–622. doi: 10.1016/j.jmoldx.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 6.Cheng D.T., Mitchell T.N., Zehir A., Shah R.H., Benayed R., Syed A., Chandramohan R., Liu Z.Y., Won H.H., Scott S.N., Brannon A.R., O'Reilly C., Sadowska J., Casanova J., Yannes A., Hechtman J.F., Yao J., Song W., Ross D.S., Oultache A., Dogan S., Borsu L., Hameed M., Nafa K., Arcila M.E., Ladanyi M., Berger M.F. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn. 2015;17:251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lih C.J., Sims D.J., Harrington R.D., Polley E.C., Zhao Y., Mehaffey M.G., Forbes T.D., Das B., Walsh W.D., Datta V., Harper K.N., Bouk C.H., Rubinstein L.V., Simon R.M., Conley B.A., Chen A.P., Kummar S., Doroshow J.H., Williams P.M. Analytical validation and application of a targeted next-generation sequencing mutation-detection assay for use in treatment assignment in the NCI-MPACT trial. J Mol Diagn. 2016;18:51–67. doi: 10.1016/j.jmoldx.2015.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gargis A.S., Kalman L., Berry M.W., Bick D.P., Dimmock D.P., Hambuch T. Assuring the quality of next-generation sequencing in clinical laboratory practice. Nat Biotechnol. 2012;30:1033–1036. doi: 10.1038/nbt.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bijwaard K., Dickey J.S., Kelm K., Tezak Z. The first FDA marketing authorizations of next-generation sequencing technology and tests: challenges, solutions and impact for future assays. Expert Rev Mol Diagn. 2015;15:33–40. doi: 10.1586/14737159.2015.979795. [DOI] [PubMed] [Google Scholar]
- 10.Hovelson D.H., McDaniel A.S., Cani A.K., Johnson B., Rhodes K., Williams P.D., Bandla S., Bien G., Choppa P., Hyland F., Gottimukkala R., Liu G., Manivannan M., Schageman J., Ballesteros-Villagrana E., Grasso C.S., Quist M.J., Yadati V., Amin A., Siddiqui J., Betz B.L., Knudsen K.E., Cooney K.A., Feng F.Y., Roh M.H., Nelson P.S., Liu C.J., Beer D.G., Wyngaard P., Chinnaiyan A.M., Sadis S., Rhodes D.R., Tomlins S.A. Development and validation of a scalable next-generation sequencing system for assessing relevant somatic variants in solid tumors. Neoplasia. 2015;17:385–399. doi: 10.1016/j.neo.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iyer G., Hanrahan A.J., Milowsky M.I., Al-Ahmadie H., Scott S.N., Janakiraman M., Pirun M., Sander C., Socci N.D., Ostrovnaya I., Viale A., Heguy A., Peng L., Chan T.A., Bochner B., Bajorin D.F., Berger M.F., Taylor B.S., Solit D.B. Genome sequencing identifies a basis for everolimus sensitivity. Science. 2012;338:221. doi: 10.1126/science.1226344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pont-Kingdon G., Gedge F., Wooderchak-Donahue W., Schrijver I., Weck K.E., Kant J.A., Oglesbee D., Bayrak-Toydemir P., Lyon E. Design and analytical validation of clinical DNA sequencing assays. Arch Pathol Lab Med. 2012;136:41–46. doi: 10.5858/arpa.2010-0623-OA. [DOI] [PubMed] [Google Scholar]
- 13.Clopper C.J., Pearson E.S. The use of confidence or fiducial limits illustrated in the case of the binomial. Biometrika. 1934;26:404–413. [Google Scholar]
- 14.1000 Genomes Project Consortium. Abecasis G.R., Altshuler D., Auton A., Brooks L.D., Durbin R.M., Gibbs R.A., Hurles M.E., McVean G.A. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robinson J.T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knutsen T., Gobu V., Knaus R., Padilla-Nash H., Augustus M., Strausberg R.L., Kirsch I.R., Sirotkin K., Ried T. The interactive online SKY/M-FISH & CGH database and the Entrez cancer chromosomes search database: linkage of chromosomal aberrations with the genome sequence. Genes Chromosomes Cancer. 2005;44:52–64. doi: 10.1002/gcc.20224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rehm H.L., Bale S.J., Bayrak-Toydemir P., Berg J.S., Brown K.K., Deignan J.L., Friez M.J., Funke B.H., Hegde M.R., Lyon E., Working Group of the American College of Medical Genetics and Genomics Laboratory Quality Assurance Commitee ACMG clinical laboratory standards for next-generation sequencing. Genet Med. 2013;15:733–747. doi: 10.1038/gim.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xuan J., Yu Y., Qing T., Guo L., Shi L. Next-generation sequencing in the clinic: promises and challenges. Cancer Lett. 2013;340:284–295. doi: 10.1016/j.canlet.2012.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Variant distribution in sensitivity assessment. A: Number of variants in each type and total used in each laboratory. B: Number of unique variants in each type. n = 265 total variants; n = 149 unique variants. CNV, copy number variant; FNLCR, Frederick National Laboratory for Cancer Research; indel, insertion and deletion; MDACC, MD Anderson Cancer Center; MGH, Massachusetts General Hospital; SNV, single-nucleotide variant; YSM, Yale University School of Medicine.
Examination of variants not detected in sensitivity assessment by the Integrative Genomics Viewer (IGV). A: Indel in PTEN (c.821delG) in sample BT549; a strand bias is noted in the IGV in this amplicon, with reads from only the reverse strand (blue reads). This variant was verified by orthogonal assay and Sanger sequencing and also noted in the Catalogue of Somatic Mutations in Cancerdatabase. Thus, this variant was determined to be a false-negative variant. B: Indel in TP53 (c.233_234insT) in sample 297527; an insertion in TP53 c.233_234insT was not detected in sample 297527. The IGV image shows the low counts of the variant at the position indicated by the two lines. The orthogonal next-generation sequencing assay detected this variant at 15% variant allele fraction. Thus, this variant was determined to be a false-negative variant. C: Large indel in the EGFR gene (c.2236_2248 delGAATTAAGAGAAGinsCAAC) in sample 54; variant was not detected by the data analysis pipeline because this variant is composed of a large (approximately 9 bp) deletion and also the introduction of two single-base substitutions. indel, insertion and deletion.
Examination of variants not detected in specificity assessment by the Integrative Genomics Viewer. False-positive gene fusion FGFR3-TACC3.F15T1 detected in the HG00403 hapmap cell line showing the mispriming event, as denoted by the arrow.